circRNA from APP Gene Changes in Alzheimer’s Disease Human Brain
Abstract
1. Introduction
2. Results
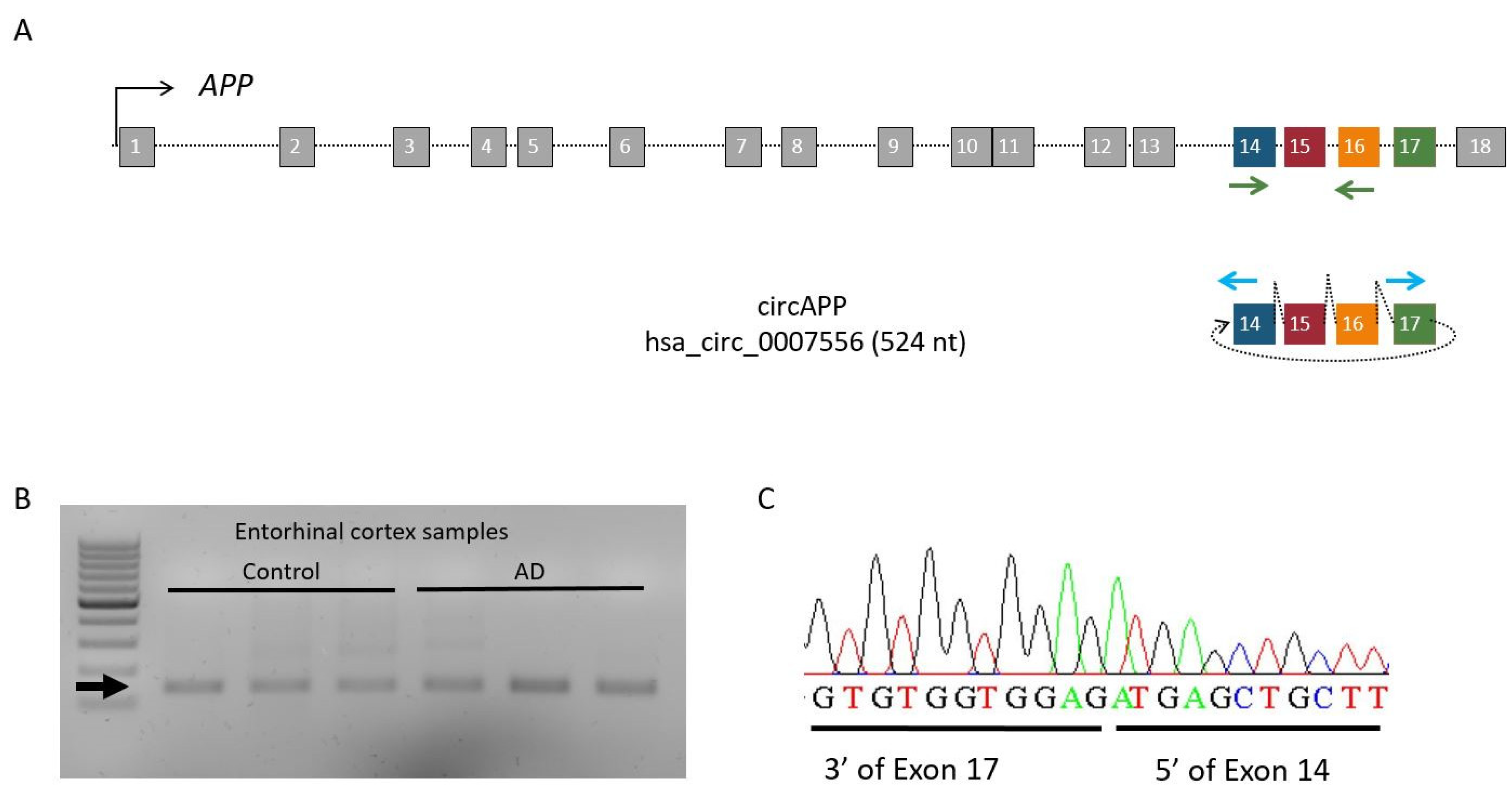
2.1. circAPP (hsa_circ_0007556) Identification in Entorhinal Human Brain
2.2. Differential Expression of circAPP (hsa_circ_0007556) and APP mRNA in AD Entorhinal Cortex
2.3. Correlation between circAPP (hsa_circ_0007556) and APP mRNA Expression and Aβ Deposits
2.4. In Silico Prediction of Biological Function
3. Discussion
4. Methods
4.1. Human Entorhinal Cortex Samples
4.2. RNA Isolation and Reverse Transcription—Polymerase Chain Reaction (RT-PCR)
4.3. Candidate Band Selection and Sanger Sequencing
4.4. Real Time Quantitative PCR (RT-qPCR) Assay
4.5. Quantitative Assessment of Aβ Deposits in Brain Tissues
4.6. Prediction of circAPP (hsa_circ_0007556) Interaction with miRNAs and Biological Function
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tortajada-Soler, M.; Sánchez-Valdeón, L.; Blanco-Nistal, M.; Benítez-Andrades, J.A.; Liébana-Presa, C.; Bayón-Darkistade, E. Prevalence of Comorbidities in Individuals Diagnosed and Undiagnosed with Alzheimer’s Disease in León, Spain and a Proposal for Contingency Procedures to Follow in the Case of Emergencies Involving People with Alzheimer’s Disease. Int. J. Environ. Res. Public Health 2020, 17, 3398. [Google Scholar] [CrossRef] [PubMed]
- Reitz, C.; Mayeux, R. Alzheimer disease: Epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem. Pharmacol. 2014, 88, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s Association. 2021 Alzheimer’s disease facts and figures. Alzheimers Dement. 2021, 17, 327–406. [Google Scholar] [CrossRef]
- O’Brien, R.J.; Wong, P.C. Amyloid precursor protein processing and Alzheimer’s disease. Annu. Rev. Neurosci. 2011, 34, 185–204. [Google Scholar] [CrossRef] [PubMed]
- Gralle, M.; Ferreira, S.T. Structure and functions of the human amyloid precursor protein: The whole is more than the sum of its parts. Prog. Neurobiol. 2007, 82, 11–32. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Álvarez-Álvarez, I.; Guillén-Grima, F.; Aguinaga-Ontoso, I. Prevalence and incidence of Alzheimer’s disease in Europe: A meta-analysis. Neurologia 2017, 32, 523–532. [Google Scholar] [CrossRef]
- Morris, G.P.; Clark, I.A.; Vissel, B. Inconsistencies and controversies surrounding the amyloid hypothesis of Alzheimer’s disease. Acta Neuropathol. Commun. 2014, 2, 135. [Google Scholar] [CrossRef]
- Del-Aguila, J.L.; Benitez, B.A.; Li, Z.; Dube, U.; Mihindukulasuriya, K.A.; Budde, J.P.; Farias, F.H.G.; Fernandez, M.V.; Ibanez, L.; Jiang, S.; et al. TREM2 brain transcript-specific studies in AD and TREM2 mutation carriers. Mol. Neurodegener. 2019, 14, 18. [Google Scholar] [CrossRef]
- Mo, D.; Li, X.; Raabe, C.A.; Rozhdestvensky, T.S.; Skryabin, B.V.; Brosius, J. Circular RNA Encoded Amyloid Beta peptides-A Novel Putative Player in Alzheimer’s Disease. Cells 2020, 9, 2196. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Xiao, M.S.; Ai, Y.; Wilusz, J.E. Biogenesis and Functions of Circular RNAs Come into Focus. Trends Cell Biol. 2020, 30, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Piwecka, M.; Glazar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef] [PubMed]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef] [PubMed]
- Hanan, M.; Soreq, H.; Kadener, S. CircRNAs in the brain. RNA Biol. 2017, 14, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Schuman, E. Circular RNAs in Brain and Other Tissues: A Functional Enigma. Trends Neurosci. 2016, 39, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Lipscombe, D.; Lopez Soto, E.J. Alternative splicing of neuronal genes: New mechanisms and new therapies. Curr. Opin. Neurobiol. 2019, 57, 26–31. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610. [Google Scholar] [CrossRef]
- Martínez-Iglesias, O.; Naidoo, V.; Cacabelos, N.; Cacabelos, R. Epigenetic Biomarkers as Diagnostic Tools for Neurodegenerative Disorders. Int. J. Mol. Sci. 2021, 23, 13. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.S.; Vasanthakumar, A.; Davis, J.W.; Idler, K.B.; Nho, K.; Waring, J.F.; Saykin, A.J. Association of peripheral blood DNA methylation level with Alzheimer’s disease progression. Clin. Epig. 2021, 13, 191. [Google Scholar] [CrossRef] [PubMed]
- Mano, T.; Sato, K.; Ikeuchi, T.; Toda, T.; Iwatsubo, T.; Iwata, A. Peripheral Blood BRCA1 Methylation Positively Correlates with Major Alzheimer’s Disease Risk Factors. J. Prev. Alzheimers Dis. 2021, 8, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Zhu, M.; Yang, J.; Pang, Y.; Wang, Q.; Li, Y.; Li, T.; Li, F.; Wei, Y. Prediction of P-tau/Aβ42 in the cerebrospinal fluid with blood microRNAs in Alzheimer’s disease. BMC Med. 2021, 19, 264. [Google Scholar] [CrossRef] [PubMed]
- Yuen, S.C.; Liang, X.; Zhu, H.; Jia, Y.; Leung, S.W. Prediction of differentially expressed microRNAs in blood as potential biomarkers for Alzheimer’s disease by meta-analysis and adaptive boosting ensemble learning. Alzheimers Res. Ther. 2021, 13, 126. [Google Scholar] [CrossRef] [PubMed]
- Dube, U.; Del-Aguila, J.L.; Li, Z.; Budde, J.P.; Jiang, S.; Hsu, S.; Ibanez, L.; Fernandez, M.V.; Farias, F.; Norton, J.; et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat. Neurosci. 2019, 22, 1903–1912. [Google Scholar] [CrossRef]
- Cervera-Carles, L.; Dols-Icardo, O.; Molina-Porcel, L.; Alcolea, D.; Cervantes-Gonzalez, A.; Muñoz-Llahuna, L.; Clarimon, J. Assessing circular RNAs in Alzheimer’s disease and frontotemporal lobar degeneration. Neurobiol. Aging 2020, 92, 7–11. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.; Liu, Y.; Wang, M.; Yu, W.; Zhang, L. Exploring the regulatory roles of circular RNAs in Alzheimer’s disease. Transl. Neurodegener. 2020, 9, 35. [Google Scholar] [CrossRef]
- Shi, Z.; Chen, T.; Yao, Q.; Zheng, L.; Zhang, Z.; Wang, J.; Hu, Z.; Cui, H.; Han, Y.; Han, X.; et al. The circular RNA ciRS-7 promotes APP and BACE1 degradation in an NF-kappaB-dependent manner. FEBS J. 2017, 284, 1096–1109. [Google Scholar] [CrossRef]
- Lukiw, W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet. 2013, 4, 307. [Google Scholar] [CrossRef]
- Zhao, Y.; Alexandrov, P.N.; Jaber, V.; Lukiw, W.J. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7). Genes 2016, 7, 116. [Google Scholar] [CrossRef]
- Lu, Y.; Tan, L.; Wang, X. Circular HDAC9/microRNA-138/Sirtuin-1 Pathway Mediates Synaptic and Amyloid Precursor Protein Processing Deficits in Alzheimer’s Disease. Neurosci. Bull. 2019, 35, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Diling, C.; Yinrui, G.; Longkai, Q.; Xiaocui, T.; Yadi, L.; Xin, Y.; Guoyan, H.; Ou, S.; Tianqiao, Y.; Dongdong, W.; et al. Circular RNA NF1-419 enhances autophagy to ameliorate senile dementia by binding Dynamin-1 and Adaptor protein 2 B1 in AD-like mice. Aging 2019, 11, 12002–12031. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, H.; Shang, H.; Chen, X.; Yang, S.; Qu, Y.; Ding, J.; Li, X. Circular RNA circ_0000950 promotes neuron apoptosis, suppresses neurite outgrowth and elevates inflammatory cytokines levels via directly sponging miR-103 in Alzheimer’s disease. Cell Cycle 2019, 18, 2197–2214. [Google Scholar] [CrossRef] [PubMed]
- Cochran, K.R.; Veeraraghavan, K.; Kundu, G.; Mazan-Mamczarz, K.; Coletta, C.; Thambisetty, M.; Gorospe, M.; De, S. Systematic Identification of circRNAs in Alzheimer’s Disease. Genes 2021, 12, 1258. [Google Scholar] [CrossRef]
- Welden, J.R.; van Doorn, J.; Nelson, P.T.; Stamm, S. The human MAPT locus generates circular RNAs. Biochim. Biophys Acta 2018, 1864, 2753–2760. [Google Scholar] [CrossRef]
- Lee, E.G.; Tulloch, J.; Chen, S.; Leong, L.; Saxton, A.D.; Kraemer, B.; Darvas, M.; Keene, C.D.; Shutes-David, A.; Todd, K.; et al. Redefining transcriptional regulation of the APOE gene and its association with Alzheimer’s disease. PLoS ONE 2020, 15, e0227667. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X.; Chen, Y.H.; Zhang, K. Identification of Circular RNA hsa_Circ_0003391 in Peripheral Blood Is Potentially Associated With Alzheimer’s Disease. Front. Aging Neurosci. 2020, 12, 601965. [Google Scholar] [CrossRef]
- Li, Y.; Lv, Z.; Zhang, J.; Ma, Q.; Li, Q.; Song, L.; Gong, L.; Zhu, Y.; Li, X.; Hao, Y.; et al. Profiling of differentially expressed circular RNAs in peripheral blood mononuclear cells from Alzheimer’s disease patients. Metab. Brain Dis. 2020, 35, 201–213. [Google Scholar] [CrossRef]
- Li, Y.; Fan, H.; Sun, J.; Ni, M.; Zhang, L.; Chen, C.; Hong, X.; Fang, F.; Zhang, W.; Ma, P. Circular RNA expression profile of Alzheimer’s disease and its clinical significance as biomarkers for the disease risk and progression. Int. J. Biochem. Cell Biol. 2020, 123, 105747. [Google Scholar] [CrossRef]
- Lo, I.; Hill, J.; Vilhjálmsson, B.J.; Kjems, J. Linking the association between circRNAs and Alzheimer’s disease progression by multi-tissue circular RNA characterization. RNA Biol. 2020, 17, 1789–1797. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Gómez-Isla, T.; Price, J.L.; McKeel, D.W.; Morris, J.C.; Growdon, J.H.; Hyman, B.T. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J. Neurosci. 1996, 16, 4491–4500. [Google Scholar] [CrossRef] [PubMed]
- Bevilaqua, L.R.; Rossato, J.I.; Bonini, J.S.; Myskiw, J.C.; Clarke, J.R.; Monteiro, S.; Lima, R.H.; Medina, J.H.; Cammarota, M.; Izquierdo, I. The role of the entorhinal cortex in extinction: Influences of aging. Neural Plast. 2008, 2008, 595282. [Google Scholar] [CrossRef]
- Liang, W.S.; Dunckley, T.; Beach, T.G.; Grover, A.; Mastroeni, D.; Walker, D.G.; Caselli, R.J.; Kukull, W.A.; McKeel, D.; Morris, J.C.; et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol. Genom. 2007, 28, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef]
- Lugli, G.; Cohen, A.M.; Bennett, D.A.; Shah, R.C.; Fields, C.J.; Hernandez, A.G.; Smalheiser, N.R. Plasma Exosomal miRNAs in Persons with and without Alzheimer Disease: Altered Expression and Prospects for Biomarkers. PLoS ONE 2015, 10, e0139233. [Google Scholar] [CrossRef]
- Riancho, J.; Vázquez-Higuera, J.L.; Pozueta, A.; Lage, C.; Kazimierczak, M.; Bravo, M.; Calero, M.; Gonalezález, A.; Rodríguez, E.; Lleó, A.; et al. MicroRNA Profile in Patients with Alzheimer’s Disease: Analysis of miR-9-5p and miR-598 in Raw and Exosome Enriched Cerebrospinal Fluid Samples. J. Alzheimers Dis. 2017, 57, 483–491. [Google Scholar] [CrossRef]
- Guévremont, D.; Tsui, H.; Knight, R.; Fowler, C.J.; Masters, C.L.; Martins, R.N.; Abraham, W.C.; Tate, W.P.; Cutfield, N.J.; Williams, J.M. Plasma microRNA vary in association with the progression of Alzheimer’s disease. Alzheimers Dement. 2022, 14, e12251. [Google Scholar] [CrossRef]
- Ravanidis, S.; Bougea, A.; Papagiannakis, N.; Koros, C.; Simitsi, A.M.; Pachi, I.; Breza, M.; Stefanis, L.; Doxakis, E. Validation of differentially expressed brain-enriched microRNAs in the plasma of PD patients. Ann. Clin. Transl. Neurol. 2020, 7, 1594–1607. [Google Scholar] [CrossRef]
- Rahmani, S.; Kadkhoda, S.; Ghafouri-Fard, S. Synaptic plasticity and depression: The role of miRNAs dysregulation. Mol. Biol. Rep. 2022, 49, 9759–9765. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Xiong, J.; Ji, L.; Xue, X. MiR-421 Aggravates Neurotoxicity and Promotes Cell Death in Parkinson’s Disease Models by Directly Targeting MEF2D. Neurochem. Res. 2021, 46, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Zhang, W. The Neuroprotective Effect of miR-136 on Pilocarpine-Induced Temporal Lobe Epilepsy Rats by Inhibiting Wnt/. Comput. Math. Methods Med. 2022, 2022, 1938205. [Google Scholar] [CrossRef]
- Raheja, R.; Regev, K.; Healy, B.C.; Mazzola, M.A.; Beynon, V.; Von Glehn, F.; Paul, A.; Diaz-Cruz, C.; Gholipour, T.; Glanz, B.I.; et al. Correlating serum micrornas and clinical parameters in amyotrophic lateral sclerosis. Muscle Nerve 2018, 58, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Boese, A.S.; Saba, R.; Campbell, K.; Majer, A.; Medina, S.; Burton, L.; Booth, T.F.; Chong, P.; Westmacott, G.; Dutta, S.M.; et al. MicroRNA abundance is altered in synaptoneurosomes during prion disease. Mol. Cell Neurosci. 2016, 71, 13–24. [Google Scholar] [CrossRef]
- Ben Halima, S.; Siegel, G.; Rajendran, L. miR-186 in Alzheimer’s disease: A big hope for a small RNA? J. Neurochem. 2016, 137, 308–311. [Google Scholar] [CrossRef]
- Rahman, M.R.; Islam, T.; Zaman, T.; Shahjaman, M.; Karim, M.R.; Huq, F.; Quinn, J.M.W.; Holsinger, R.M.D.; Gov, E.; Moni, M.A. Identification of molecular signatures and pathways to identify novel therapeutic targets in Alzheimer’s disease: Insights from a systems biomedicine perspective. Genomics 2020, 112, 1290–1299. [Google Scholar] [CrossRef]
- You, J.; Qian, F.; Huang, Y.; Guo, Y.; Lv, Y.; Yang, Y.; Lu, X.; Guo, T.; Wang, J.; Gu, B. lncRNA WT1-AS attenuates hypoxia/ischemia-induced neuronal injury during cerebral ischemic stroke via miR-186-5p/XIAP axis. Open Med. 2022, 17, 1338–1349. [Google Scholar] [CrossRef]
- Cai, S.C.; Li, X.P.; Li, X.; Tang, G.Y.; Yi, L.M.; Hu, X.S. Oleanolic Acid Inhibits Neuronal Pyroptosis in Ischaemic Stroke by Inhibiting miR-186-5p Expression. Exp. Neurobiol. 2021, 30, 401–414. [Google Scholar] [CrossRef]
- Kim, J.; Yoon, H.; Chung, D.E.; Brown, J.L.; Belmonte, K.C. miR-186 is decreased in aged brain and suppresses BACE1 expression. J. Neurochem. 2016, 137, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, D.; Brager, D.H.; Rymer, J.K.; Bunk, A.T.; White, A.R.; Elsayed, N.A.; Krzeski, J.C.; Snider, A.; Schroeder Carter, L.M.; Danzer, S.C.; et al. MicroRNA inhibition upregulates hippocampal A-type potassium current and reduces seizure frequency in a mouse model of epilepsy. Neurobiol. Dis. 2019, 130, 104508. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; He, X.; Yang, L.; Li, Q.; Xu, Y. Inhibition of miR-421 Preserves Mitochondrial Function and Protects against Parkinson’s Disease Pathogenesis via Pink1/Parkin-Dependent Mitophagy. Dis. Markers 2022, 2022, 5186252. [Google Scholar] [CrossRef]
- Hu, F.; Shao, L.; Zhang, J.; Zhang, H.; Wen, A.; Zhang, P. Knockdown of ZFAS1 Inhibits Hippocampal Neurons Apoptosis and Autophagy by Activating the PI3K/AKT Pathway via Up-regulating miR-421 in Epilepsy. Neurochem. Res. 2020, 45, 2433–2441. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Han, X.R.; Wang, Y.J.; Wang, S.; Shen, M.; Zhang, Z.F.; Fan, S.H.; Shan, Q.; Wang, L.; Li, M.Q.; et al. MicroRNA-421 suppresses the apoptosis and autophagy of hippocampal neurons in epilepsy mice model by inhibition of the TLR/MYD88 pathway. J. Cell. Physiol. 2018, 233, 7022–7034. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, H.; Huang, Y.; Li, Y.; Fan, J.; Wang, R.; Han, Z.; Yang, Z.; Wu, L.; Wu, D.; et al. Dysregulation of Principal Circulating miRNAs in Non-human Primates Following Ischemic Stroke. Front. Neurosci. 2021, 15, 738576. [Google Scholar] [CrossRef]
- Li, N.; Zhang, D.; Guo, H.; Yang, Q.; Li, P.; He, Y. Inhibition of circ_0004381 improves cognitive function via miR-647/PSEN1 axis in an Alzheimer disease mouse model. J. Neuropathol. Exp. Neurol. 2022, 82, 84–92. [Google Scholar] [CrossRef]
- Li, K.; Shen, L.; Zheng, P.; Wang, Y.; Wang, L.; Meng, X.; Lv, Y.; Xue, Z.; Guo, X.; Zhang, A.; et al. Identification of MicroRNAs as potential biomarkers for detecting ischemic stroke. Genes Genom. 2022, 44, 9–17. [Google Scholar] [CrossRef]
- Rademakers, R.; Eriksen, J.L.; Baker, M.; Robinson, T.; Ahmed, Z.; Lincoln, S.J.; Finch, N.; Rutherford, N.J.; Crook, R.J.; Josephs, K.A.; et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum. Mol. Genet. 2008, 17, 3631–3642. [Google Scholar] [CrossRef]
- Piscopo, P.; Grasso, M.; Fontana, F.; Crestini, A.; Puopolo, M.; Del Vescovo, V.; Venerosi, A.; Calamandrei, G.; Vencken, S.F.; Greene, C.M.; et al. Reduced miR-659-3p Levels Correlate with Progranulin Increase in Hypoxic Conditions: Implications for Frontotemporal Dementia. Front. Mol. Neurosci. 2016, 9, 31. [Google Scholar] [CrossRef]
- Ibrahim, H.M.; AlZahrani, A.; Hanieh, H.; Ahmed, E.A.; Thirugnanasambantham, K. MicroRNA-7188-5p and miR-7235 regulates Multiple sclerosis in an experimental mouse model. Mol. Immunol. 2021, 139, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Miller-Delaney, S.F.; Bryan, K.; Das, S.; McKiernan, R.C.; Bray, I.M.; Reynolds, J.P.; Gwinn, R.; Stallings, R.L.; Henshall, D.C. Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy. Brain 2015, 138, 616–631. [Google Scholar] [CrossRef]
- Karisetty, B.C.; Bhatnagar, A.; Armour, E.M.; Beaver, M.; Zhang, H.; Elefant, F. Amyloid-β Peptide Impact on Synaptic Function and Neuroepigenetic Gene Control Reveal New Therapeutic Strategies for Alzheimer’s Disease. Front. Mol. Neurosci. 2020, 13, 577622. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Huang, Z.S.; Yu, C.C.; Wang, H.H.; Zhou, H.; Kong, L.H. Epigenetic Regulation of Amyloid-beta Metabolism in Alzheimer’s Disease. Curr. Med. Sci. 2020, 40, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Huang, X.; Su, Y.; Yin, G.; Wang, S.; Yu, B.; Li, H.; Qi, J.; Chen, H.; Zeng, W.; et al. Activation of Wnt/β-catenin pathway mitigates blood-brain barrier dysfunction in Alzheimer’s disease. Brain 2022, 145, 4474–4488. [Google Scholar] [CrossRef] [PubMed]
- Hegde, A.N.; Smith, S.G.; Duke, L.M.; Pourquoi, A.; Vaz, S. Perturbations of Ubiquitin-Proteasome-Mediated Proteolysis in Aging and Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 324. [Google Scholar] [CrossRef]
- Zhang, L.; Qi, Z.; Li, J.; Li, M.; Du, X.; Wang, S.; Zhou, G.; Xu, B.; Liu, W.; Xi, S.; et al. Roles and Mechanisms of Axon-Guidance Molecules in Alzheimer’s Disease. Mol. Neurobiol. 2021, 58, 3290–3307. [Google Scholar] [CrossRef]
- Skaper, S.D.; Facci, L.; Zusso, M.; Giusti, P. Synaptic Plasticity, Dementia and Alzheimer Disease. CNS Neurol. Disord. Drug Targets 2017, 16, 220–233. [Google Scholar] [CrossRef]
- Snow, A.D.; Cummings, J.A.; Lake, T. The Unifying Hypothesis of Alzheimer’s Disease: Heparan Sulfate Proteoglycans/Glycosaminoglycans Are Key as First Hypothesized Over 30 Years Ago. Front. Aging Neurosci. 2021, 13, 710683. [Google Scholar] [CrossRef]
- Sandwall, E.; O’Callaghan, P.; Zhang, X.; Lindahl, U.; Lannfelt, L.; Li, J.P. Heparan sulfate mediates amyloid-beta internalization and cytotoxicity. Glycobiology 2010, 20, 533–541. [Google Scholar] [CrossRef]
- Beckman, M.; Holsinger, R.M.; Small, D.H. Heparin activates beta-secretase (BACE1) of Alzheimer’s disease and increases autocatalysis of the enzyme. Biochemistry 2006, 45, 6703–6714. [Google Scholar] [CrossRef] [PubMed]
- Bukke, V.N.; Archana, M.; Villani, R.; Romano, A.D.; Wawrzyniak, A.; Balawender, K.; Orkisz, S.; Beggiato, S.; Serviddio, G.; Cassano, T. The Dual Role of Glutamatergic Neurotransmission in Alzheimer’s Disease: From Pathophysiology to Pharmacotherapy. Int. J. Mol. Sci. 2020, 21, 7452. [Google Scholar] [CrossRef]
- Bell, J.E.; Alafuzoff, I.; Al-Sarraj, S.; Arzberger, T.; Bogdanovic, N.; Budka, H.; Dexter, D.T.; Falkai, P.; Ferrer, I.; Gelpi, E.; et al. Management of a twenty-first century brain bank: Experience in the BrainNet Europe consortium. Acta Neuropathol. 2008, 115, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Montine, T.J.; Phelps, C.H.; Beach, T.G.; Bigio, E.H.; Cairns, N.J.; Dickson, D.W.; Duyckaerts, C.; Frosch, M.P.; Masliah, E.; Mirra, S.S.; et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: A practical approach. Acta Neuropathol. 2012, 123, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Alafuzoff, I.; Arzberger, T.; Kretzschmar, H.; Del Tredici, K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006, 112, 389–404. [Google Scholar] [CrossRef]
- Mirra, S.S.; Heyman, A.; McKeel, D.; Sumi, S.M.; Crain, B.J.; Brownlee, L.M.; Vogel, F.S.; Hughes, J.P.; van Belle, G.; Berg, L. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 1991, 41, 479–486. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Navarro Gonzalez, J.; Zweig, A.S.; Speir, M.L.; Schmelter, D.; Rosenbloom, K.R.; Raney, B.J.; Powell, C.C.; Nassar, L.R.; Maulding, N.D.; Lee, C.M.; et al. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 2021, 49, D1046–D1057. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Method of Thal | Braak and Braak Classification | Method of CERAD | ABC Score | Global Average Area of Aβ Deposits | ||
|---|---|---|---|---|---|---|
| log(circAPP) | Correlation coeficient (Spearman’s Rho) | −0.496 ** | −0.503 ** | −0.499 ** | −0.515 ** | −0.480 * |
| Sig. (bilateral) | 0.001 | 0.001 | 0.001 | 0.000 | 0.024 | |
| N | 43 | 44 | 43 | 43 | 22 | |
| log(APP mRNA) | Correlation coeficient (Spearman’s Rho) | −0.339 * | −0.415 ** | −0.243 | −0.384 * | −0.652 ** |
| Sig. (bilateral) | 0.026 | 0.005 | 0.116 | 0.011 | 0.001 | |
| N | 43 | 44 | 43 | 43 | 22 |
| miRNA | Neurological Disorders Related | Expression in AD | Tissue/Fluid Studied | References |
|---|---|---|---|---|
| hsa-miR-1200 | - | |||
| hsa-miR-1208 | - | |||
| hsa-miR-1270 | - | |||
| hsa-miR-1272 | - | |||
| hsa-miR-136 | Prion disease, epilepsy, depression, PD | [50,51,53,56] | ||
| hsa-miR-186 | AD, ischemic stroke | - | Neuro-2a cells, 7PA2 cells | [57,58,59,60,61] |
| hsa-miR-324-5p | AD | ↓ | plasma | [49,62] |
| hsa-miR-421 | PD, epilepsy, ischemic stroke | [63,64,65,66] | ||
| hsa-miR-518a-5p | - | |||
| hsa-miR-527 | - | |||
| hsa-miR-549 | - | |||
| hsa-miR-598 | AD | No detected | CSF | [48] |
| ↑(tendency) | CSF exosomes | |||
| hsa-miR-620 | - | |||
| hsa-miR-646 | - | |||
| hsa-miR-647 | AD | [67] | ||
| hsa-miR-659 | AD, ischemic stroke, frontotemporal dementia, MS | ↓ | hippocampal neurons AD mouse model | [47,68,69,70,71] |
| hsa-miR-876-3p | Epilepsy | ↑ | Plasma exosomes | [72] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urdánoz-Casado, A.; Sánchez-Ruiz de Gordoa, J.; Robles, M.; Roldan, M.; Macías Conde, M.; Acha, B.; Blanco-Luquin, I.; Mendioroz, M. circRNA from APP Gene Changes in Alzheimer’s Disease Human Brain. Int. J. Mol. Sci. 2023, 24, 4308. https://doi.org/10.3390/ijms24054308
Urdánoz-Casado A, Sánchez-Ruiz de Gordoa J, Robles M, Roldan M, Macías Conde M, Acha B, Blanco-Luquin I, Mendioroz M. circRNA from APP Gene Changes in Alzheimer’s Disease Human Brain. International Journal of Molecular Sciences. 2023; 24(5):4308. https://doi.org/10.3390/ijms24054308
Chicago/Turabian StyleUrdánoz-Casado, Amaya, Javier Sánchez-Ruiz de Gordoa, Maitane Robles, Miren Roldan, Mónica Macías Conde, Blanca Acha, Idoia Blanco-Luquin, and Maite Mendioroz. 2023. "circRNA from APP Gene Changes in Alzheimer’s Disease Human Brain" International Journal of Molecular Sciences 24, no. 5: 4308. https://doi.org/10.3390/ijms24054308
APA StyleUrdánoz-Casado, A., Sánchez-Ruiz de Gordoa, J., Robles, M., Roldan, M., Macías Conde, M., Acha, B., Blanco-Luquin, I., & Mendioroz, M. (2023). circRNA from APP Gene Changes in Alzheimer’s Disease Human Brain. International Journal of Molecular Sciences, 24(5), 4308. https://doi.org/10.3390/ijms24054308






