Brassica napus BnaC9.DEWAX1 Negatively Regulates Wax Biosynthesis via Transcriptional Suppression of BnCER1-2
Abstract
1. Introduction
2. Results
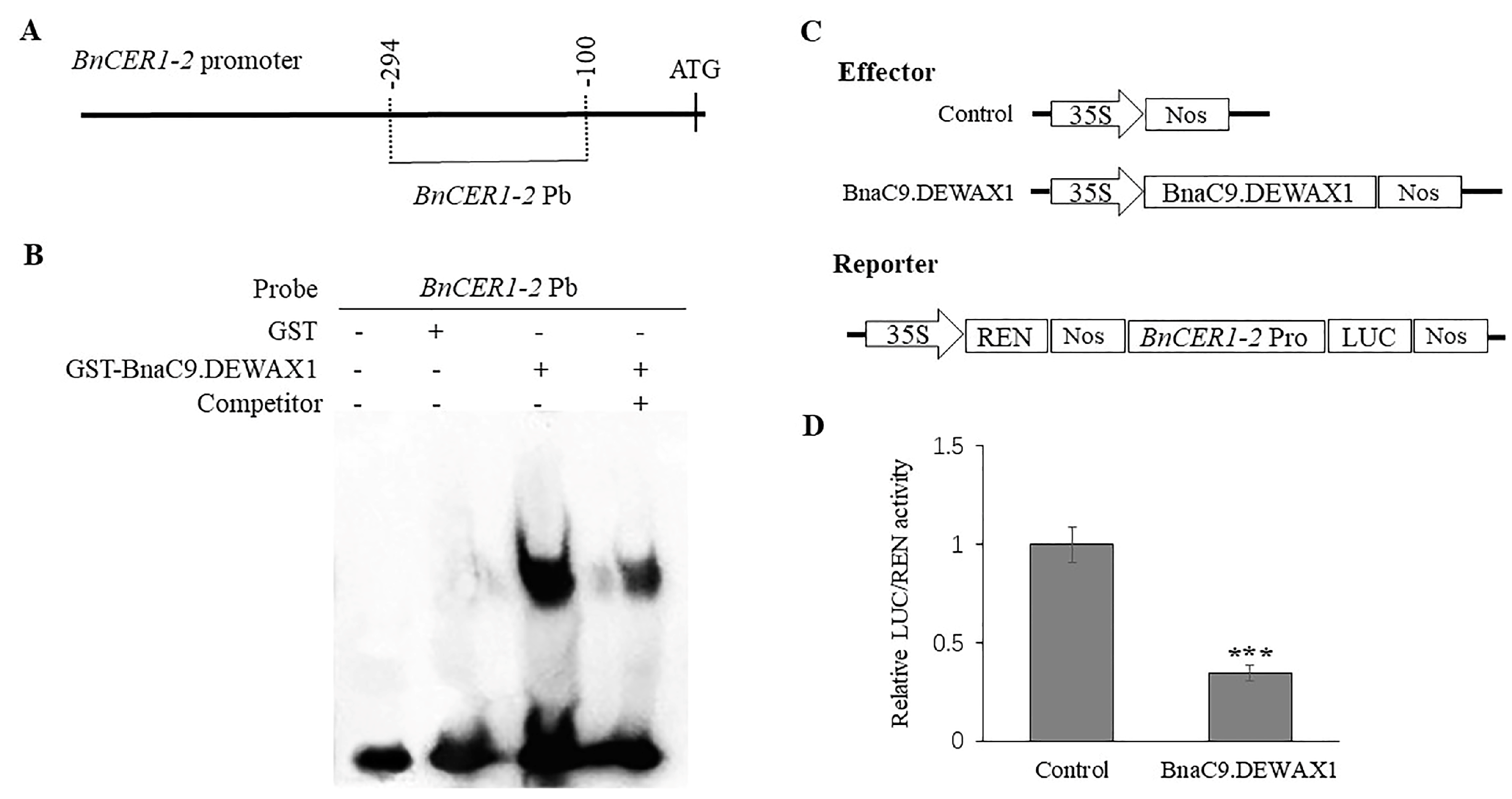
2.1. Transcription Factor BnaC9.DEWAX1 Binds to and Represses the BnCER1-2 Promoter
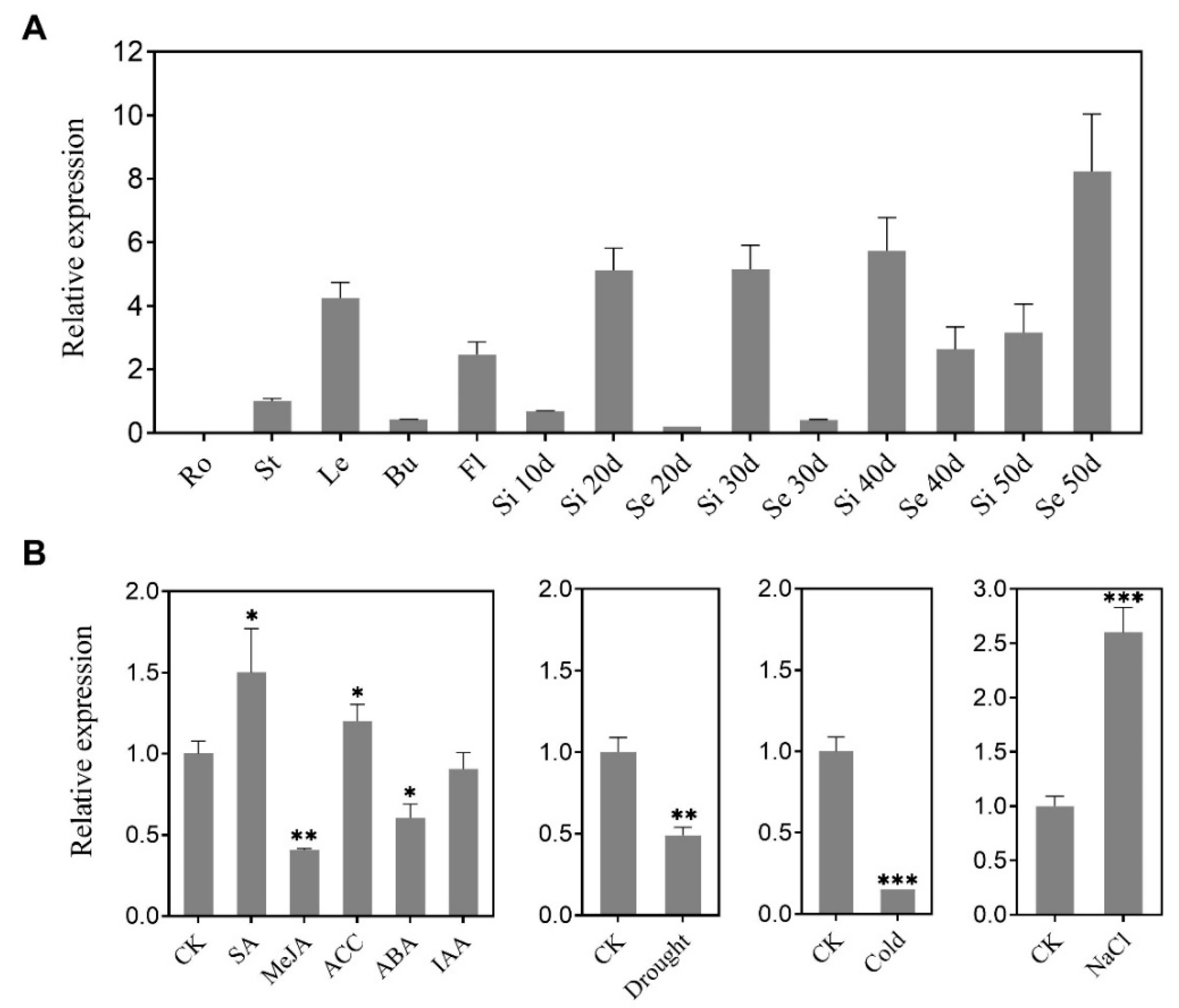
2.2. BnaC9.DEWAX1 Is Predominately Expressed in the Leaf and Exhibits Altered Levels of Expression in Response to Abiotic Stress
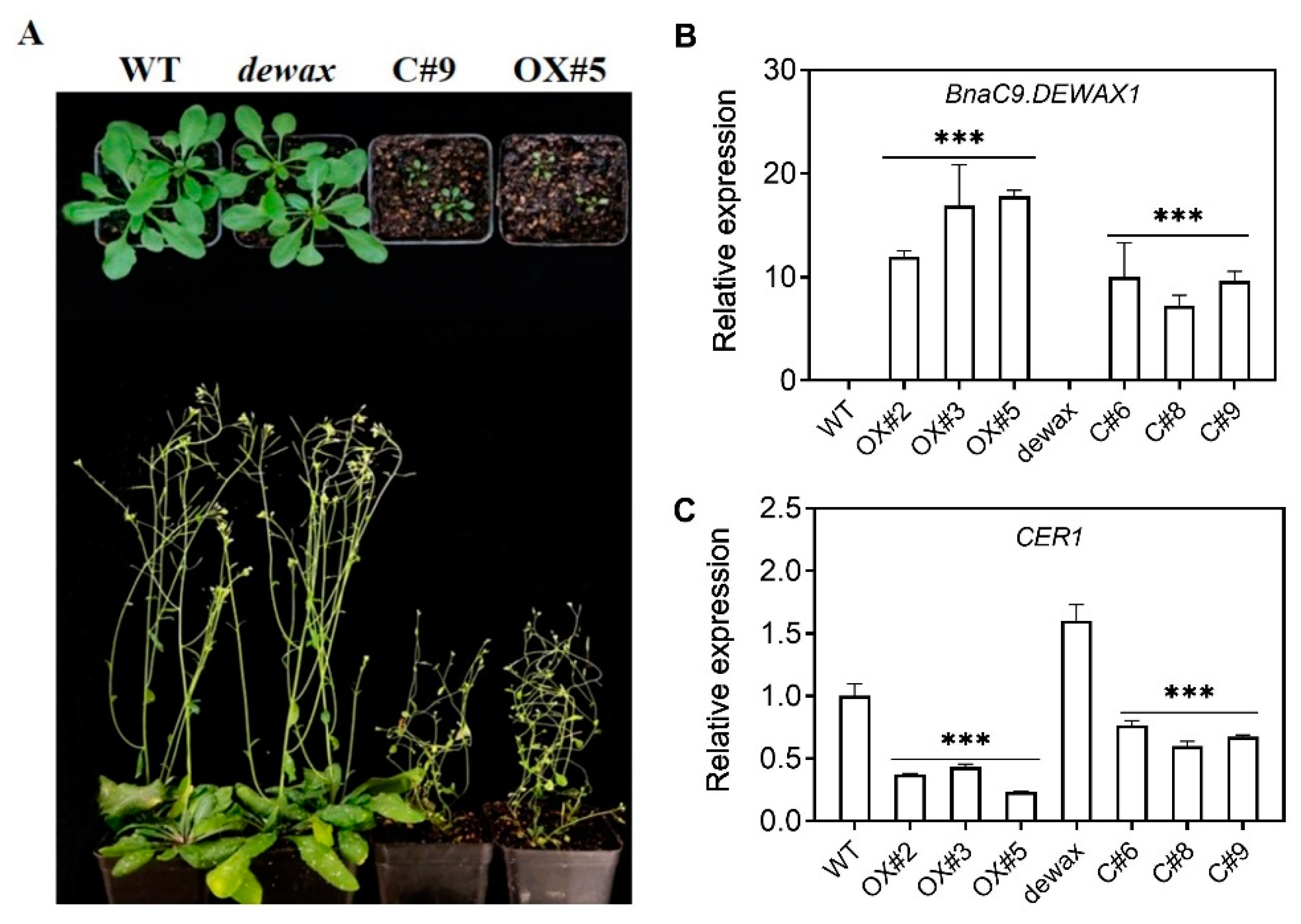
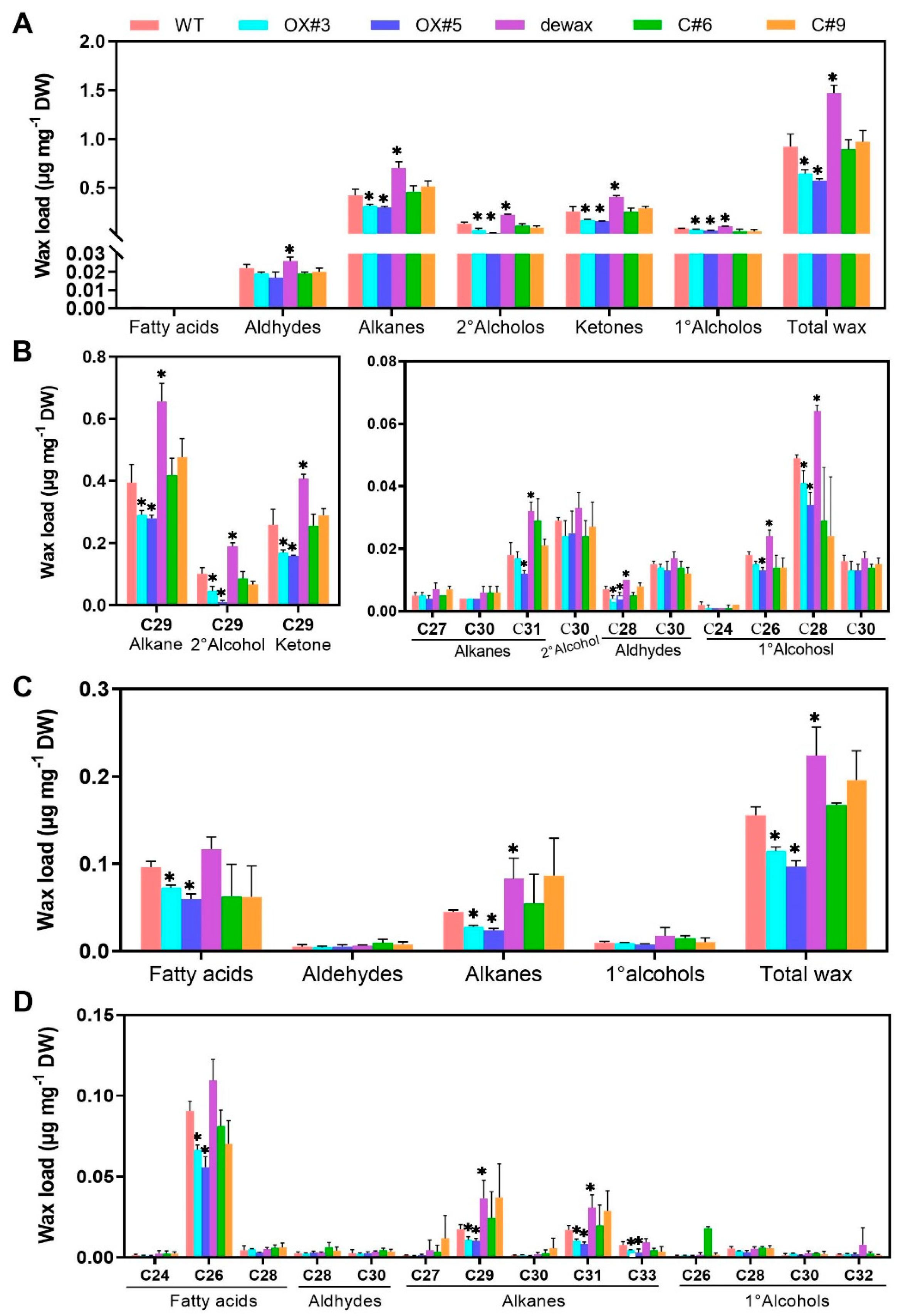
2.3. Cuticular Wax Deposition Is Negatively Regulated by BnaC9.DEWAX1
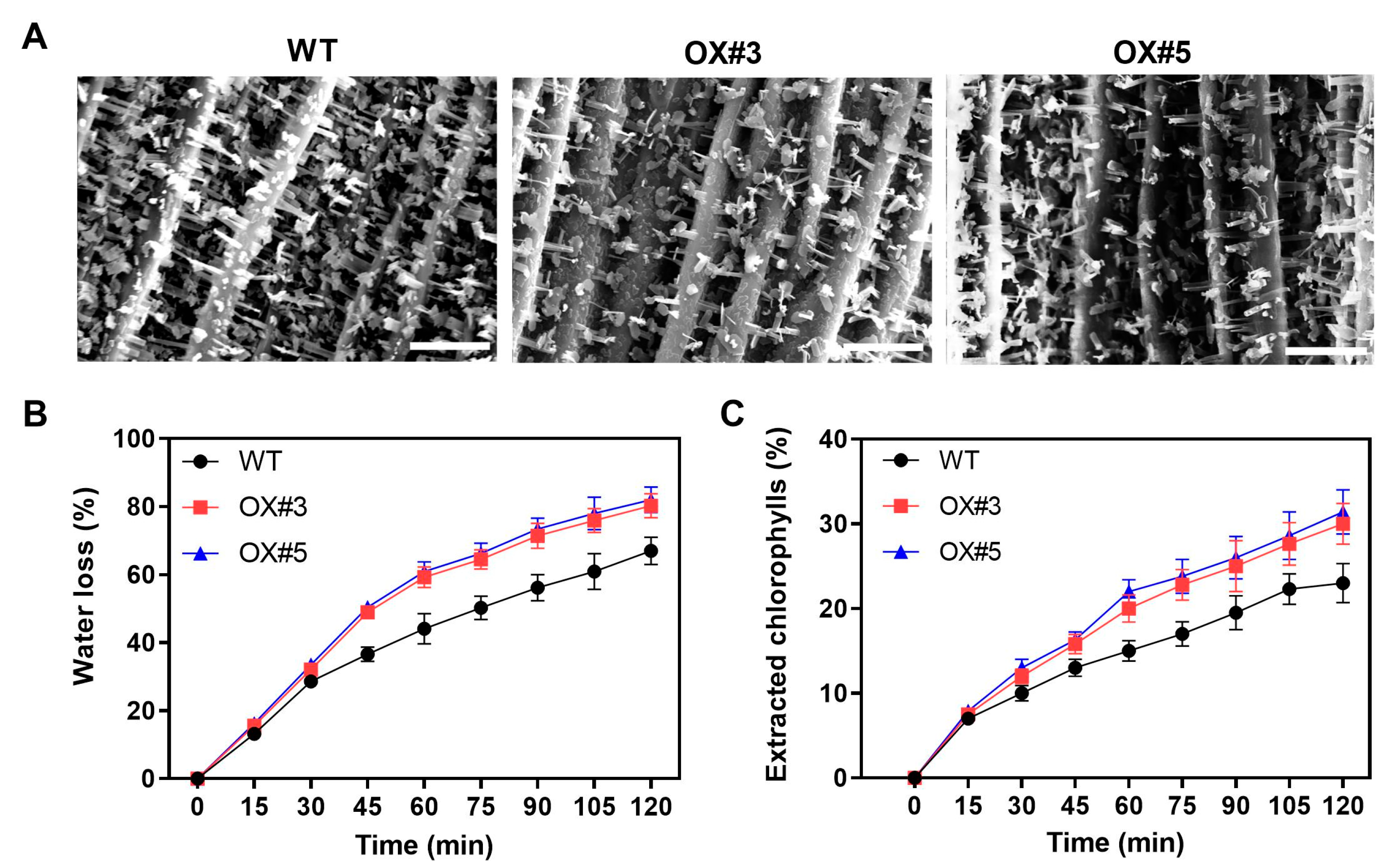
2.4. BnaC9.DEWAX1 Negatively Regulates Cuticle Permeability in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Generation of Transgenic Plants
4.3. qRT-PCR Analysis
4.4. Yeast One-Hybrid Assays
4.5. Subcellular Localization
4.6. Transcriptional Repression Assay
4.7. Transient Dual-LUC Assay
4.8. Electrophoretic Mobility Shift Assays (EMSA)
4.9. Cuticular Wax Analysis
4.10. Scanning Electron Microscope (SEM)
4.11. Cuticle Permeability Analysis
4.12. Phylogenetic Analysis
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dietz, K.J.; Zorb, C.; Geilfus, C.M. Drought and crop yield. Plant Biol. 2021, 23, 881–893. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rico-Medina, A.; Cano-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; van Arkel, G.; Pereira, A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 2004, 16, 2463–2480. [Google Scholar] [CrossRef]
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytol. 2020, 227, 698–713. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, T.; Griffiths, D.W. The effects of stress on plant cuticular waxes. New Phytol. 2006, 171, 469–499. [Google Scholar] [CrossRef] [PubMed]
- Al-Abdallat, A.M.; Al-Debei, H.S.; Ayad, J.Y.; Hasan, S. Over-Expression of SlSHN1 Gene Improves Drought Tolerance by Increasing Cuticular Wax Accumulation in Tomato. Int. J. Mol. Sci. 2014, 15, 19499–19515. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Broeckling, C.D.; Sumner, L.W.; Wang, Z.-Y. Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 2007, 64, 265–278. [Google Scholar] [CrossRef]
- Zhou, X.; Jenks, M.A.; Liu, J.; Liu, A.; Zhang, X.; Xiang, J.; Zou, J.; Peng, Y.; Chen, X. Overexpression of Transcription Factor OsWR2 Regulates Wax and Cutin Biosynthesis in Rice and Enhances its Tolerance to Water Deficit. Plant Mol. Biol. Report. 2014, 32, 719–731. [Google Scholar] [CrossRef]
- Zhu, L.; Guo, J.; Zhu, J.; Zhou, C. Enhanced expression of EsWAX1 improves drought tolerance with increased accumulation of cuticular wax and ascorbic acid in transgenic Arabidopsis. Plant Physiol. Biochem. 2014, 75, 24–35. [Google Scholar] [CrossRef]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef]
- Jetter, R.; Kunst, L.; Samuels, A.L. Composition of Plant Cuticular Waxes. In Annual Plant Reviews Volume 23: Biology of the Plant Cuticle; Blackwell Publishing: Oxford, UK, 2006; pp. 145–181. [Google Scholar]
- Kunst, L.; Samuels, A.L. Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 2003, 42, 51–80. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Suh, M.C. Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 2015, 34, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Bourdenx, B.; Bernard, A.; Domergue, F.; Pascal, S.; Léger, A.; Roby, D.; Pervent, M.; Vile, D.; Haslam, R.P.; Napier, J.A.; et al. Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 2011, 156, 29–45. [Google Scholar] [CrossRef]
- Li, H.; Guo, Y.; Cui, Q.; Zhang, Z.; Yan, X.; Ahammed, G.J.; Yang, X.; Yang, J.; Wei, C.; Zhang, X. Alkanes (C29 and C31)-Mediated Intracuticular Wax Accumulation Contributes to Melatonin- and ABA-Induced Drought Tolerance in Watermelon. J. Plant Growth Regul. 2020, 39, 1441–1450. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, Y.; Xu, C.; Ren, J.; Liu, X.; Black, K.; Gai, X.; Wang, Q.; Ren, H. Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol. Biol. 2015, 87, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Domergue, F.; Pascal, S.; Jetter, R.; Renne, C.; Faure, J.D.; Haslam, R.P.; Napier, J.A.; Lessire, R.; Joubès, J. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 2012, 24, 3106–3118. [Google Scholar] [CrossRef]
- Pascal, S.; Bernard, A.; Deslous, P.; Gronnier, J.; Fournier-Goss, A.; Domergue, F.; Rowland, O.; Joubès, J. Arabidopsis CER1-LIKE1 Functions in a Cuticular Very-Long-Chain Alkane-Forming Complex. Plant Physiol. 2019, 179, 415–432. [Google Scholar] [CrossRef]
- Go, Y.S.; Kim, H.; Kim, H.J.; Suh, M.C. Arabidopsis Cuticular Wax Biosynthesis Is Negatively Regulated by the DEWAX Gene Encoding an AP2/ERF-Type Transcription Factor. Plant Cell 2014, 26, 1666–1680. [Google Scholar] [CrossRef]
- Kim, H.; Go, Y.S.; Suh, M.C. DEWAX2 Transcription Factor Negatively Regulates Cuticular Wax Biosynthesis in Arabidopsis Leaves. Plant Cell Physiol. 2018, 59, 966–977. [Google Scholar] [CrossRef]
- Liu, Q.; Huang, H.; Chen, Y.; Yue, Z.; Wang, Z.; Qu, T.; Xu, D.; Lü, S.; Hu, H. Two Arabidopsis MYB-SHAQKYF transcription repressors regulate leaf wax biosynthesis via transcriptional suppression on DEWAX. New Phytol. 2022, 236, 2115–2130. [Google Scholar] [CrossRef]
- Yang, S.U.; Kim, H.; Kim, R.J.; Kim, J.; Suh, M.C. AP2/DREB Transcription Factor RAP2.4 Activates Cuticular Wax Biosynthesis in Arabidopsis Leaves Under Drought. Front. Plant Sci. 2020, 11, 895. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Go, Y.S.; Park, C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Kim, J.; Suh, M.C.; Seo, P.J. The MIEL1 E3 Ubiquitin Ligase Negatively Regulates Cuticular Wax Biosynthesis in Arabidopsis Stems. Plant Cell Physiol. 2017, 58, 2040. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.; Zhao, L.; Eveleigh, N.; Yu, Y.; Chen, X.; Kunst, L. The exosome and trans-acting small interfering RNAs regulate cuticular wax biosynthesis during Arabidopsis inflorescence stem development. Plant Physiol. 2015, 167, 323–336. [Google Scholar] [CrossRef]
- Lam, P.; Zhao, L.; McFarlane, H.E.; Aiga, M.; Lam, V.; Hooker, T.S.; Kunst, L. RDR1 and SGS3, Components of RNA-Mediated Gene Silencing, Are Required for the Regulation of Cuticular Wax Biosynthesis in Developing Inflorescence Stems of Arabidopsis. Plant Physiol. 2012, 159, 1385–1395. [Google Scholar] [CrossRef]
- Kim, H.; Yu, S.-I.; Jung, S.H.; Lee, B.-H.; Suh, M.C. The F-Box Protein SAGL1 and ECERIFERUM3 Regulate Cuticular Wax Biosynthesis in Response to Changes in Humidity in Arabidopsis. Plant Cell 2019, 31, 2223–2240. [Google Scholar] [CrossRef]
- Ménard, R.; Verdier, G.; Ors, M.; Erhardt, M.; Beisson, F.; Shen, W.-H. Histone H2B monoubiquitination is involved in the regulation of cutin and wax composition in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 455–466. [Google Scholar] [CrossRef]
- Lee, S.; Fu, F.; Xu, S.; Lee, S.Y.; Yun, D.J.; Mengiste, T. Global Regulation of Plant Immunity by Histone Lysine Methyl Transferases. Plant Cell 2016, 28, 1640–1661. [Google Scholar] [CrossRef]
- Wang, T.; Xing, J.; Liu, X.; Yao, Y.; Hu, Z.; Peng, H.; Xin, M.; Zhou, D.X.; Zhang, Y.; Ni, Z. GCN5 contributes to stem cuticular wax biosynthesis by histone acetylation of CER3 in Arabidopsis. J. Exp. Bot. 2018, 69, 2911–2922. [Google Scholar] [CrossRef]
- Batool, M.; El-Badri, A.M.; Hassan, M.U.; Haiyun, Y.; Chunyun, W.; Zhenkun, Y.; Jie, K.; Wang, B.; Zhou, G. Drought Stress in Brassica napus: Effects, Tolerance Mechanisms, and Management Strategies. J. Plant Growth Regul. 2023, 42, 21–45. [Google Scholar] [CrossRef]
- Khanzada, H.; Wassan, G.M.; He, H.; Mason, A.S.; Keerio, A.A.; Khanzada, S.; Faheem, M.; Solangi, A.M.; Zhou, Q.; Fu, D.; et al. Differentially evolved drought stress indices determine the genetic variation of Brassica napus at seedling traits by genome-wide association mapping. J. Adv. Res. 2020, 24, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, S.; Xu, Y.; Li, S.; Zhang, S.; Yuan, Z.; Li, J.; Ni, Y. Overexpression of BnKCS1-1, BnKCS1-2, and BnCER1-2 promotes cuticular wax production and increases drought tolerance in Brassica napus. Crop J. 2020, 8, 26–37. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Kosma, D.K.; Bourdenx, B.; Bernard, A.; Parsons, E.P.; Lü, S.; Joubès, J.; Jenks, M.A. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 2009, 151, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- Barthlott, W.; Mail, M.; Bhushan, B.; Koch, K. Plant surfaces: Structures and functions for biomimetic innovations. Nano Micro Lett. 2017, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Jach, G.; Binot, E.; Frings, S.; Luxa, K.; Schell, J. Use of red fluorescent protein from Discosoma sp. (dsRED) as a reporter for plant gene expression. Plant J. 2001, 28, 483–491. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L.; et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef]
- Tang, J.; Yang, X.; Xiao, C.; Li, J.; Chen, Y.; Li, R.; Li, S.; Lü, S.; Hu, H. GDSL lipase occluded stomatal pore 1 is required for wax biosynthesis and stomatal cuticular ledge formation. New Phytol. 2020, 228, 1880–1896. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Bai, C.; Luo, N.; Jiang, Y.; Wang, Y.; Liu, Y.; Chen, C.; Wang, Y.; Gan, Q.; Jin, S.; et al. Brassica napus BnaC9.DEWAX1 Negatively Regulates Wax Biosynthesis via Transcriptional Suppression of BnCER1-2. Int. J. Mol. Sci. 2023, 24, 4287. https://doi.org/10.3390/ijms24054287
Wang S, Bai C, Luo N, Jiang Y, Wang Y, Liu Y, Chen C, Wang Y, Gan Q, Jin S, et al. Brassica napus BnaC9.DEWAX1 Negatively Regulates Wax Biosynthesis via Transcriptional Suppression of BnCER1-2. International Journal of Molecular Sciences. 2023; 24(5):4287. https://doi.org/10.3390/ijms24054287
Chicago/Turabian StyleWang, Saiyu, Chengcheng Bai, Na Luo, Youwei Jiang, Yulu Wang, Yu Liu, Chunjie Chen, Yuxin Wang, Qiaoqiao Gan, Shurong Jin, and et al. 2023. "Brassica napus BnaC9.DEWAX1 Negatively Regulates Wax Biosynthesis via Transcriptional Suppression of BnCER1-2" International Journal of Molecular Sciences 24, no. 5: 4287. https://doi.org/10.3390/ijms24054287
APA StyleWang, S., Bai, C., Luo, N., Jiang, Y., Wang, Y., Liu, Y., Chen, C., Wang, Y., Gan, Q., Jin, S., & Ni, Y. (2023). Brassica napus BnaC9.DEWAX1 Negatively Regulates Wax Biosynthesis via Transcriptional Suppression of BnCER1-2. International Journal of Molecular Sciences, 24(5), 4287. https://doi.org/10.3390/ijms24054287




