Metabolomic Insights of Biosurfactant Activity from Bacillus niabensis against Planktonic Cells and Biofilm of Pseudomonas stutzeri Involved in Marine Biofouling
Abstract
1. Introduction
2. Results
2.1. Isolation and Identification of Marine Forming Biofilm Bacteria
2.2. Biosurfactant Production from Bacillus niabensis
2.3. Effects of Biosurfactant in Growth and Biofilm Inhibition
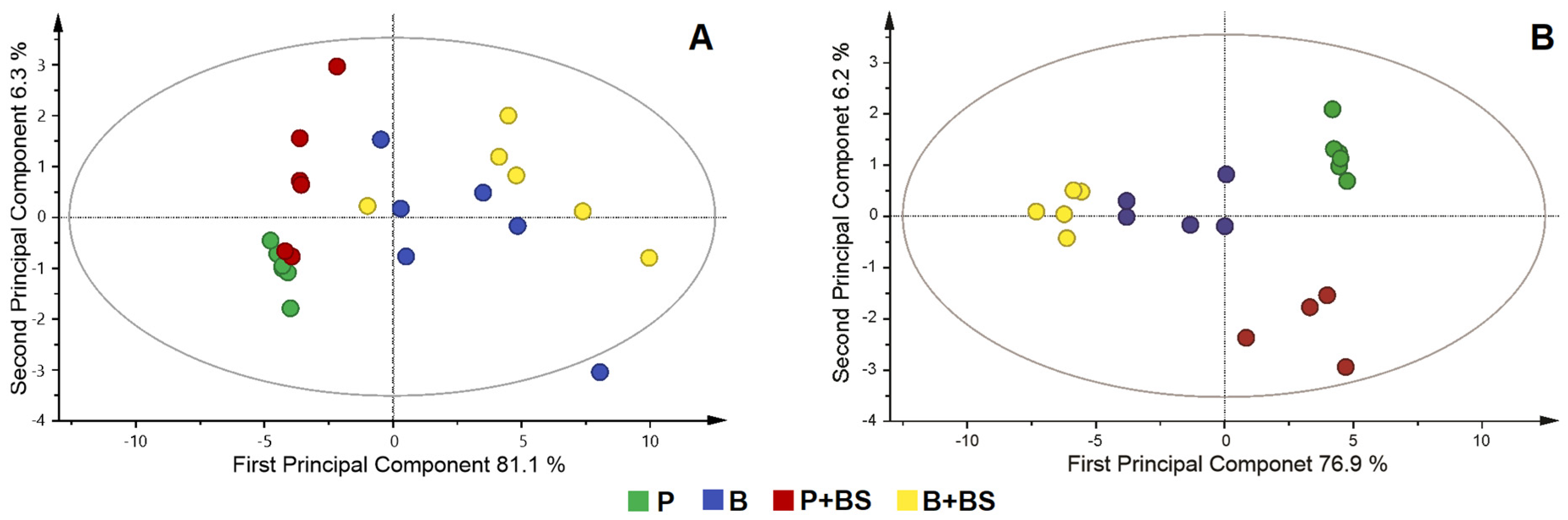
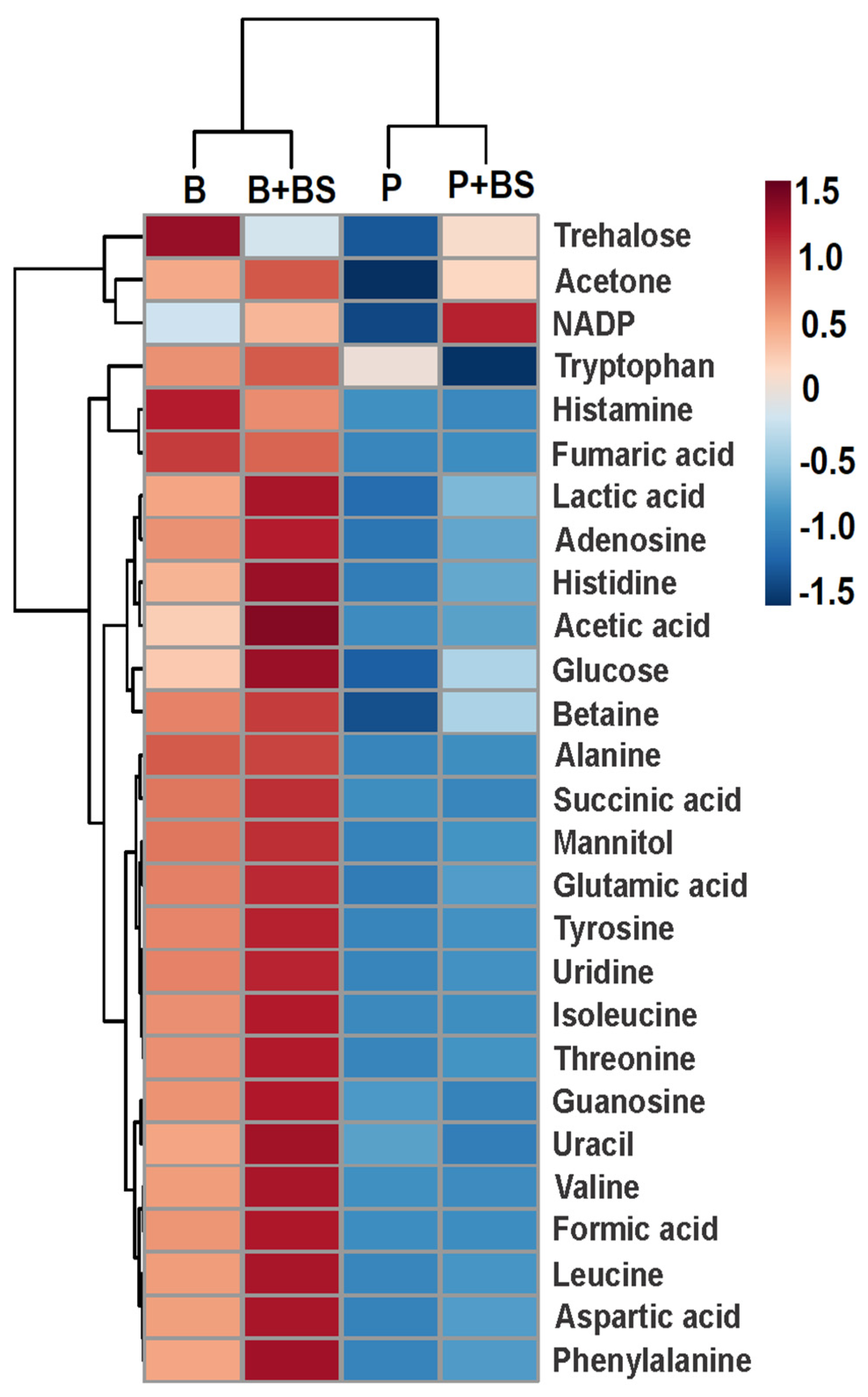
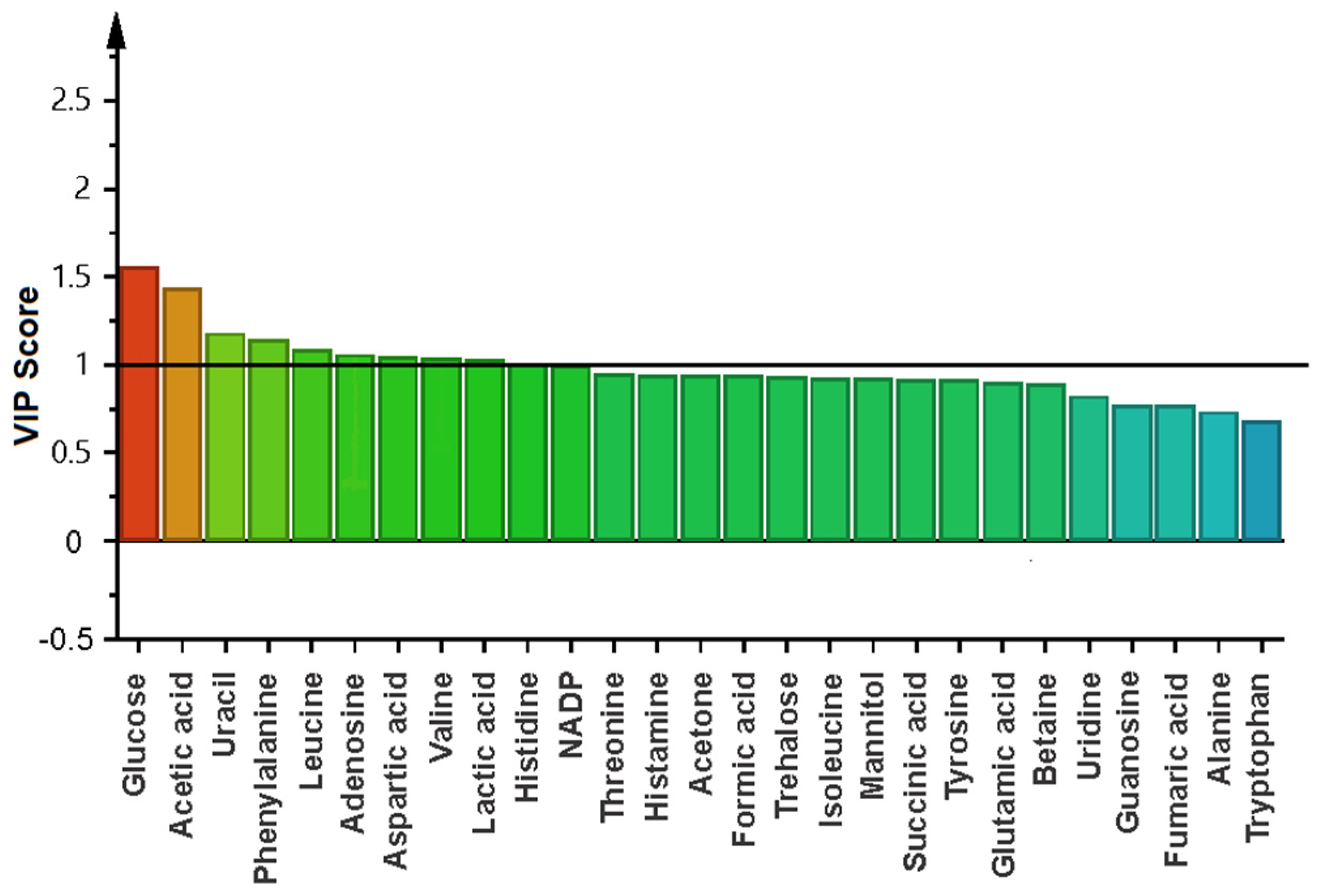
2.4. Metabolomic Changes in Planktonic Cells and Biofilm by Effect of Biosurfactant
3. Discussion
4. Materials and Methods
4.1. Chemical and Strain Identification
4.2. Culture and Biosurfactant Properties of Bacillus niabensis
4.2.1. Inoculum and Culture Conditions
4.2.2. Drop Collapse Test
4.2.3. Oil Displacement Test
4.2.4. Emulsification Assay
4.3. Assay of Effect of BS in the Growth and Biofilm Formation
4.3.1. Production and Extraction of Crude Biosurfactant
4.3.2. Antibacterial and Antibiofilm Activity
Antibacterial Assay by Agar Well Diffusion Method
Biofilm Formation
Quantitation of Biofilm Protein
Effect of BS in Growth Rates of P. stutzeri
Growth and Biofilm Inhibition
4.4. 1H NMR Metabolic Analysis
4.4.1. Biofilm and Planktonic Cells Source and Sample Preparation
4.4.2. Nuclear Magnetic Resonance (NMR) Experiments and Data Analysis
4.4.3. Identification and Quantification of Metabolites
4.4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dobrestov, S.; Teplistki, M.; Paul, V. Mini-review: Quorum sensing in the marine environment and its relationship to biofouling. Biofouling 2009, 25, 413–427. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Panda, A.K.; Mandal, D.S.; Shakeel, M.; Bisht, S.S.; Khan, J. Natural Anti-biofilm Agents: Strategies to control biofilm-Forming Pathogens. Front. Microbiol. 2020, 11, 566325. [Google Scholar] [CrossRef] [PubMed]
- Padmavathi, A.R.; Pandian, S.K. Antibiofilm activity of biosurfactant producing coral associated bacteria isolated from gulf of Mannar. Indian. J. Microbiol. 2014, 54, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, M.G. Biofilms and marine invertebrate larvae: What bacteria produce that larvae use to choose settlement sites. Annu. Rev. Mar. Sci. 2011, 3, 453–470. [Google Scholar] [CrossRef] [PubMed]
- Dobrestov, S.; Rittschof, D. Love at first taste: Induction of larval settlement by Marine Microbes. Int. J. Mol. Sci. 2020, 21, 731. [Google Scholar] [CrossRef]
- Flemming, H.C. Microbial Biofouling: Unsolved problems, insufficient approaches, and possible solutions. In Biofilm Highlights; Springer Series on Biofilms; Springer: Berlin/Heidelberg, Germany, 2011; Volume 5, pp. 81–109. [Google Scholar] [CrossRef]
- Little, B.J.; Lee, J.S. Microbiologically influenced corrosion: An update. Int. Mater. Rev. 2014, 59, 384–393. [Google Scholar] [CrossRef]
- Amara, I.; Miled, W.; Slama, R.B.; Ladhari, N. Antifouling processes and toxicity effects of antifouling paints on marine environment. A review. Environ. Toxicol. Pharmacol. 2018, 57, 115–130. [Google Scholar] [CrossRef]
- Cournet, A.; Bergé, M.; Roques, C.; Bergel, A.; Délia, M.L. Electrochemical reduction of oxygen catalyzed by Pseudomonas aeruginosa. Electrochim. Acta 2010, 5, 4902–4908. [Google Scholar] [CrossRef]
- Zhou, E.; Li, H.; Yang, C.; Wang, J.; Xu, D.; Zhang, D.; Gu, T. Accelerated corrosion of 2304 duplex stainless steel bye marine Pseudomonas aeruginosa biofilm. Int. Biodeterior. Biodegrad. 2018, 127, 1–9. [Google Scholar] [CrossRef]
- Lalucat, J.; Bennasar, A.; Bosch, R.; García-Valdés, E.; Palleroni, N.J. Biology of Pseudomonas stutzeri. Microbiol. Mol. Biol. Rev. 2006, 70, 510–547. [Google Scholar] [CrossRef] [PubMed]
- Agostini, V.O.; Macedo, A.J.; Muxagata, E.; da Silva, M.V.; Lopes, G. Natural and non-toxic products from Fabaceae Brazilian plants as a replacement for traditional antifouling biocides: An inhibition potential against initial biofouling. Environ. Sci. Pollut. Res. 2019, 26, 27112–27127. [Google Scholar] [CrossRef] [PubMed]
- Jude, F.; Arpin, C.; Brachet-Castang, C.; Capdepuv, M.; Caumette, P.; Quentin, C. TbtABM, a multidrug efflux pump associated with tributyltin resistance in Pseudomonas stutzeri. FEMS Microbiol. Lett. 2004, 232, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Favre, L.; Ortalo-Magné, A.; Pichereaux, C.; Gargaros, A.; Burlet-Schiltz, O.; Cotelle, V.; Culioli, G. Metabolome and proteome changes between biofilm and planktonic phenotypes od the marine bacterium Pseudoalteromonas lipolytica TC8. Biofouling 2018, 34, 132–148. [Google Scholar] [CrossRef]
- Gjersing, E.L.; Herberg, J.L.; Horn, J.; Schaldach, C.M.; Maxwell, R.S. NMR Metabolomics of Planktonic and Biofilm Modes of Growth in Pseudomonas aeruginosa. Anal. Chem. 2007, 79, 8037–8045. [Google Scholar] [CrossRef]
- Leggett, A.; Li, D.-W.; Bruschweiler-Li, L.; Sullivan, A.; Stoodley, P.; Brüschweiler, R. Differential metabolism between biofilm and suspended Pseudomonas aeruginosa cultures in bovine synovial fluid by 2D NMR-Based Metabolomics. Sci. Rep. 2022, 12, 17317. [Google Scholar] [CrossRef]
- García-Méndez, D.F.; Rengifo-Herrera, J.A.; Sanabria, J.; Wist, J. Analysis of the metabolic response of planktonic cells and biofilms of Klebsiella pneumoniae to sublethal disinfection with sodium hypochlorite measured by NMR. Microorganisms 2022, 10, 1323. [Google Scholar] [CrossRef]
- Koutsaftis, A.; Aoyama, I. Toxicity of four antifouling biocides and their mixtures on the brine shrimp Artemia salina. Sci. Total Environ. 2007, 387, 166–174. [Google Scholar] [CrossRef]
- Bao, W.Y.; Lee, O.O.; Chung, H.C.; Li, M.; Qian, P.Y. Copper affects biofilm inductiveness to larval settlement of the serpulid polychaete Hydroides elegans (Hasswell). Biofouling 2010, 29, 119–128. [Google Scholar] [CrossRef]
- Dahms, H.U.; Dobretsov, S. Antifouling compounds from marine macroalgae. Mar. Drugs 2017, 15, 265. [Google Scholar] [CrossRef]
- Bovio, E.; Fauchon, M.; Toueix, Y.; Mehiri, M.; Varese, G.C.; Hellio, C. The sponge-associated fungus Eurotium chevalier MUT 2316 and its bioactive molecules: Potential applications in the field of antifouling. Mar. Biotechnol. 2019, 21, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Lozano, I.; Hernández-Guerrero, C.J.; Muñoz-Ochoa, M.; Hellio, C. Biomimetic approaches for the development of new antifouling solutions: Study of incorporation of macroalgae and sponge extracts for the development of new environmentally-friendly coatings. Int. J. Mol. Sci. 2019, 20, 4863. [Google Scholar] [CrossRef] [PubMed]
- Alemán-Vega, M.; Sánchez-Lozano, I.; Hernández-Guerrero, C.J.; Hellio, C.; Quintana, E.T. Exploring antifouling activity of biosurfactants producing marine bacterial isolated from Gulf of California. Int. J. Mol. Sci. 2020, 21, 6068. [Google Scholar] [CrossRef] [PubMed]
- Satpute, S.K.; Banpurkar, A.G.; Banat, I.M.; Sangshetti, J.N.; Patil, R.H.; Gade, W.N. Multiple roles of biosurfactants in biofilm. Curr. Pharm. Des. 2016, 22, 1429–1448. [Google Scholar] [CrossRef]
- Ortega-Morales, B.O.; Chan-Bacab, M.J.; Miranda-Tello, E.; Fardeau, M.L.; Cerrero, J.C.; Stein, T. Antifouling activity of sessile bacilli derived from marine surfaces. J. Ind. Microbiol. Biotechnol. 2008, 35, 9–15. [Google Scholar] [CrossRef]
- Nastro, R.A.; Arguelles-Arias, A.; Ongena, M.; Di Constanzo, A.; Trifuoggi, M.; Guida, M.; Fickers, P. Antimicrobial activity of Bacillus amyloquefaciens ANT1 toward pathogenic bacteria and mold: Effects on biofilm formation. Probiotics Antimicrob. Proteins 2013, 5, 252–258. [Google Scholar] [CrossRef]
- Giri, S.S.; Ryu, E.C.; Sukumaran, V.; Park, S.C. Antioxidant, antibacterial and anti-adhesive activities of biosurfactants isolated from Bacillus strains. Microb. Pathog. 2019, 132, 66–72. [Google Scholar] [CrossRef]
- Berlanga, M.; Guerrero, R. Living together in biofilms: The microbial cell factory and its biotechnological implications. Microb. Cell. Fact. 2016, 15, 165. [Google Scholar] [CrossRef]
- Xu, Y.J.; Wang, C.; Ho, W.W.; Ong, C.N. Recent developments and applications of metabolomics in microbiological investigations. Trends. Anal. Chem. 2014, 56, 37–48. [Google Scholar] [CrossRef]
- Shen, F.; Ge, C.; Yuan, P. Metabolomics study reveals inhibition and metabolomic dysregulation in Staphylococcus aureus planktonic cells and biofilms induced by carnosol. Front. Microbiol. 2020, 11, 538572. [Google Scholar] [CrossRef]
- Gao, Y.; Meng, Q.; Zhou, X.; Luo, X.; Su, Z.; Chen, Z.; Huang, R.; Liu, Y.; Zhang, X. How do environmentally friendly antifouling alkaloids affect marine fouling microbial communities? Sci. Total Environ. 2022, 820, 152910. [Google Scholar] [CrossRef] [PubMed]
- Hoiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Waite, C.C.d.C.; da Silva, G.O.A.; Bitencourt, J.A.P.; Chequer, L.P.T.; Pennafirme, S.; Jurelevicius, D.d.A.; Seldin, L.; Crapez, M.A.C. Potential application of Pseudomonas stutzeri W228 for removal of copper and lead from marine environments. PLoS ONE 2020, 15, e0240486. [Google Scholar] [CrossRef]
- Das, P.; Mukherjee, S.; Sen, R. Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans. J. Appl. Microbiol. 2008, 104, 1675–1684. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, E.Z. Antimicrobial Activity of a biosurfactant produced by Bacillus licheniformis Strain M104 Grown on Whey. Braz. Arch. Biol. Technol. 2013, 56, 259–268. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Li, Q.; Fan, X.; Gao, J.; Luo, Y. Rapid biosynthesis of gold nanoparticles by the extracellular secretion of Bacillus niabensis 45: Characterization and antibiofilm activity. J. Chem. 2016, 2781347. [Google Scholar] [CrossRef]
- Purwasena, I.A.; Astuti, D.I.; Fauziyyah, N.A.; Putri, D.A.S.; Sugai, Y. Inhibition of microbial influenced corrosion on carbon steel ST37 using biosurfactant produced by Bacillus sp. Mater. Res. Expresss 2019, 6, 115405. [Google Scholar] [CrossRef]
- Abdel-Mawgoud, A.M.; Aboulwafa, M.M.; Hassouna, N.A.H. Opmization of Surfactin Production by Bacillus subtilis Isolate BS5. Appl. Biochem. Biotechnol. 2008, 150, 305–325. [Google Scholar] [CrossRef]
- Floris, R.; Rizzo, C.; Giudice, A.L. Biosurfactants from Marine Microorganisms. In Metabolomics-New Insights into Biology and Medicine; Hozzein, W.N., Ed.; IntechOpen: London, UK, 2018; pp. 1–16. [Google Scholar] [CrossRef]
- Narendrakumar, L.; Das, B.; Paramasivan, B.; Rasu, J.; Thomas, S. Quorum quenching and biofilm inhibition: Alternative imminent strategies to control the disease cholera. In Biotechnological Applications of Quorum Sensing Inhibitors; Springer: Berlin, Germany, 2018; pp. 66–85. [Google Scholar]
- Nitschke, M.; Pastore, G.M. Production and properties of a surfactant obtained from Bacillus subtilis grown on cassava wastewater. Bioresour. Technol. 2005, 97, 336–341. [Google Scholar] [CrossRef]
- Lawrance, A.; Balakrishnan, M.; Joseph, T.C.; Sukumaran, D.P.; Valsalan, V.N.; Gopal, D.; Ramalingam, K. Functional and molecular characterization of a lipopeptide surfactant from the marine sponge-associated eubacteria Bacillus licheniformis IOT-AMKV06 of Adaman and Nicobar Islands, India. Mar. Pollut. Bull. 2014, 82, 76–85. [Google Scholar] [CrossRef]
- Plaza, G.; Chojniak, J.; Rudnicka, K.; Paeaszkiewicz, K.; Bernat, P. Detection of biosurfactants in Bacillus species: Genes and products identification. J. Appl. Microbiol. 2015, 119, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.X.; Zhang, Y.; Shan, H.H.; Tong, Y.H.; Chen, X.J.; Liu, F.Q. Isolation and identification of antifungal peptides from Bacillus amyliquefaciens Wqwo. Environ. Sci. Pollut. Res. 2017, 24, 25000–25009. [Google Scholar] [CrossRef] [PubMed]
- He, C.P.; Fan, L.Y.; Wu, W.H.; Liang, Y.Q.; Li, R.; Tang, W.; Zheng, X.L.; Xiao, Y.N.; Liu, Z.X.; Zheng, F.C. Identification of lipopeptides produced by Bacillus subtilis strain producing multiple types of cyclic lipopeptides and evaluation of their surface-tension-lowering activities. J. Oleo Sci. 2017, 66, 785–790. [Google Scholar] [CrossRef]
- Englerova, K.; Bedlovicõva, Z.; Nemcová, R.; Király, J.; Mad’ar, M.; Hajducková, Z.; Styková, E.; Mucha, R.; Reiffová, K. Bacillus amyloliquefaciens- derived lipopeptide biosurfactants inhibit biofilm formation and expression of biofilm-related genes of Staphylococcus aureus. Antibiotics 2021, 10, 1252. [Google Scholar] [CrossRef]
- Gudiña, E.J.; Fernandes, E.C.; Rodrigues, A.I.; Teixeira, J.A.; Rodrigues, L.R. Biosurfactant production by Bacillus subtilis using corn steep liquor as culture medium. Front. Microbiol. 2015, 6, 59. [Google Scholar] [CrossRef]
- Coronel-León, J.; Grau, G.; Grau-Campistany, A.; Farfan, M.; Rabanal, F.; Manresa, A.; Marqués, A.M. Biosurfactant production by AL 1.1 a Bacillus licheniformis strain isolated from Antartica: Production, chemical characterization, and properties. Ann. Microbiol. 2015, 65, 2065–2078. [Google Scholar] [CrossRef]
- Wood, T.L.; Gong, T.; Zhu, L.; Miller, J.; Miller, D.S.; Yin, B.; Wood, T.K. Rhamnolipids from Pseudomonas aruginosa disperse the biofilms of sulfate-reducing bacteria. NPJ Biofilms Microbiomes 2018, 4, 22. [Google Scholar] [CrossRef]
- Rodrigues, L.R.; Banat, I.M.; Van Der Mei, H.C.; Teixeira, J.A.; Oliveira, R. Interference in adhesion of bacteria and yeasts isolated from explanted voice prostheses to silicone rubber by rhamnolipid biosurfactants. J. Appl. Microbiol. 2006, 100, 470–480. [Google Scholar] [CrossRef]
- Yan, X.; Gu, S.; Cui, X.; Shi, Y.; Wen, S.; Chen, H.; Ge, J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Ataphylococcus aureus CMCC26003. Microb. Pathog. 2019, 127, 12–20. [Google Scholar] [CrossRef]
- Horn, J.N.; Sengillo, J.D.; Lin, D.; Romo, T.D.; Grossfield, A. Characterization of a potent antimicrobial lipopeptide via coarse-gained molecular dynamics. Biochim. Biophys. Acta BBA Biomembr. 2012, 1818, 212–218. [Google Scholar] [CrossRef]
- Price, A.C.; Choi, K.H.; Heath, R.J.; Li, Z.; White, S.W.; Rock, C.O. Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. structure and mechanism. J. Biol. Chem. 2001, 276, 6551–6559. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.T.; Solanki, J.; Patel, K.; Nataraj, M. Chapter 13—Application of biosurfactants as antifouling agent. In Green Sustainable Process for Chemical and Environmental Engineering and Science; Inamuddin, C., Oluwaseun, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 275–289. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, L.; Shen, J.; Cao, B.; Cheng, T.; Zhao, T.; Liu, X.; Zhang, H. Metabolomic signatures of esophageal cancer, NMR-based metabolomics and UHPLC-based focused metabolomics of blood serum. Biochim. Biophys. Acta 2013, 1832, 1207–1216. [Google Scholar] [CrossRef]
- Wan, N.; Wang, H.; Ng, C.K.; Mukherjee, M.; Ren, D.; Cao, B.; Tang, J.Y. Bacterial metabolism during biofilm growth investigated by 13C tracing. Front. Microbiol. 2018, 9, 2657. [Google Scholar] [CrossRef] [PubMed]
- Stipetic, L.H.; Dalby, M.J.; Davies, R.L.; Morton, F.R.; Ramage, G.; Burgess, K.E.V. A novel metabolomic approach for the comparison of Staphyloccocus aureus planktonic cells and biofilm samples. Metabolomics 2016, 12, 75. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Zheng, X.; Wei, Y.; Zhou, X.; Zhang, K.; Wang, S.; Cheng, L.; Li, Y.; Ren, B.; Xu, X.; et al. d- Alanine metabolism is essential for growth and biofilm formation of Streptococcus mutans. Mol. Oral. Microbiol. 2016, 31, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Yeom, J.; Shin, J.H.; Yang, J.Y.; Kim, J.; Hwang, G.S. 1H NMR-Based metabolite profiling of planktonic and biofilm cells in Acinetobacter baumannii 1656-2. PLoS ONE 2013, 8, ee57730. [Google Scholar] [CrossRef] [PubMed]
- Djonovic, S.; Urbach, J.M.; Drenkard, E.; Bush, J.; Feinbaun, R.; Ausubel, J.L.; Traficante, D.; Risech, M.; Kocks, C.; Fischbach, M.A.; et al. Trehalose biosynthesis promotes Pseudomonas aeruginosa pathogenicity in plants. PloS Pathog. 2013, 9, e1003217. [Google Scholar] [CrossRef]
- Casaité, V.; Bruzyté, S.; Meskys, R. Effect of glycine betaine on osmoadaptation of Arthrobacter strain. Biologija 2006, 4, 46–48. [Google Scholar]
- Lee, J.J.; Lee, S.-K.; Song, N.; Nathan, T.O.; Swarts, B.M.; Eum, S.-Y.; Ehrt, S.; Cho, S.-N.; Eoh, H. Transient drug-tolerance and permanent drug-resistance rely on the trehalose-catalytic shift in Mycobacterium tuberculosis. Nat. Commun. 2019, 10, 2928. [Google Scholar] [CrossRef] [PubMed]
- Kart, D.; Recber, T.; Nemutlu, E.; Sagiroglu, M. Sub-Inhibitory concentration of Ciprofloxacin alone and combinations with plant-derived compounds against P. aeruginosa biofilms and their effects on the metabolomic profile of P. aeruginosa biofilms. Antibiotics 2021, 10, 414. [Google Scholar] [CrossRef]
- Beaume, M.; Kohler, T.; Fontana, T.; Tognon, M.; Renzoni, A.; van Delden, C. Metabolic pathways of Pseudomonas aeruginosa involved in competition with respiratory bacterial pathogens. Front. Microbiol. 2015, 6, 321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pei, G.; Chen, L.; Zhang, W. Metabolic dynamics of Desulfovibrio vulgaris biofilm grown on a steel surface. Biofouling 2016, 32, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.K.; Rao, R.S. An improved crystal violet assay for biofilm quantification in 96-well microtiter plate. BioRxiv 2017, 100214. [Google Scholar] [CrossRef]
- Stach, J.E.M.; Bathe, S.; Clapp, J.P.; Burns, R.G. PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiol. 2001, 36, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, P.G.; Mardaraz, C.; Pitta-Álvarez, S.I.; Giulietti, A.M. Isolation and selection of biosurfactant-producing bacteria. World J. Microbiol. Biotechnol 1996, 12, 82–84. [Google Scholar] [CrossRef]
- Youssef, N.H.; Duncan, K.E.; Nagle, D.P.; Savage, K.N.; Knapp, R.M.; Mclnerney, M.J. Comparison methods to detect biosurfactant production by diverse microorganism. J. Microbio. Methods. 2004, 56, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, M.; Hirata, Y.; Imanaka, T. A study on the structure-function relationship of lipopeptide biosurfactants. Biochim. Biophys. Acta 2000, 1488, 211–218. [Google Scholar] [CrossRef]
- Das, M.; Das, S.K.; Mukherjee, R.K. Surface active properties of the culture filtrates of a Micrococcus species grown on n-alkanes and sugars. Bioresour. Technol. 1998, 63, 231–235. [Google Scholar] [CrossRef]
- Ghribi, D.; Ellouze-Chaabouni, S. Enhancement of Bacillus subtilis lipopeptide biosurfactants production through optimization of medium composition and adequate control of aeration. Biotechnol. Res. Int. 2011, 2011, 653654. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye biding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Widdel, F. Theory and measurement of bacterial growth. Di Dalam Grund. Mikrobiol. 2007, 4, 1–11. [Google Scholar]
- Mikkelsen, H.; Duck, Z.; Lilley, K.S.; Welch, M. Interrelationships between colonies, biofilm and planktonic cells of Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 2411–2416. [Google Scholar] [CrossRef] [PubMed]
- Villa-Ruano, N.; Velásquez-Valle, R.; Zepeda-Vallejo, L.G.; Pérez-Hernández, N.; Velázquez-Ponce, M.; Arcos-Adame, V.M.; Becerra-Martínez, E. 1H NMR-based metabolomic profiling for identification of metabolites in Capsicum annuum cv. Mirasol infected by beet mild curly top virus (BMCTV). Food. Res. Int. 2018, 106, 870–877. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, G.; Zhang, J.; Zhang, Y.; Li, J.; Xiong, C.; Zhang, Q.; Li, X.; Lai, X. Metabolic discrimination of sea buckthorn from different Hippophaë species by 1H NMR based metabolomics. Sci. Rep. 2017, 7, 1585. [Google Scholar] [CrossRef]
- Villa-Ruano, N.; Ramírez-Meraz, M.; Méndez-Aguilar, R.; Zepeda-Vallejo, L.G.; Álvarez-Bravo, A.; Pérez-Hernández, N.; Becerra-Martínez, E. 1H NMR-based metabolomics profiling of ten new races from Capsium annuum cv. Serrano produced in Mexico. Food. Res. Int. 2019, 119, 785–792. [Google Scholar] [CrossRef]
- Cerceau, C.I.; Barbosa, L.C.A.; Filomeno, C.A.; Alvarenga, E.S.; Demuner, A.J.; Fidencio, P.H. An optimized and validated 1H NMR method for the quantification of α-pinene in essential soils. Talanta 2016, 150, 97–103. [Google Scholar] [CrossRef]
- Farag, M.A.; Fekry, M.I.; Al-Hammady, M.A.; Khalil, M.N.; El-Seedi, H.R.; Meyer, A.; Porzel, A.; Westphal, H.; Wessjohann, L.A. Cytotoxic effects of Sarcophyton sp. soft corals-Is there a correlation to their NMR fingerprints? Mar. Drugs 2017, 15, 2011. [Google Scholar] [CrossRef]
- Al-Khelaifi, F.; Diboun, I.; Donati, F.; Botré, F.; Alsayrafi, M.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; Elrayess, M.A. A pilot study comparing the metabolic profiles of elite-level athletes from different sporting disciplines. Sports Med. 2018, 4, 1–15. [Google Scholar] [CrossRef]
- Kim, J.; Jung, Y.; Song, B.; Yeon-Sik, B.; Ryu, D.H.; Kwang-Sik, L.; Geum-Sook, H. Discrimination of cabbage (Brassica rapa ssp. pekinesis) cultivars grown in different geographical areas using 1H NMR-based metabolomics. Food Chem. 2013, 137, 68–75. [Google Scholar] [CrossRef] [PubMed]

| Bacteria | Gram Reaction | Oil Spreading Test (mm) | Drop Collapse Test (mm) | Emulsification Index (% EI24) (Toluene) | Yield (mg/L) |
|---|---|---|---|---|---|
| Bacillus niabensis | Bacillus Gram+ | Positive 5.00 cm | Positive 9.34 mm | 69.33 ± 1.44 | 147.00 |
| Controls | |||||
| MB | - | 0 | 3.60 | 10.70 ± 1.20 | - |
| SDS (10%) | - | 5.00 cm | 11.76 | 59.00 ± 0.90 | - |
| (A) | (B) | (C) | (D) | |
|---|---|---|---|---|
| Biosurfactant Concentration (µg/mL) | Antibacterial Activity Zone Diameter (mm) | Biofilm Formation | Biofilm Total Protein (mg/mL) | Growth Rates (µ) |
| 0 |  | 0.84 ± 0.04 | 0.044 | |
| 30 | 12.53 ± 1.40 | 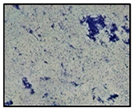 | 0.68 ± 0.07 | 0.041 |
| 50 | 9.80 ± 1.91 |  | 0.47 ± 0.18 | 0.033 |
| 100 | 0 |  | 0.35 ± 0.08 | 0.027 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Lozano, I.; Muñoz-Cruz, L.C.; Hellio, C.; Band-Schmidt, C.J.; Cruz-Narváez, Y.; Becerra-Martínez, E.; Hernández-Guerrero, C.J. Metabolomic Insights of Biosurfactant Activity from Bacillus niabensis against Planktonic Cells and Biofilm of Pseudomonas stutzeri Involved in Marine Biofouling. Int. J. Mol. Sci. 2023, 24, 4249. https://doi.org/10.3390/ijms24044249
Sánchez-Lozano I, Muñoz-Cruz LC, Hellio C, Band-Schmidt CJ, Cruz-Narváez Y, Becerra-Martínez E, Hernández-Guerrero CJ. Metabolomic Insights of Biosurfactant Activity from Bacillus niabensis against Planktonic Cells and Biofilm of Pseudomonas stutzeri Involved in Marine Biofouling. International Journal of Molecular Sciences. 2023; 24(4):4249. https://doi.org/10.3390/ijms24044249
Chicago/Turabian StyleSánchez-Lozano, Ilse, Luz Clarita Muñoz-Cruz, Claire Hellio, Christine J. Band-Schmidt, Yair Cruz-Narváez, Elvia Becerra-Martínez, and Claudia J. Hernández-Guerrero. 2023. "Metabolomic Insights of Biosurfactant Activity from Bacillus niabensis against Planktonic Cells and Biofilm of Pseudomonas stutzeri Involved in Marine Biofouling" International Journal of Molecular Sciences 24, no. 4: 4249. https://doi.org/10.3390/ijms24044249
APA StyleSánchez-Lozano, I., Muñoz-Cruz, L. C., Hellio, C., Band-Schmidt, C. J., Cruz-Narváez, Y., Becerra-Martínez, E., & Hernández-Guerrero, C. J. (2023). Metabolomic Insights of Biosurfactant Activity from Bacillus niabensis against Planktonic Cells and Biofilm of Pseudomonas stutzeri Involved in Marine Biofouling. International Journal of Molecular Sciences, 24(4), 4249. https://doi.org/10.3390/ijms24044249






