Transcriptomic Analysis Revealed Key Defense Genes and Signaling Pathways Mediated by the Arabidopsis thaliana Gene SAD2 in Response to Infection with Pseudomonas syringae pv. Tomato DC3000
Abstract
1. Introduction
2. Results
2.1. SAD2 Expression Increased Resistance to Pst DC3000 Infection
2.2. RNA-Sequencing Library Construction, Sequencing, and Read Mapping
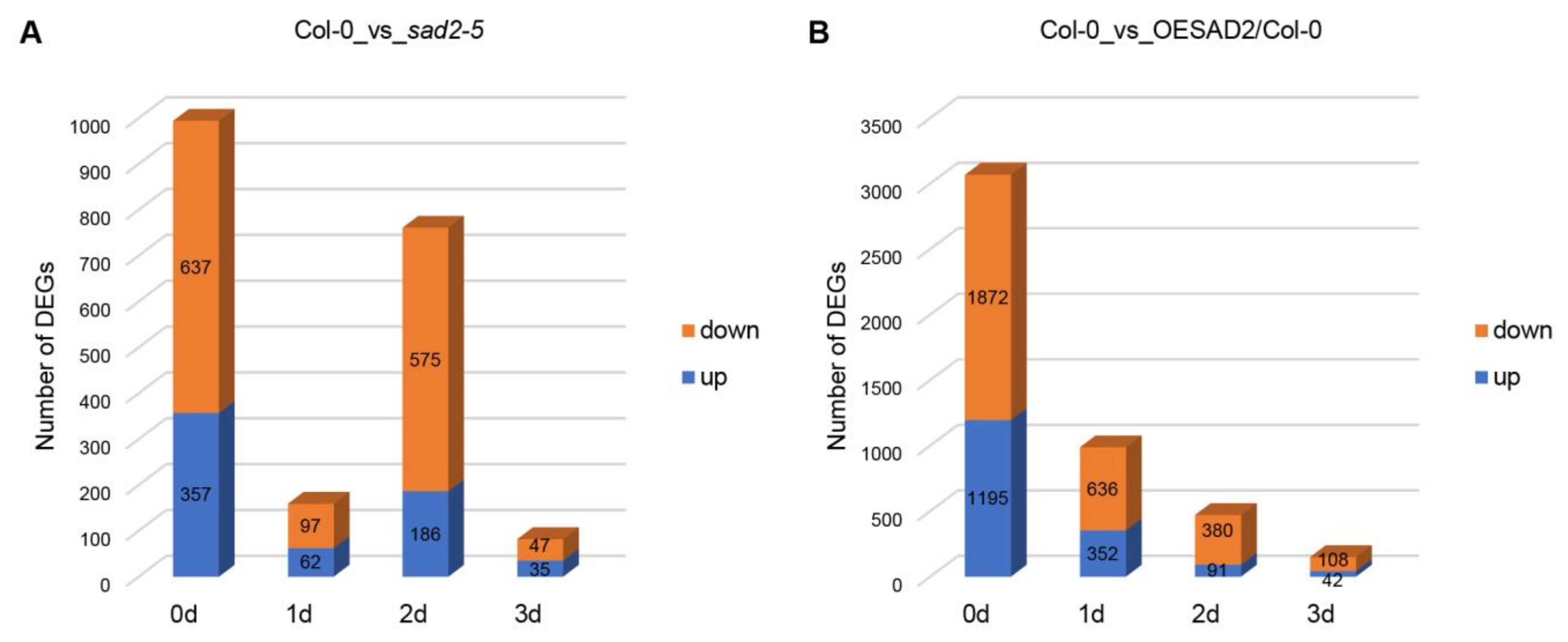
2.3. DEGs in Col-0, sad2-5, and OESAD2/Col-0 after Pst DC3000 Infection
2.4. Key DEGs Regulated by SAD2 and Involved in the Pst DC3000 Defense Response
2.5. GO Term and KEGG Pathway Enrichment among Pathogen-Induced DEGs
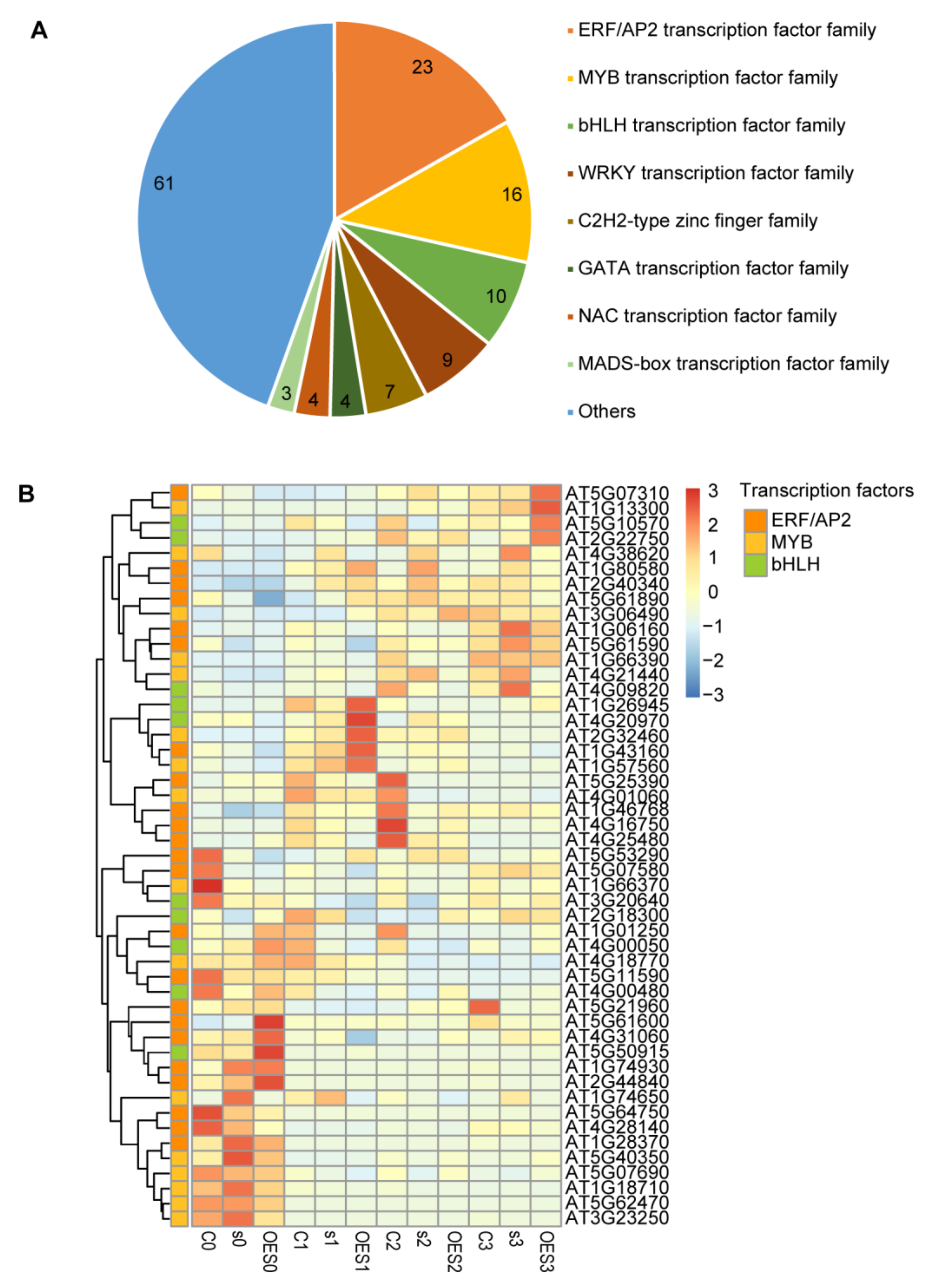
2.6. Identification of Transcription Factors (TFs) Involved in the Pathogen Response
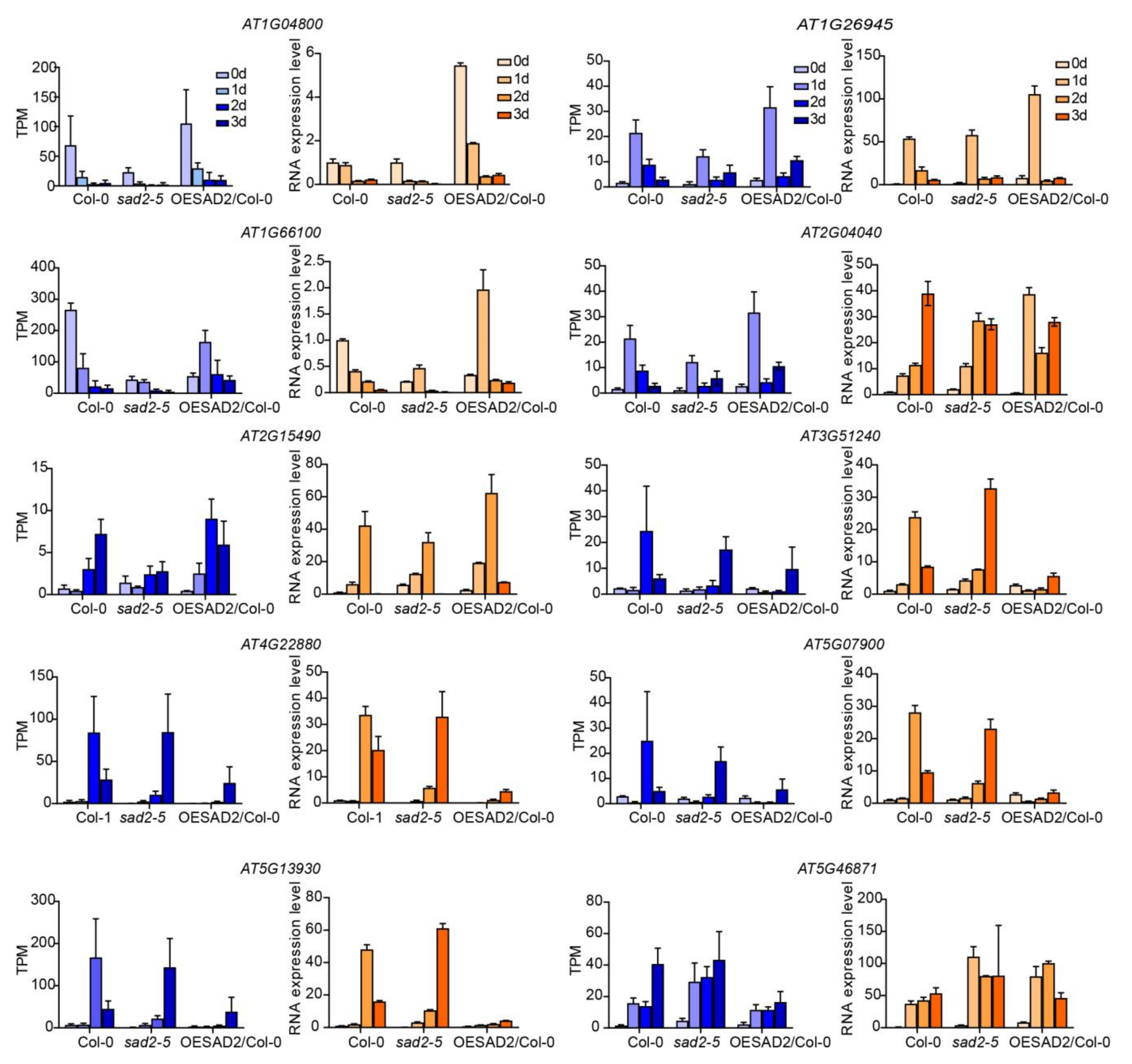
2.7. Verification of RNA-Seq Results via RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Arabidopsis Inoculation with Pst DC3000 and Leaf Sample Collection
4.3. RNA Extraction, cDNA Library Construction, and Illumina Deep Sequencing
4.4. RNA-Sequencing Data Analysis
4.5. Gene Enrichment Analysis
4.6. Validation of RNA-Seq Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Boyd, L.A.; Ridout, C.; O’Sullivan, D.M.; Leach, J.E.; Leung, H. Plant-pathogen interactions: Disease resistance in modern agriculture. Trends Genet. 2013, 29, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Ni, Y.; Liu, X.; Zhao, H.; Chen, Y.; Jia, M.; Liu, M.; Liu, H.; Tian, B. The mechanism of sesame resistance against Macrophomina phaseolina was revealed via a comparison of transcriptomes of resistant and susceptible sesame genotypes. BMC Plant. Biol. 2021, 21, 159. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T.; Hussain, H.I.; Rookes, J.E.; Cahill, D.M. Transcriptome analysis, using RNA-Seq of Lomandra longifolia roots infected with Phytophthora cinnamomi reveals the complexity of the resistance response. Plant. Biol. 2018, 20, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Feng, B.; Zhou, J.M.; Tang, D. Plant immune signaling: Advancing on two frontiers. J. Integr. Plant. Biol. 2020, 62, 2–24. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Loo, E.P. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef]
- Zhou, J.M.; Zhang, Y. Plant Immunity: Danger Perception and Signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Shi, T.; Gao, Y.; Wang, H.; Liu, J. Nucleo-cytoplasmic Transport and Transport Receptors in Plant Disease Resistance Defense Response. Chin. Bull. Bot. 2021, 56, 480–487. [Google Scholar] [CrossRef]
- Schwessinger, B.; Ronald, P.C. Plant innate immunity: Perception of conserved microbial signatures. Ann. Rev. Plant. Biol. 2012, 63, 451–482. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; He, S.Y. Innate Immunity in Plants: An Arms Race Between Pattern Recognition Receptors in Plants and Effectors in Microbial Pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Zhou, J.M. Plant-bacterial pathogen interactions mediated by type III effectors. Curr. Opin. Plant. Biol. 2012, 15, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Nishimura, M.T.; Dangl, J.L. Arabidopsis and the plant immune system. Plant. J. 2010, 61, 1053–1066. [Google Scholar] [CrossRef]
- Jacob, F.; Kracher, B.; Mine, A.; Seyfferth, C.; Blanvillain-Baufume, S.; Parker, J.E.; Tsuda, K.; Schulze-Lefert, P.; Maekawa, T. A dominant-interfering camta3 mutation compromises primary transcriptional outputs mediated by both cell surface and intracellular immune receptors in Arabidopsis thaliana. New Phytol. 2018, 217, 1667–1680. [Google Scholar] [CrossRef]
- Lolle, S.; Stevens, D.; Coaker, G. Plant NLR-triggered immunity: From receptor activation to downstream signaling. Curr. Opin. Immunol. 2020, 62, 99–105. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Li, X. A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant. Cell 2005, 17, 1306–1316. [Google Scholar] [CrossRef]
- Liu, J.; Coaker, G. Nuclear trafficking during plant innate immunity. Molecular. Plant 2008, 1, 411–422. [Google Scholar] [CrossRef]
- Park, C.J.; Ronald, P.C. Cleavage and nuclear localization of the rice XA21 immune receptor. Nat. Commun. 2012, 3, 920. [Google Scholar] [CrossRef]
- Deslandes, L.; Rivas, S. The plant cell nucleus: A true arena for the fight between plants and pathogens. Plant. Signal. Behav. 2011, 6, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Wiermer, M.; Palma, K.; Zhang, Y.; Li, X. Should I stay or should I go? Nucleocytoplasmic trafficking in plant innate immunity. Cell Microbiol. 2007, 9, 1880–1890. [Google Scholar] [CrossRef] [PubMed]
- Teh, O.K.; Hofius, D. Membrane trafficking and autophagy in pathogen-triggered cell death and immunity. J. Exp. Bot. 2014, 65, 1297–1312. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zebell, S.G.; Liang, Z.; Wang, S.; Kang, B.H.; Dong, X. Nuclear Pore Permeabilization Is a Convergent Signaling Event in Effector-Triggered Immunity. Cell 2016, 166, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.M.; Liu, P.Q.; Xu, Y.J.; Xiao, S. Protein trafficking during plant innate immunity. J. Integr. Plant. Biol. 2016, 58, 284–298. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, Z.; Jing, B.; Gannon, P.; Ding, J.; Xu, F.; Li, X.; Zhang, Y. Transportin-SR is required for proper splicing of resistance genes and plant immunity. PLoS Genet. 2011, 7, e1002159. [Google Scholar] [CrossRef]
- Xu, K.; Tao, T.; Jie, J.; Lu, X.D.; Li, X.Z.; Mehmood, M.A.; He, H.; Liu, Z.; Xiao, X.Y.; Yang, J.; et al. Increased importin 13 activity is associated with the pathogenesis of pterygium. Mol. Vis. 2013, 19, 604–613. [Google Scholar]
- Kimura, M.; Imamoto, N. Biological significance of the importin-beta family-dependent nucleocytoplasmic transport pathways. Traffic 2014, 15, 727–748. [Google Scholar] [CrossRef]
- Verslues, P.E.; Guo, Y.; Dong, C.H.; Ma, W.; Zhu, J.K. Mutation of SAD2, an importin beta-domain protein in Arabidopsis, alters abscisic acid sensitivity. Plant. J. 2006, 47, 776–787. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, W.; Zhao, Y.; Gong, X.; Guo, L.; Zhu, G.; Wang, X.; Gong, Z.; Schumaker, K.S.; Guo, Y. SAD2, an importin -like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant. Cell 2007, 19, 3805–3818. [Google Scholar] [CrossRef]
- Gao, Y.; Gong, X.; Cao, W.; Zhao, J.; Fu, L.; Wang, X.; Schumaker, K.S.; Guo, Y. SAD2 in Arabidopsis functions in trichome initiation through mediating GL3 function and regulating GL1, TTG1 and GL2 expression. J. Integr. Plant. Biol. 2008, 50, 906–917. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhan, Q.; Shi, T.; Liu, J.; Zhao, K.; Gao, Y. The nuclear transporter SAD2 plays a role in calcium- and H2 O2 -mediated cell death in Arabidopsis. Plant. J. 2020, 101, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Smarda, P.; Bures, P.; Horova, L.; Leitch, I.J.; Mucina, L.; Pacini, E.; Tichy, L.; Grulich, V.; Rotreklova, O. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proc. Natl. Acad. Sci. USA 2014, 111, E4096–E4102. [Google Scholar] [CrossRef] [PubMed]
- Ng, D.W.; Abeysinghe, J.K.; Kamali, M. Regulating the Regulators: The Control of Transcription Factors in Plant Defense Signaling. Int. J. Mol. Sci. 2018, 19, 3737. [Google Scholar] [CrossRef]
- Amorim, L.L.B.; da Fonseca Dos Santos, R.; Neto, J.P.B.; Guida-Santos, M.; Crovella, S.; Benko-Iseppon, A.M. Transcription Factors Involved in Plant Resistance to Pathogens. Curr. Protein Pept. Sci. 2017, 18, 335–351. [Google Scholar] [CrossRef]
- Molina, A.; García-Olmedo, F. Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J. 2003, 12, 669–675. [Google Scholar] [CrossRef]
- Li, B.; Zhang, Y.; Qiu, D.; Francis, F.; Wang, S. Comparative Proteomic Analysis of Sweet Orange Petiole Provides Insights Into the Development of Huanglongbing Symptoms. Front. Plant. Sci. 2021, 12, 656997. [Google Scholar] [CrossRef]
- Dong, S.; Yin, W.; Kong, G.; Yang, X.; Qutob, D.; Chen, Q.; Kale, S.D.; Sui, Y.; Zhang, Z.; Dou, D.; et al. Phytophthora sojae avirulence effector Avr3b is a secreted NADH and ADP-ribose pyrophosphorylase that modulates plant immunity. PLoS Pathog. 2011, 7, e1002353. [Google Scholar] [CrossRef]
- Hou, H.M.; Li, H.E.; Gao, M.; Wang, H.; Jiao, C.; Wang, X.P. Expression of a GDP-L-galactose phosphorylase-like gene in a Chinese wild Vitis species induces responses to Erysiphe necator and defense signaling molecules. Genet. Mol. Res. 2013, 12, 3830–3844. [Google Scholar] [CrossRef]
- Upadhyay, N.; Kar, D.; Deepak Mahajan, B.; Nanda, S.; Rahiman, R.; Panchakshari, N.; Bhagavatula, L.; Datta, S. The multitasking abilities of MATE transporters in plants. J. Exp. Bot. 2019, 70, 4643–4656. [Google Scholar] [CrossRef]
- Devanna, B.N.; Jaswal, R.; Singh, P.K.; Kapoor, R.; Jain, P.; Kumar, G.; Sharma, Y.; Samantaray, S.; Sharma, T.R. Role of transporters in plant disease resistance. Physiol. Plant. 2021, 171, 849–867. [Google Scholar] [CrossRef] [PubMed]
- Nawrath, C.; Heck, S.; Parinthawong, N.; Metraux, J.P. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant. Cell 2002, 14, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, T.; Sekine, K.T.; Hase, S.; Kanayama, Y.; Seo, S.; Ohashi, Y.; Kusano, T.; Shibata, D.; Shah, J.; Takahashi, H. Overexpression of the Arabidopsis thaliana EDS5 gene enhances resistance to viruses. Plant. Biol. 2008, 10, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Dobritzsch, M.; Lubken, T.; Eschen-Lippold, L.; Gorzolka, K.; Blum, E.; Matern, A.; Marillonnet, S.; Bottcher, C.; Drager, B.; Rosahl, S. MATE Transporter-Dependent Export of Hydroxycinnamic Acid Amides. Plant. Cell 2016, 28, 583–596. [Google Scholar] [CrossRef]
- Sun, X.; Gilroy, E.M.; Chini, A.; Nurmberg, P.L.; Hein, I.; Lacomme, C.; Birch, P.R.; Hussain, A.; Yun, B.W.; Loake, G.J. ADS1 encodes a MATE-transporter that negatively regulates plant disease resistance. New Phytol. 2011, 192, 471–482. [Google Scholar] [CrossRef]
- Tiwari, M.; Sharma, D.; Singh, M.; Tripathi, R.D.; Trivedi, P.K. Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis. Sci. Rep. 2014, 4, 3964. [Google Scholar] [CrossRef]
- Shah, J. Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu. Rev. Phytopathol. 2005, 43, 229–260. [Google Scholar] [CrossRef]
- Pelegrini, P.B.; Franco, O.L. Plant gamma-thionins: Novel insights on the mechanism of action of a multi-functional class of defense proteins. Int. J. Biochem. Cell Biol. 2005, 37, 2239–2253. [Google Scholar] [CrossRef]
- Oard, S.V. Deciphering a mechanism of membrane permeabilization by alpha-hordothionin peptide. Biochim. Biophys. Acta 2011, 1808, 1737–1745. [Google Scholar] [CrossRef]
- Asano, T.; Miwa, A.; Maeda, K.; Kimura, M.; Nishiuchi, T. The secreted antifungal protein thionin 2.4 in Arabidopsis thaliana suppresses the toxicity of a fungal fruit body lectin from Fusarium graminearum. PLoS Pathog. 2013, 9, e1003581. [Google Scholar] [CrossRef]
- Hao, G.; Stover, E.; Gupta, G. Overexpression of a Modified Plant Thionin Enhances Disease Resistance to Citrus Canker and Huanglongbing (HLB). Front. Plant. Sci. 2016, 7, 1078. [Google Scholar] [CrossRef] [PubMed]
- Muramoto, N.; Tanaka, T.; Shimamura, T.; Mitsukawa, N.; Hori, E.; Koda, K.; Otani, M.; Hirai, M.; Nakamura, K.; Imaeda, T. Transgenic sweet potato expressing thionin from barley gives resistance to black rot disease caused by Ceratocystis fimbriata in leaves and storage roots. Plant. Cell Rep. 2012, 31, 987–997. [Google Scholar] [CrossRef] [PubMed]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant. Sci. 2012, 3, 222. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation Is a Major Regulator of Phenylpropanoid Availability and Biological Activity in Plants. Front. Plant. Sci. 2016, 7, 735. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant. J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Jan, R.; Aaqil Khan, M.; Asaf, S.; Lubna; Park, J.R.; Lee, I.J.; Kim, K.M. Flavonone 3-hydroxylase Relieves Bacterial Leaf Blight Stress in Rice via Overaccumulation of Antioxidant Flavonoids and Induction of Defense Genes and Hormones. Int. J. Mol. Sci. 2021, 22, 6152. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defence responses. Plant. Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, C.; Li, Q.; Zhang, K.; Lou, Q.; Li, J.; Chen, J. Multi-omics analysis revealed that MAPK signaling and flavonoid metabolic pathway contributed to resistance against Meloidogyne incognita in the introgression line cucumber. J. Proteomics 2020, 220, 103675. [Google Scholar] [CrossRef]
- Wu, J.; Deng, Y.; Hu, J.; Jin, C.; Zhu, X.; Li, D. Genome-wide analyses of direct target genes of an ERF11 transcription factor involved in plant defense against bacterial pathogens. Biochem. Biophys Res. Commun. 2020, 532, 76–81. [Google Scholar] [CrossRef]
- Zhao, Y.; Chang, X.; Qi, D.; Dong, L.; Wang, G.; Fan, S.; Jiang, L.; Cheng, Q.; Chen, X.; Han, D.; et al. A Novel Soybean ERF Transcription Factor, GmERF113, Increases Resistance to Phytophthora sojae Infection in Soybean. Front. Plant. Sci. 2017, 8, 299. [Google Scholar] [CrossRef]
- Li, Z.; Tian, Y.; Xu, J.; Fu, X.; Gao, J.; Wang, B.; Han, H.; Wang, L.; Peng, R.; Yao, Q. A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against Pseudomonas syringae pv. tomato DC3000. Plant. Physiol. Biochem. 2018, 132, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Ma, H.; Zhang, Y.; Zhang, X.; Ji, M.; van Nocker, S.; Ahmad, B.; Zhao, Z.; Wang, X.; et al. Overexpression of the Apple (Malus x domestica) MdERF100 in Arabidopsis Increases Resistance to Powdery Mildew. Int. J. Mol. Sci. 2021, 22, 5713. [Google Scholar] [CrossRef]
- He, J.; Liu, Y.; Yuan, D.; Duan, M.; Liu, Y.; Shen, Z.; Yang, C.; Qiu, Z.; Liu, D.; Wen, P.; et al. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc. Natl. Acad. Sci. USA 2020, 117, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, X.; He, Q.; Guo, D.; Liu, C.; Cao, J.; Wu, Z.; Kang, Z.; Wang, X. TaMYB29: A Novel R2R3-MYB Transcription Factor Involved in Wheat Defense Against Stripe Rust. Front. Plant. Sci. 2021, 12, 783388. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Yang, C.; Cao, J.; Chen, H.; Pang, J.; Zhao, Q.; Wang, Z.; Qing Fu, Z.; Liu, J. A bHLH transcription activator regulates defense signaling by nucleo-cytosolic trafficking in rice. J. Integr. Plant. Biol. 2020, 62, 1552–1573. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.G.; Mudgett, M.B. Tomato bHLH132 Transcription Factor Controls Growth and Defense and Is Activated by Xanthomonas euvesicatoria Effector XopD During Pathogenesis. Mol. Plant. Microbe Interact. 2019, 32, 1614–1622. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Samples | Replicate | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Q20 (%) | Q30 (%) | GC (%) | Total_ Mapping (%) |
|---|---|---|---|---|---|---|---|---|---|
| Col-0_0d | 1 | 35,015,991 | 5.25 G | 34,168,202 | 5.13 G | 98.01 | 94.20 | 45.34 | 97.79 |
| 2 | 35,996,956 | 5.39 G | 34,874,632 | 5.23 G | 97.97 | 94.14 | 45.48 | 98.02 | |
| 3 | 39,606,195 | 5.94 G | 38,314,248 | 5.75 G | 98.06 | 94.37 | 45.27 | 97.93 | |
| Col-0_1d | 1 | 39,189,239 | 5.87 G | 38,014,575 | 5.7 G | 98.07 | 94.38 | 44.98 | 97.98 |
| 2 | 38,505,552 | 5.77 G | 37,168,785 | 5.58 G | 98.07 | 94.35 | 45.05 | 98.05 | |
| 3 | 40,696,320 | 6.1 G | 39,371,453 | 5.91 G | 97.95 | 94.12 | 45.25 | 97.90 | |
| Col-0_2d | 1 | 38,202,509 | 5.73 G | 37,115,329 | 5.57 G | 97.98 | 94.12 | 44.75 | 98.03 |
| 2 | 39,131,200 | 5.86 G | 37,617,017 | 5.64 G | 97.96 | 94.09 | 45.11 | 98.00 | |
| 3 | 42,585,194 | 6.38 G | 41,201,001 | 6.18 G | 97.98 | 94.17 | 45.11 | 97.06 | |
| Col-0_3d | 1 | 38,249,317 | 5.73 G | 37,029,249 | 5.55 G | 98.09 | 94.41 | 44.87 | 98.04 |
| 2 | 35,949,275 | 5.39 G | 34,517,786 | 5.18 G | 98.02 | 94.22 | 45.01 | 97.92 | |
| 3 | 34,922,101 | 5.23 G | 33,703,571 | 5.06 G | 98.14 | 94.54 | 44.98 | 98.08 | |
| sad2-5_0d | 1 | 37,867,490 | 5.68 G | 36,537,734 | 5.48 G | 98.11 | 94.48 | 45.32 | 97.33 |
| 2 | 40,558,326 | 6.08 G | 39,307,108 | 5.9 G | 98.08 | 94.43 | 45.26 | 97.29 | |
| 3 | 44,222,634 | 6.63 G | 42,603,821 | 6.39 G | 98.06 | 94.44 | 45.37 | 96.85 | |
| sad2-5_1d | 1 | 38,518,272 | 5.77 G | 37,239,684 | 5.59 G | 98.11 | 94.48 | 45.13 | 97.60 |
| 2 | 39,760,691 | 5.96 G | 38,544,874 | 5.78 G | 97.98 | 94.12 | 45.05 | 97.60 | |
| 3 | 38,946,660 | 5.84 G | 37,622,323 | 5.64 G | 98.00 | 94.18 | 45.00 | 97.37 | |
| sad2-5_2d | 1 | 35,927,275 | 5.38 G | 34,755,848 | 5.21 G | 98.16 | 94.58 | 44.97 | 97.56 |
| 2 | 38,878,184 | 5.83 G | 37,212,316 | 5.58 G | 97.73 | 93.36 | 45.23 | 97.34 | |
| 3 | 36,917,936 | 5.53 G | 35,742,305 | 5.36 G | 97.99 | 94.20 | 45.10 | 97.27 | |
| sad2-5_3d | 1 | 34,058,842 | 5.1 G | 32,974,801 | 4.95 G | 98.07 | 94.38 | 45.16 | 97.47 |
| 2 | 38,813,455 | 5.82 G | 37,614,495 | 5.64 G | 98.12 | 94.45 | 44.95 | 97.34 | |
| 3 | 41,067,633 | 6.16 G | 40,173,877 | 6.03 G | 97.79 | 93.66 | 45.37 | 96.55 | |
| OESAD2_0d | 1 | 33,942,065 | 5.09 G | 33,246,869 | 4.99 G | 98.03 | 94.25 | 45.89 | 98.06 |
| 2 | 30,279,425 | 4.54 G | 29,684,633 | 4.45 G | 98.09 | 94.36 | 45.80 | 98.17 | |
| 3 | 39,884,234 | 5.98 G | 38,968,579 | 5.85 G | 98.10 | 94.43 | 46.15 | 98.11 | |
| OESAD2_1d | 1 | 38,695,180 | 5.8 G | 37,829,383 | 5.67 G | 97.83 | 93.78 | 45.30 | 97.81 |
| 2 | 41,162,035 | 6.17 G | 40,342,339 | 6.05 G | 98.03 | 94.23 | 45.24 | 98.05 | |
| 3 | 27,256,874 | 4.08 G | 26,714,782 | 4.01 G | 97.90 | 93.90 | 45.18 | 98.00 | |
| OESAD2_2d | 1 | 32,949,733 | 4.94 G | 32,275,938 | 4.84 G | 97.91 | 93.95 | 45.19 | 97.85 |
| 2 | 41,170,080 | 6.17 G | 40,515,870 | 6.08 G | 98.05 | 94.28 | 45.34 | 97.57 | |
| 3 | 35,475,771 | 5.32 G | 34,878,026 | 5.23 G | 98.12 | 94.42 | 45.18 | 98.03 | |
| OESAD2_3d | 1 | 39,301,446 | 5.89 G | 38,509,059 | 5.78 G | 97.87 | 93.82 | 45.15 | 96.70 |
| 2 | 40,545,374 | 6.08 G | 39,523,760 | 5.93 G | 97.97 | 94.14 | 45.36 | 97.15 | |
| 3 | 40,018,743 | 6 G | 39,135,547 | 5.87 G | 98.01 | 94.19 | 45.34 | 97.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Shi, T.; Lyu, M.; Wang, R.; Xu, A.; Chen, L.; Luo, R.; Sun, Y.; Guo, X.; Liu, J.; et al. Transcriptomic Analysis Revealed Key Defense Genes and Signaling Pathways Mediated by the Arabidopsis thaliana Gene SAD2 in Response to Infection with Pseudomonas syringae pv. Tomato DC3000. Int. J. Mol. Sci. 2023, 24, 4229. https://doi.org/10.3390/ijms24044229
Li S, Shi T, Lyu M, Wang R, Xu A, Chen L, Luo R, Sun Y, Guo X, Liu J, et al. Transcriptomic Analysis Revealed Key Defense Genes and Signaling Pathways Mediated by the Arabidopsis thaliana Gene SAD2 in Response to Infection with Pseudomonas syringae pv. Tomato DC3000. International Journal of Molecular Sciences. 2023; 24(4):4229. https://doi.org/10.3390/ijms24044229
Chicago/Turabian StyleLi, Sha, Tiantian Shi, Mingjie Lyu, Rui Wang, Andi Xu, Luoying Chen, Rong Luo, Yinglu Sun, Xiaoying Guo, Jun Liu, and et al. 2023. "Transcriptomic Analysis Revealed Key Defense Genes and Signaling Pathways Mediated by the Arabidopsis thaliana Gene SAD2 in Response to Infection with Pseudomonas syringae pv. Tomato DC3000" International Journal of Molecular Sciences 24, no. 4: 4229. https://doi.org/10.3390/ijms24044229
APA StyleLi, S., Shi, T., Lyu, M., Wang, R., Xu, A., Chen, L., Luo, R., Sun, Y., Guo, X., Liu, J., Wang, H., & Gao, Y. (2023). Transcriptomic Analysis Revealed Key Defense Genes and Signaling Pathways Mediated by the Arabidopsis thaliana Gene SAD2 in Response to Infection with Pseudomonas syringae pv. Tomato DC3000. International Journal of Molecular Sciences, 24(4), 4229. https://doi.org/10.3390/ijms24044229








