New Biomarkers Based on Dendritic Cells for Breast Cancer Treatment and Prognosis Diagnosis
Abstract
1. Introduction
2. Results
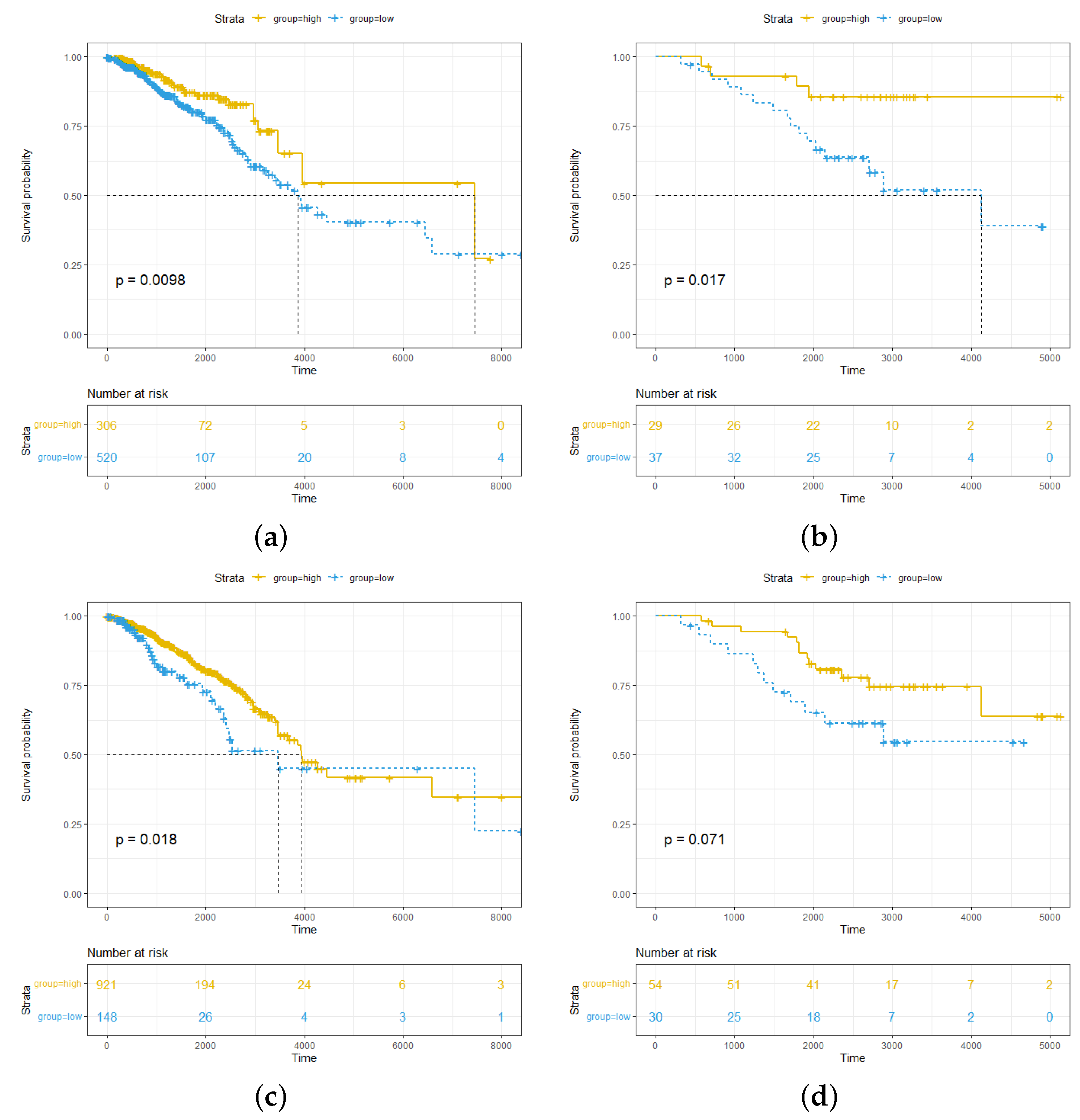
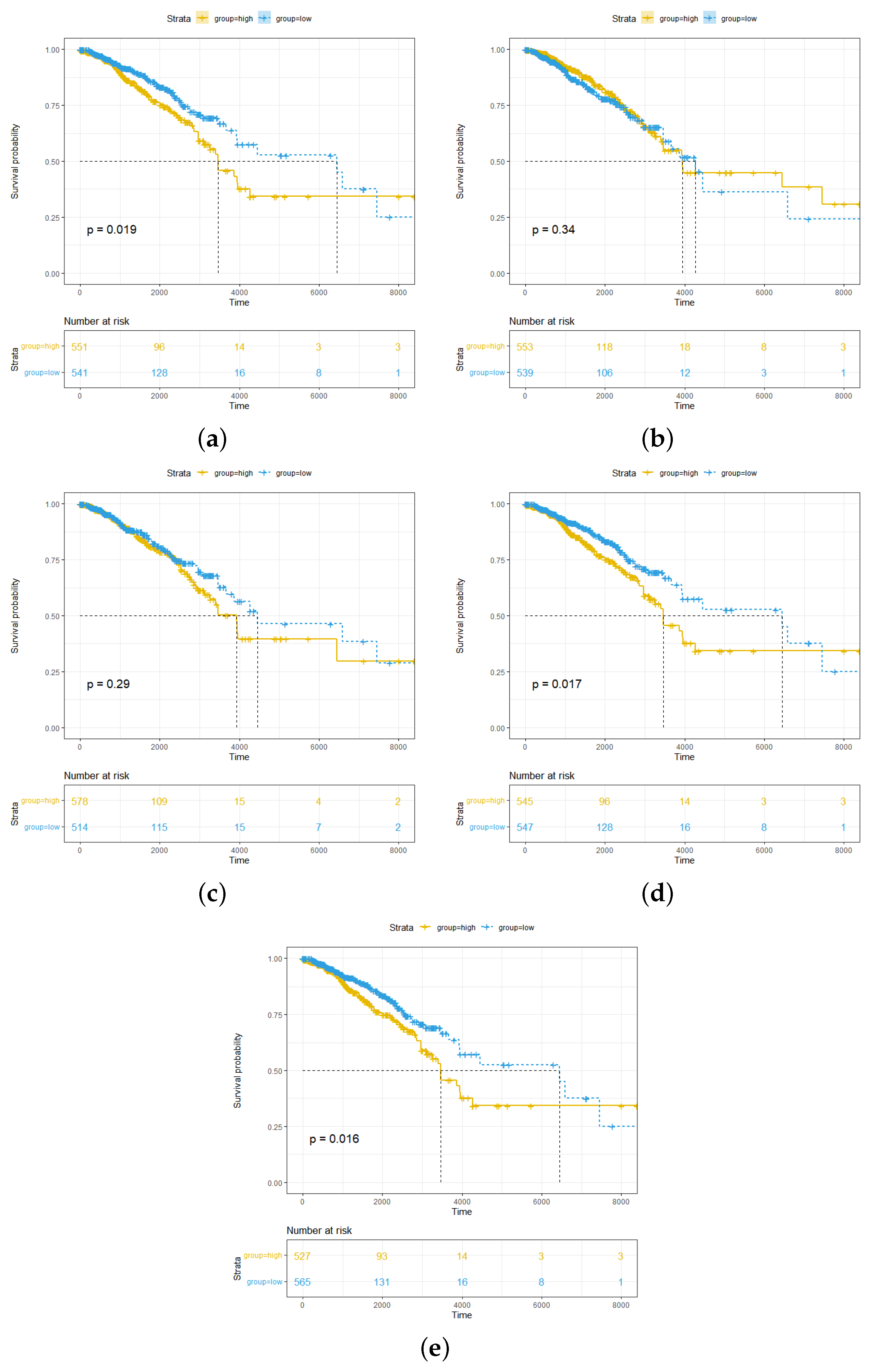
2.1. High Abundance of pDCs and cDCs in Breast Cancer Is Associated with Better Survival Outcomes for pDCs More Significantly than for cDCs
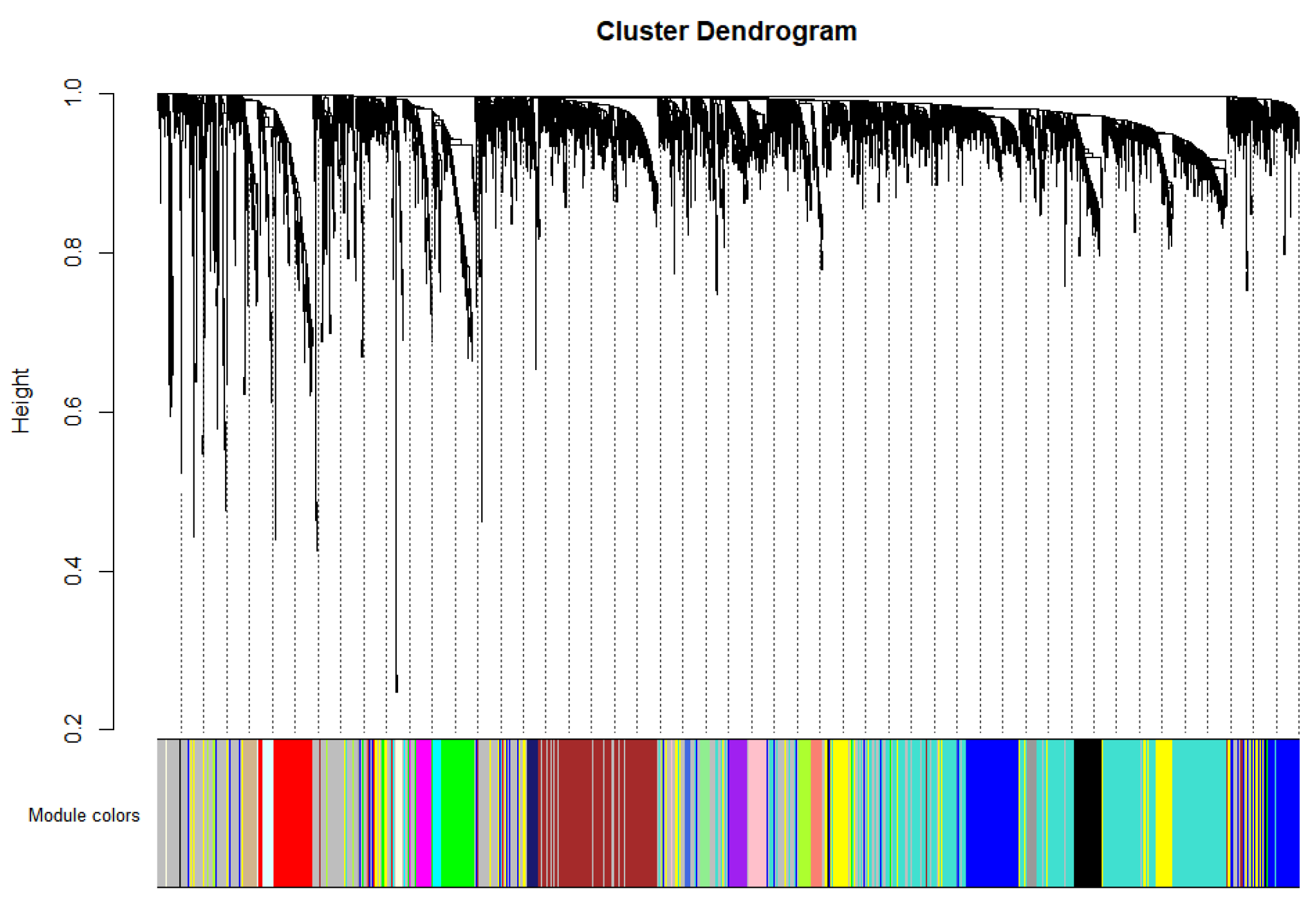
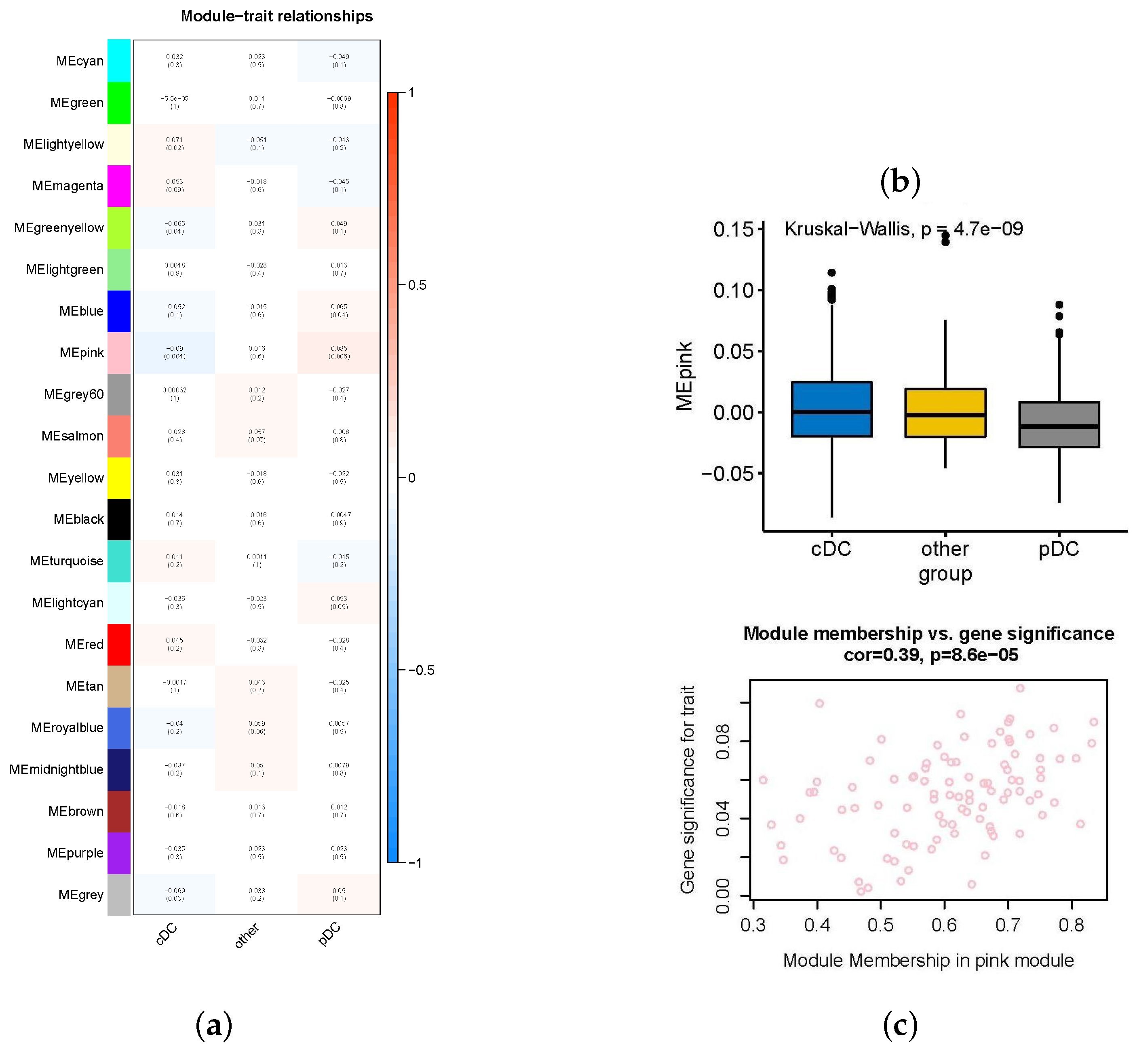
2.2. Important Expression Modules for High-Abundance pDCs and cDCs
2.3. Enrichment Analysis of Important Module Genes
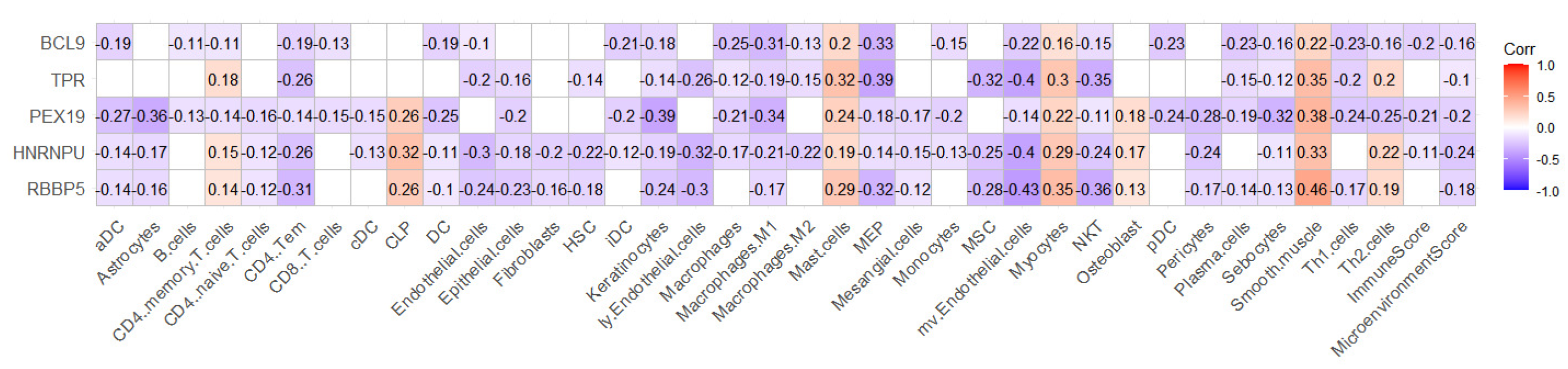
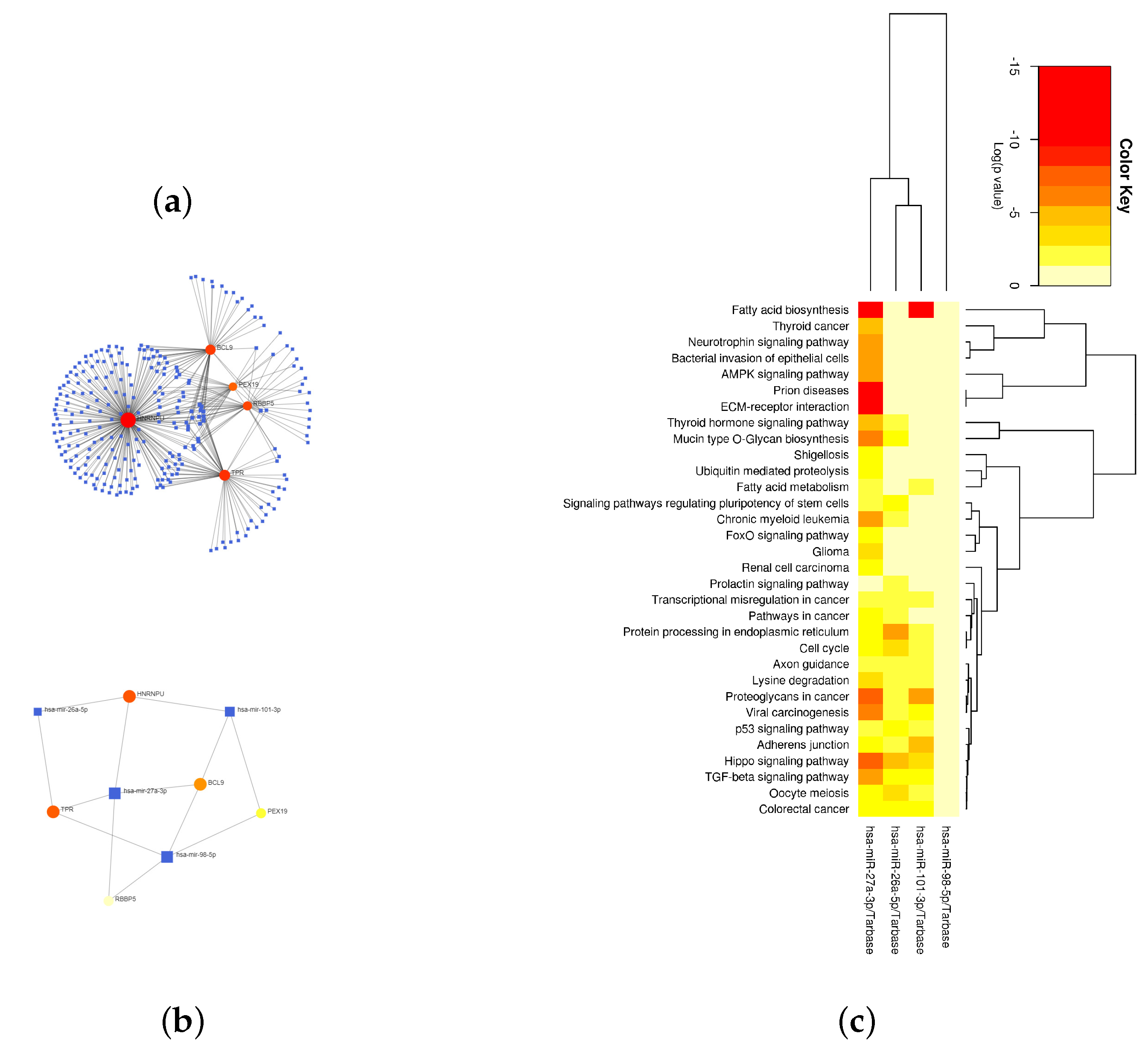
2.4. RBBP5, HNRNPU, PEX19, TPR, and BCL9 Are Closely Related to Immunity
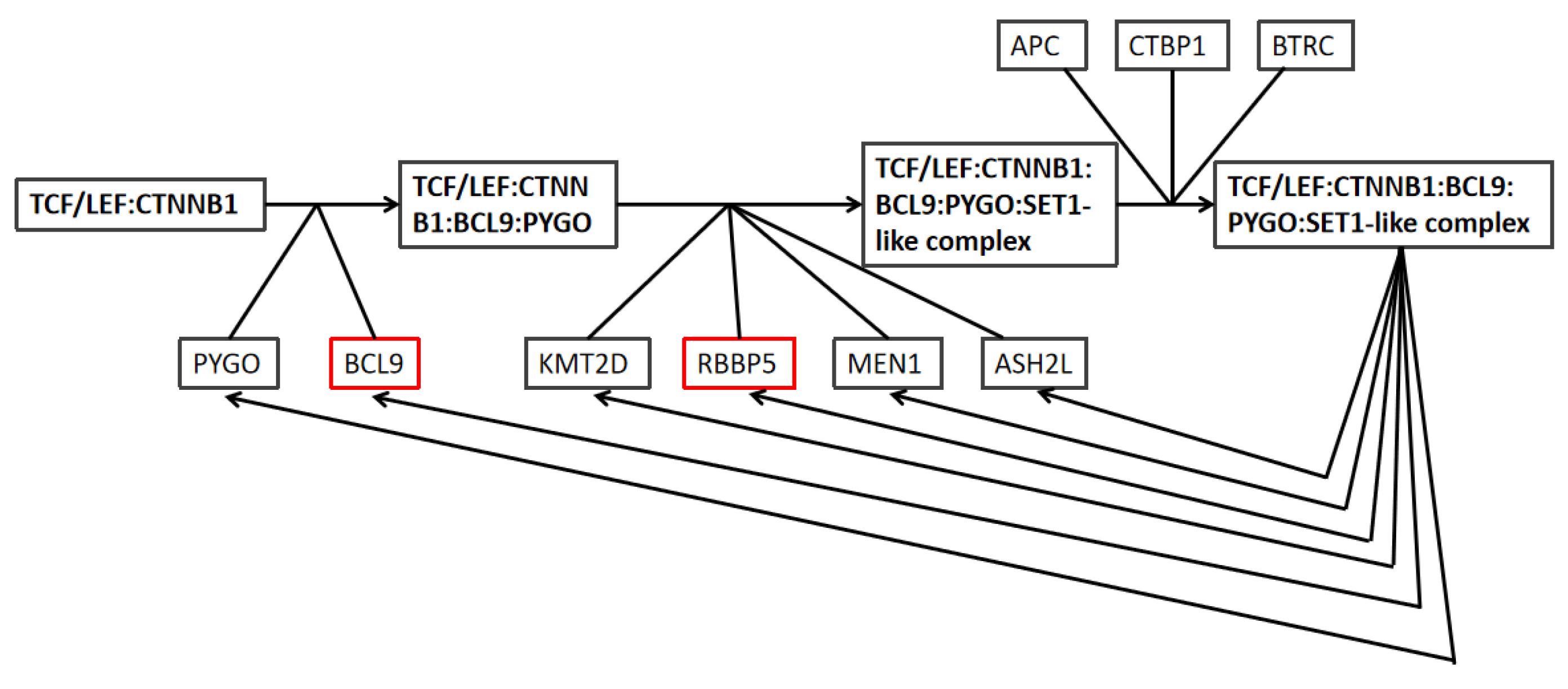
2.5. Overexpression of RBBP5, TPR, and BCL9 Is Associated with Poor Clinical Breast Cancer Outcomes and the Wnt Pathway
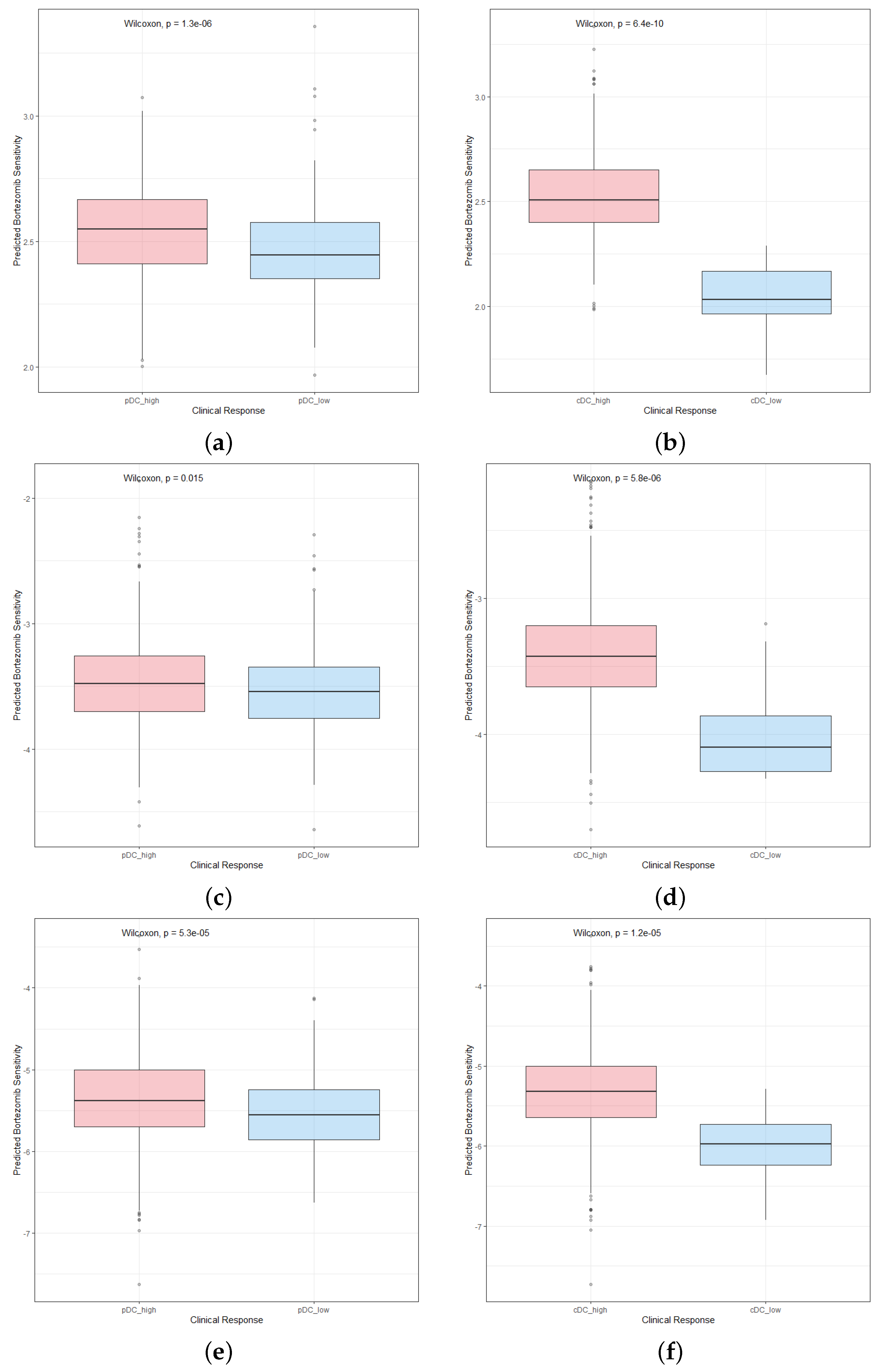
2.6. Patients with High Abundance of pDCs and cDCs Are More Sensitive to Chemotherapy Drugs
3. Discussion
4. Materials and Methods
4.1. Database
4.2. Calculation of Immune-Activity-Related Cellular Abundance
4.3. Survival Analysis of pDC and cDC Infiltration
4.4. Searching for Co-Expressed Gene Modules with WGCNA
4.5. Enrichment Analysis of Important Modules
4.6. Looking for Hub Genes of Important Modules and Their Associated miRNAs
4.7. Survival Analysis and Pathway Analysis of Hub Genes
4.8. Chemotherapy Response Prediction and Drug Screening
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DC | Dendritic Cell |
| cDC | Conventional Dendritic Cell |
| pDC | Plasmacytoid Dendritic Cell |
| WGCNA | Weighted Correlation Network Analysis |
| PPI | Protein–Protein Interaction |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Li, Z.; Zheng, Z.; Wei, Z.; Zhang, L.; Li, F.; Lin, L.; Liu, R.; Huang, X.; Lv, J.; Chen, F.; et al. Comprehensive characterization of the alternative splicing landscape in head and neck squamous cell carcinoma reveals novel events associated with tumorigenesis and the immune microenvironment. Theranostics 2019, 9, 7648–7665. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zhang, J.; Zeng, D.; Sun, H.; Rong, X.; Shi, M.; Bin, J.; Liao, Y.; Liao, W. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I-III colon cancer. Cancer Immunol. Immunother. 2019, 68, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Ma, Y.; Yu, S.; An, R.; He, J.; Zhang, H. Development of an Immune-Related Prognostic Signature in Breast Cancer. Front. Genet. 2020, 10, 1390. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.A. Breast cancer immunotherapy: Facts and hopes. Clin. Cancer Res. 2018, 24, 511–520. [Google Scholar] [CrossRef]
- Steven, A.; Fisher, S.A.; Robinson, B.W. Immunotherapy for lung cancer. Respirology 2016, 21, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Rachel, S.R.; Carl, H.J.; Robert, L.; Michael, J.M. Delivery technologies for cancer immunotherapy. Nat. Rev. Drug Discov. 2019, 18, 175–196. [Google Scholar]
- Gavin, P.D.; Allen, T.B.; Hiroaki, I.; Lloyd, J.O.; Robert, D.S. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar]
- Bell, D.; Chomarat, P.; Broyles, D.; Netto, G.; Harb, G.M.; Lebecque, S.; Valladeau, J.; Davoust, J.; Palucka, K.A.; Banchereau, J. In Breast Carcinoma Tissue, Immature Dendritic Cells Reside within the Tumor, Whereas Mature Dendritic Cells Are Located in Peritumoral Areas. J. Exp. Med. 1999, 190, 1417–1426. [Google Scholar] [CrossRef]
- Topalian, S.; Drake, C.; Pardoll, D. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015, 27, 450. [Google Scholar] [CrossRef]
- Chen, D.; Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef]
- Bordon, Y. Dendritic cells: Sorting, sorted! Nat. Rev. Immunol. 2016, 16, 657. [Google Scholar] [CrossRef] [PubMed]
- Swiecki, M.; Colonna, M. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 2015, 15, 471–485. [Google Scholar] [CrossRef] [PubMed]
- Poropatich, K.O.; Dominguez, D.; Chan, W.C.; Andrade, J.; Zhang, B. OX40+ plasmacytoid dendritic cells in the tumor microenvironment promote antitumor immunity. J. Clin. Investig. 2020, 130, 3528–3542. [Google Scholar] [CrossRef] [PubMed]
- Paula, M.; Floriane, N.; Eve, Z.; Urszula, C.; Phil’emon, S.; Omar, A.; Christel, G.; Alix, S.D.; Anne, V.S.; Fabien, R. Adjustment of dendritic cells to the breast-cancer microenvironment is subset specific. Nat. Immunol. 2018, 19, 885–897. [Google Scholar]
- Tian, S.; Yan, L.; Fu, L.; Zhang, Z.; Zhang, W. A Comprehensive Investigation to Reveal the Relationship Between Plasmacytoid Dendritic Cells and Breast Cancer by Multiomics Data Analysis. Front. Cell Dev. Biol. 2021, 9, 640476. [Google Scholar] [CrossRef]
- Masanori, O.; Stephanie, N.; Yoshihisa, T.; Li, Y.; Ryusei, M.; Pawel, K.; Itaru, E.; Kazuaki, T. Plasmacytoid Dendritic Cell (pDC) Infiltration Correlate with Tumor Infiltrating Lymphocytes, Cancer Immunity, and Better Survival in Triple Negative Breast Cancer (TNBC) More Strongly than Conventional Dendritic Cell (cDC). Cancers 2020, 12, E3342. [Google Scholar]
- Aberle, H.; Bauer, A.; Stappert, J.; Kispert, A.; Kemler, R. β-catenin is a target for the ubiquitin–proteasome pathway. Embo J. 1997, 16, 3797–3804. [Google Scholar] [CrossRef]
- Andtbacka, R.H.; Kaufman, H.L.; Collichio, F.; Amatruda, T.; Senzer, N.; Chesney, J.; Delman, K.A.; Spitler, L.E.; Puzanov, I.; Agarwala, S.S.; et al. Talimogene Laherparepvec Improves Durable Response Rate in Patients With Advanced Melanoma. J. Clin. Oncol. 2015, 33, 2780–2788. [Google Scholar] [CrossRef]
- Han, J.; Li, T.; Li, Y.; Li, M.; Wang, X.; Peng, C.; Su, C.; Li, N.; Li, Y.; Xu, Y. The internal interaction in RBBP5 regulates assembly and activity of MLL1 methyltransferase complex. Nuclc Acids Res. 2019, 47, 19. [Google Scholar] [CrossRef]
- Alvaro, G.A.; Praveena, S.T.; Erin, E.M.-H.; Daniel, J.S.; James, S.H.; Tyler, J.A.; Soumya, M.T.; Awad, J.; Ofer, R.; Michelle, S.L.; et al. Glioblastoma Cancer Stem Cells Evade Innate Immune Suppression of Self-Renewal through Reduced TLR4 Expression. Cell Stem Cell 2017, 20, 450–461. [Google Scholar] [CrossRef]
- Cerveny, L.; Straskova, A.; Dankova, V.; Hartlova, A.; Ceckova, M.; Staud, F.; Stulik, J. Tetratricopeptide Repeat Motifs in the World of Bacterial Pathogens: Role in Virulence Mechanisms. Infect. Immun. 2013, 81, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.H.; Jeong, Y.J.; Choi, S.H.; Kim, S.J.; Seog, D.H. Dynamin-1-like protein (Dnm1L) interaction with kinesin light chain 1 (KLC1) through the tetratricopeptide repeat (TPR) domains. Biosci. Biotechnol. Biochem. 2014, 78, 2069–2072. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.M.; Cao, X.Y.; Ping, H.; Li, J.; Feng, M.X.; Zhang, Y.L.; Zhang, X.L.; Wang, Y.H.; Qin, Y.; Lei, Z. Overexpression of Rac GTPase activating Protein 1 Contributes to Proliferation of Cancer Cells by Reducing Hippo Signaling to Promote Cytokinesis. Gastroenterology 2018, 155, 1233–1249. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Zhang, J.L.; Wang, Y.; Zhang, Y.Y.; Li, J.R. The Long Non-coding RNA LINC01705 Regulates the Development of Breast Cancer by Sponging miR-186-5p to Mediate TPR Expression as a Competitive Endogenous RNA. Front. Genet. 2020, 11, 779. [Google Scholar] [CrossRef]
- Jiang, M.; Kang, Y.; Sewastianik, T.; Wang, J.; Tanton, H.; Alder, K.; Dennis, P.; Xin, Y.; Wang, Z.; Liu, R.; et al. BCL9 provides multi-cellular communication properties in colorectal cancer by interacting with paraspeckle proteins. Nat. Commun. 2020, 11, 19. [Google Scholar] [CrossRef]
- Behbod, F.; Elsarraj, H.; Hong, Y.; Valdez, K.; Fields, T. Expression profiling of in vivo DCIS progression models identified BCL9 as a molecular driver of invasive progression. Cancer Res. 2016, 76, 128. [Google Scholar] [CrossRef]
- Wang, X.S.; Feng, M.; Xiao, T.F.; Guo, B.S.; Liu, D.Y.; Liu, C.L.; Pei, J.P.; Liu, Q.F.; Xiao, Y.; Rosin-Arbesfeld, R.; et al. BCL9/BCL9L promotes tumorigenicity through immune-dependent and independent mechanisms in triple negative breast cancer. Oncogene 2021, 16, 2982–2997. [Google Scholar] [CrossRef]
- Jean-Pierre, B.; Nehme, E.H.; Abed, E.E.; Jamilah, A.; Natasha, S.; Riam, S.; Moutih, R. Thymoproteasome-Expressing Mesenchymal Stromal Cells Confer Protective Anti-Tumor Immunity via Cross-Priming of Endogenous Dendritic Cells. Front. Immunol. 2021, 11, 596303. [Google Scholar]
- Mei, S.Y.; Liu, Y.H.; Bao, Y.; Zhang, Y.; Min, S.P.; Liu, Y.F.; Huang, Y.; Yuan, X.D.; Feng, Y.; Shi, J.D. Dendritic Cell-Associated miRNAs Are Modulated via Chromatin Remodeling in Response to Different Environments. PLoS ONE 2014, 4, e90231. [Google Scholar] [CrossRef]
- David, M.G.; Rachel, A.R.; Miryam, M.; Michael, C.H.; Ann, H.; William, C.; Joshua, D.L.; Rene, J.; Colin, N.; David, J.H.; et al. Loss of BCL9/9l suppresses Wnt driven tumourigenesis in models that recapitulate human cancer. Nat. Commun. 2019, 10, 723. [Google Scholar]
- Minesaki, A.; Kai, K.; Kuratomi, Y.; Aishima, S. Infiltration of CD1a-positive dendritic cells in advanced laryngeal cancer correlates with unfavorable outcomes post-laryngectomy. BMC Cancer 2021, 21, 973. [Google Scholar] [CrossRef] [PubMed]
- Kiyoshi, M.; Masaki, I.; Takuma, M.; James, B.W.; Hiroaki, S.; Mizuyu, O.; Ryuta, U.; Daisuke, K.; Tatsuya, T.; Satoru, T.; et al. Mature dendritic cells enriched in regulatory molecules may control regulatory T cells and the prognosis of head and neck cancer. Cancer Sci. 2022. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Dedeurwaerder, S.; Desmedt, C.; Calonne, E.; Singhal, S.K.; Haibe-Kains, B.; Luciani, J.; Putmans, P.; Lallemand, F.; Larsimont, D.; Toussaint, J.; et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol. Med. 2011, 3, 726–741. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Croft, D.; O’Kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2011, 39, D691–D697. [Google Scholar] [CrossRef]
- Geeleher, P.; Cox, N.J.; Huang, R. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014, 15, R47. [Google Scholar] [CrossRef]

| Category | Term | Genes | p Value |
|---|---|---|---|
| KEGG | hsa04146:Peroxisome | GNPAT, PEX19, PEX11B | 1.6 × 10−2 |
| KEGG | hsa03013:Nucleocytoplasmic transport | IPO9, NUP133, TPR | 2.8 × 10−4 |
| Pharmaceuticals | pDC_FoldChange | cDC_FoldChange |
|---|---|---|
| Lapatinib | 1.021542892 | 1.157434474 |
| Paclitaxel | 0.982754003 | 0.893367891 |
| Docetaxel | 0.976301981 | 0.917481043 |
| Pharmaceuticals | pDChigh_Average | cDChigh_Average |
|---|---|---|
| Bortezmib_1191 | 0.0087 | 0.0086 |
| Dactinomycin_1911 | 0.0103 | 0.0102 |
| Docetaxel_1007 | 0.0145 | 0.0141 |
| Daporinad_1248 | 0.0153 | 0.0152 |
| Sepantronium.bromide_1941 | 0.0154 | 0.0155 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, F.; Yan, F.; Liu, H. New Biomarkers Based on Dendritic Cells for Breast Cancer Treatment and Prognosis Diagnosis. Int. J. Mol. Sci. 2023, 24, 4058. https://doi.org/10.3390/ijms24044058
Zhao F, Yan F, Liu H. New Biomarkers Based on Dendritic Cells for Breast Cancer Treatment and Prognosis Diagnosis. International Journal of Molecular Sciences. 2023; 24(4):4058. https://doi.org/10.3390/ijms24044058
Chicago/Turabian StyleZhao, Fanjia, Fang Yan, and Haihong Liu. 2023. "New Biomarkers Based on Dendritic Cells for Breast Cancer Treatment and Prognosis Diagnosis" International Journal of Molecular Sciences 24, no. 4: 4058. https://doi.org/10.3390/ijms24044058
APA StyleZhao, F., Yan, F., & Liu, H. (2023). New Biomarkers Based on Dendritic Cells for Breast Cancer Treatment and Prognosis Diagnosis. International Journal of Molecular Sciences, 24(4), 4058. https://doi.org/10.3390/ijms24044058




