Microbial Enzyme Biotechnology to Reach Plastic Waste Circularity: Current Status, Problems and Perspectives
Abstract
1. Introduction
2. Plastics: From Waste Generation to Recycling
2.1. Plastic Classification
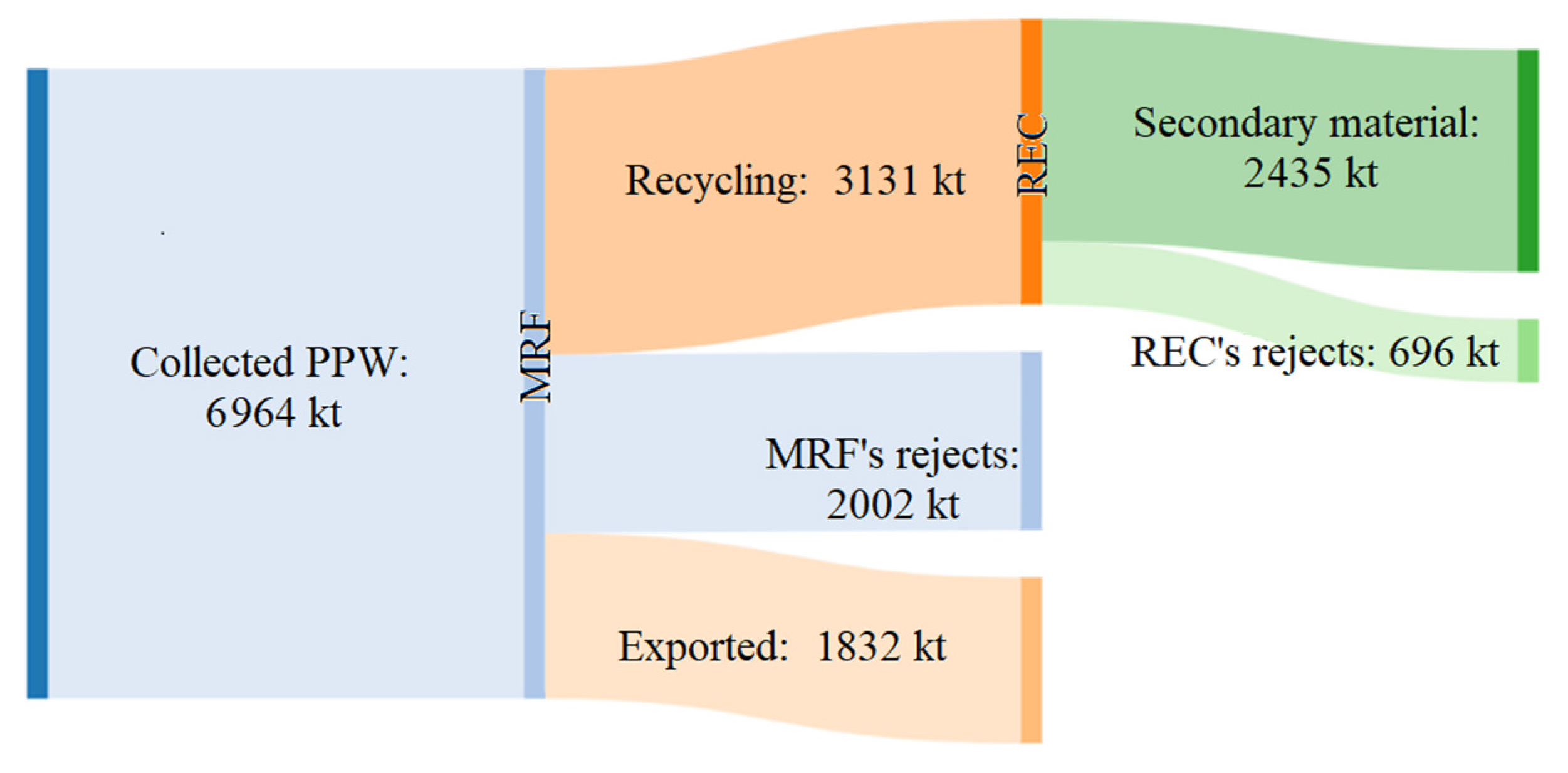
2.2. EU Plastic Waste Flows
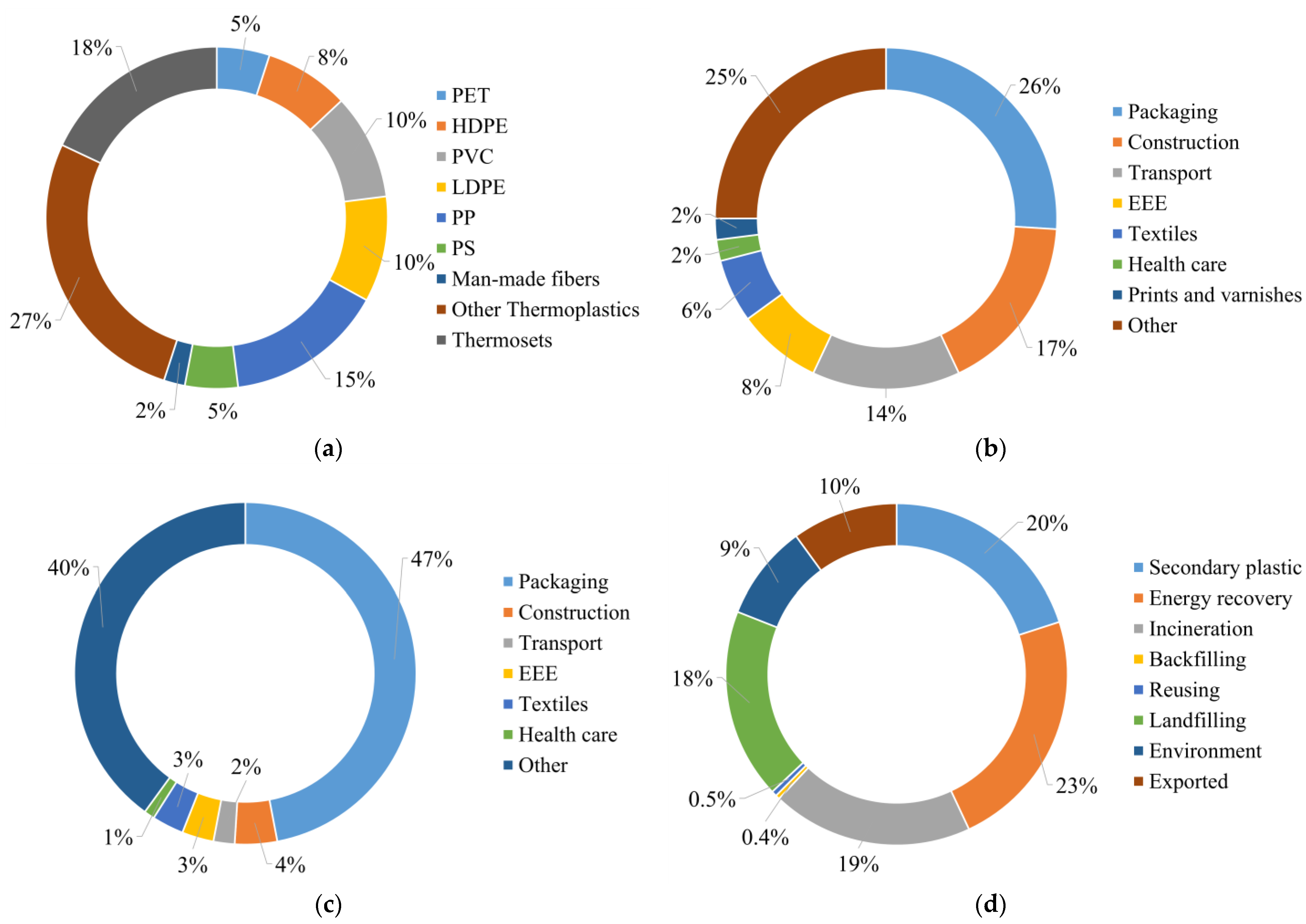
2.3. Options for Thermoplastic Waste Recycling
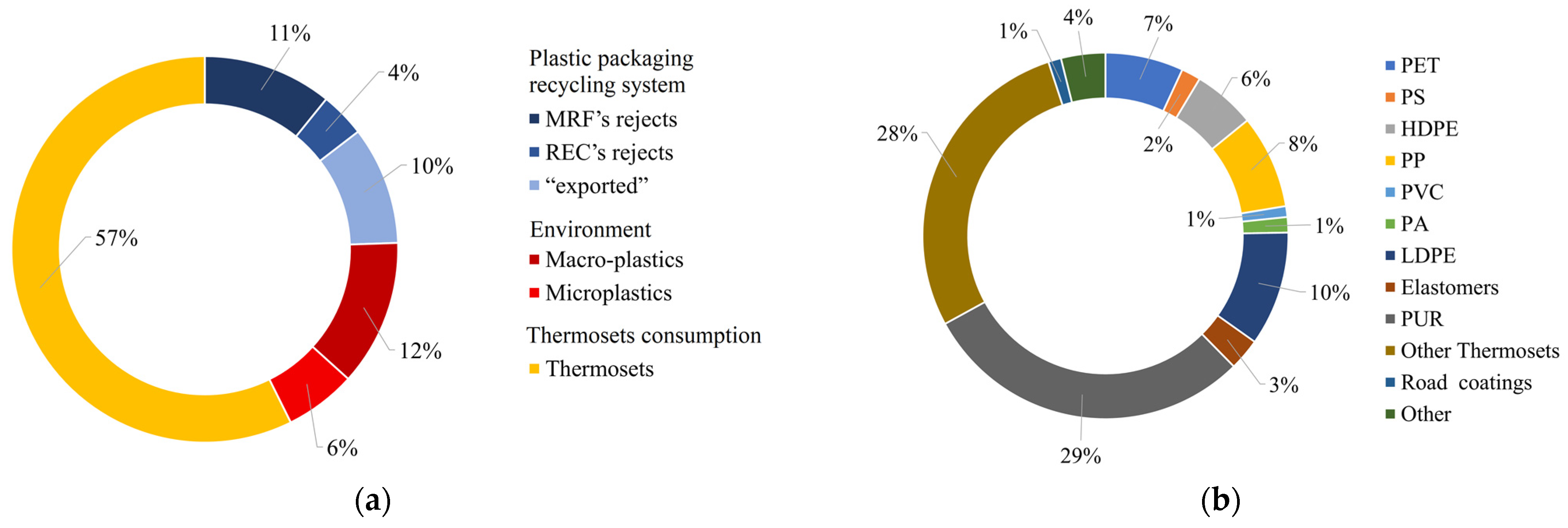
2.4. Thermoplastics Recycling Issues: The Plastic Packaging Waste Case
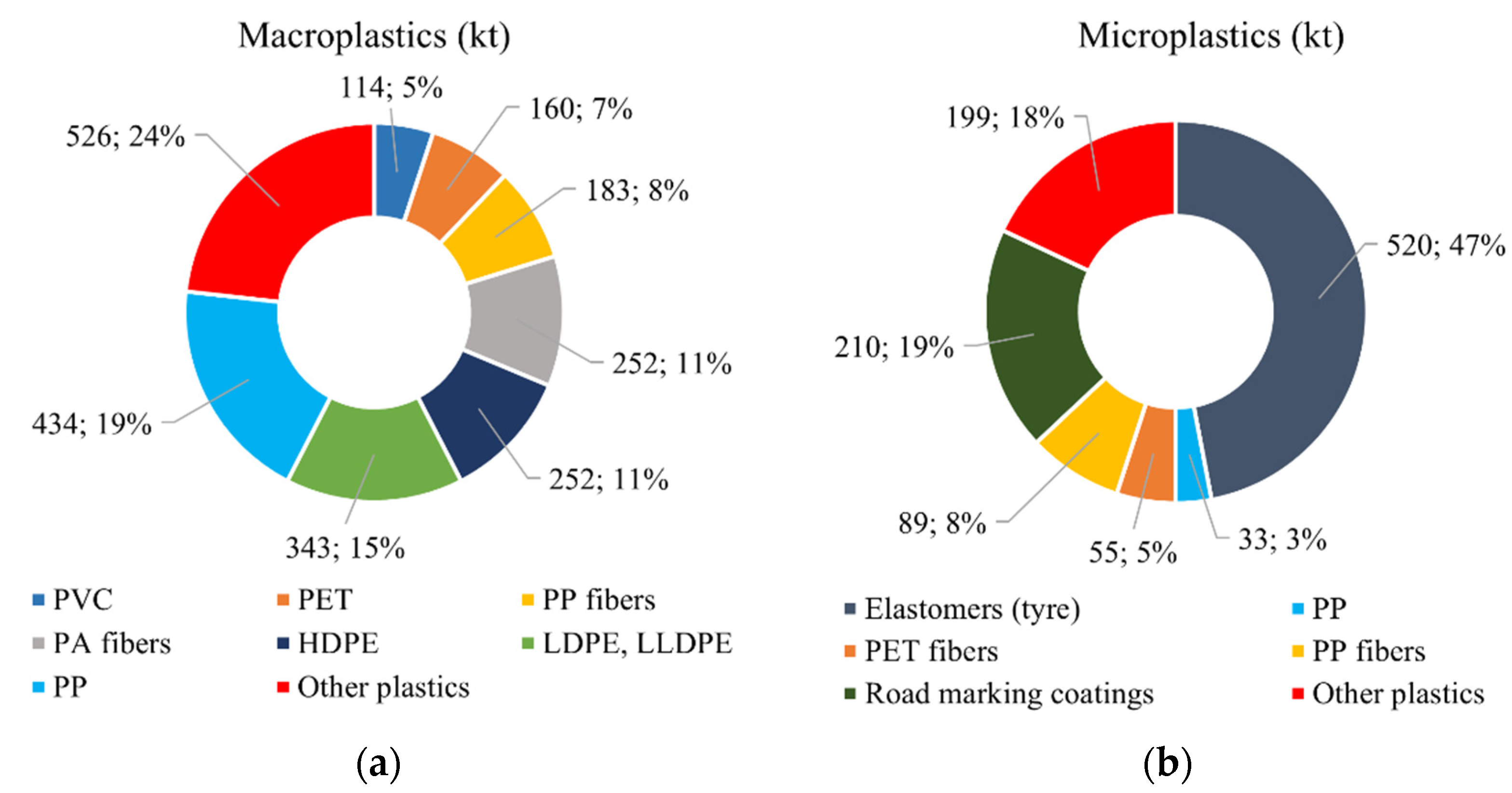
2.5. Removal of Environmental Plastic Pollution
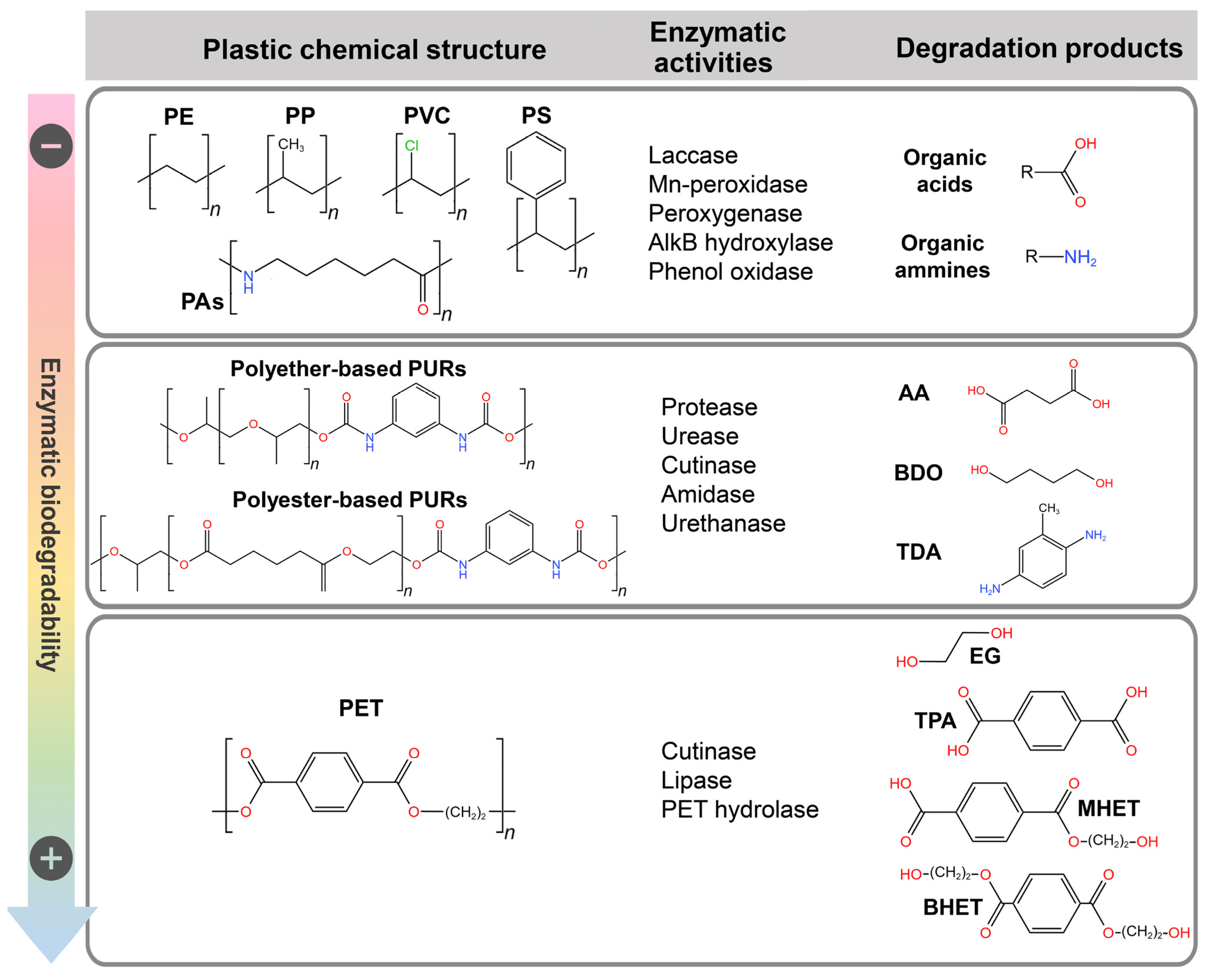
3. Biotechnological Systems Applied to Plastic Depolymerization to Target Recycling
3.1. Enzymatic PET Recycling: Potential Industrial-Scale Application
3.1.1. Enzymatic Degradation of PET
3.1.2. Closed-Loop PET Recycling by Enzymatic Depolymerization
3.1.3. Open-Loop PET Bio-Recycling
| Approach | Enzymes or Microbial Strain(s) | Plastic Monomer | Conversion Products (Yields %) | Reference |
|---|---|---|---|---|
| Separate depolymerization and conversion | Pseudomonas putida strains JM37 and KT2440 | EG | Glyoxylic acid (≈20%) | [157] |
| Three derived Pseudomonas putida KT2440 strains | AA, BDO, EG | Monorhamnolipids (<1%) | [158] | |
| Escherichia coli strain | TPA, EG | Gallic acid (≈93%), pyrogallol (≈40%), muconic acid (≈85%), vanillic acid (≈40%), glycolic acid (≈98.6%) | [149] | |
| Escherichia coli strain | TPA | 2-pyrone-4,6-dicarboxylic acid (96.08%) | [151] | |
| Pseudomonas putida KT2440 strain | BHET | β-ketoadipic acid (≈76%) | [152] | |
| Escherichia coli strain | TPA | Catechol (≈67%) | [150] | |
| Combined depolymerization and conversion | LCC 1, Pseudomonas umsongensis GO16 strain | TPA, EG | PHAs and HAA (<2%) | [153] |
| LCC 2, Escherichia coli MG1655 RARE strain | TPA, EG | Vanillin (≈79%) | [154] | |
| Consolidated bioprocessing | Yarrowia lipolytica Po1f expressing PETase 3, Pseudomonas stutzeri TPA3 | TPA, EG | PHB (≈2.5%) | [155] |
| Pseudomonas putida KT2440-tacRDL expressing LCC 4 | TPA, EG | Muconic acid (≈50%) | [156] |
3.2. PUR Biodegradation Is Restricted to Polyester-Based Polymers
3.2.1. Enzymatic Degradation of PURs
3.2.2. Microbial Degradation of PURs
3.2.3. Closed-Loop and Open-Loop PUR Bio-Recycling
3.3. Prospective for the Biological Degradation of Other Petroleum-Based Plastics
4. Perspectives
4.1. Plastic Recycling Challenges in the EU: Current Limits of the Biotechnological Approaches
4.2. Finding New Biotechnologically Degradable Plastic Materials
4.3. Study of New Polyester-Degrading Organisms and Enzymes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ayeleru, O.O.; Dlova, S.; Akinribide, O.J.; Ntuli, F.; Kupolati, W.K.; Marina, P.F.; Blencowe, A.; Olubambi, P.A. Challenges of plastic waste generation and management in sub-Saharan Africa: A review. Waste Manag. 2020, 110, 24–42. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Xiong, X.; He, M.; Tsang, D.C.; Gupta, J.; Khan, E.; Harrad, S.; Hou, D.; Ok, Y.S.; Bolan, N.S. Microplastics as pollutants in agricultural soils. Environ. Pollut. 2020, 265, 114980. [Google Scholar] [CrossRef]
- Arp, H.P.H.; Kühnel, D.; Rummel, C.; MacLeod, M.; Potthoff, A.; Reichelt, S.; Rojo-Nieto, E.; Schmitt-Jansen, M.; Sonnenberg, J.; Toorman, E.; et al. Weathering plastics as a planetary boundary threat: Exposure, fate, and hazards. Environ. Sci. Technol. 2021, 55, 7246–7255. [Google Scholar] [CrossRef] [PubMed]
- Borrelle, S.B.; Ringma, J.; Law, K.L.; Monnahan, C.C.; Lebreton, L.; McGivern, A.; Murphy, E.; Jambeck, J.; Leonard, G.H.; Hilleary, M.A.; et al. Predicted growth in plastic waste exceeds efforts to mitigate plastic pollution. Science 2020, 369, 1515–1518. [Google Scholar] [CrossRef] [PubMed]
- Ferronato, N.; Torretta, V. Waste mismanagement in developing countries: A review of global issues. Int. J. Environ. Res. Public Health 2019, 16, 1060. [Google Scholar] [CrossRef] [PubMed]
- Brahney, J.; Hallerud, M.; Heim, E.; Hahnenberger, M.; Sukumaran, S. Plastic rain in protected areas of the United States. Science 2020, 368, 1257–1260. [Google Scholar] [CrossRef]
- Li, L.; Zuo, J.; Duan, X.; Wang, S.; Hu, K.; Chang, R. Impacts and mitigation measures of plastic waste: A critical review. Environ. Impact Assess. Rev. 2021, 90, 106642. [Google Scholar] [CrossRef]
- Bergmann, M.; Collard, F.; Fabres, J.; Gabrielsen, G.W.; Provencher, J.F.; Rochman, C.M.; van Sebille, E.; Tekman, M.B. Plastic pollution in the Arctic. Nat. Rev. Earth Environ. 2022, 3, 323–337. [Google Scholar] [CrossRef]
- Bhatt, P.; Pathak, V.M.; Bagheri, A.R.; Bilal, M. Microplastic contaminants in the aqueous environment, fate, toxicity consequences, and remediation strategies. Environ. Res. 2021, 200, 111762. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, J.; Xing, B. Environmental source, fate, and toxicity of microplastics. J. Hazard. Mater. 2021, 407, 124357. [Google Scholar] [CrossRef]
- Isobe, A.; Iwasaki, S. The fate of missing ocean plastics: Are they just a marine environmental problem? Sci. Total Environ. 2022, 825, 153935. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Zhou, Y.; Wang, Y.; Zhang, D.; Pan, X. Transfer of Micro (nano) plastics in animals: A mini-review and future research recommendation. J. Hazard. Mater. Adv. 2022, 7, 100101. [Google Scholar] [CrossRef]
- Leslie, H.A.; van Velzen, M.J.; Brandsma, S.H.; Vethaak, A.D.; Garcia-Vallejo, J.J.; Lamoree, M.H. Discovery and quantification of plastic particle pollution in human blood. Environ. Int. 2022, 163, 107199. [Google Scholar] [CrossRef]
- De Souza Machado, A.A.; Kloas, W.; Zarfl, C.; Hempel, S.; Rillig, M.C. Microplastics as an emerging threat to terrestrial ecosystems. Glob. Chang. Biol. 2018, 24, 1405–1416. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.; An, Y.-J. Effects of micro-and nanoplastics on aquatic ecosystems: Current research trends and perspectives. Mar. Pollut. Bull. 2017, 124, 624–632. [Google Scholar] [CrossRef]
- Sridharan, S.; Kumar, M.; Bolan, N.S.; Singh, L.; Kumar, S.; Kumar, R.; You, S. Are microplastics destabilizing the global network of terrestrial and aquatic ecosystem services? Environ. Res. 2021, 198, 111243. [Google Scholar] [CrossRef]
- Pirillo, V.; Baranzini, N. Current research on the effects of plastics pollution in marine and freshwater aquatic invertebrates. Invertebr. Surviv. J. 2022, 19, 136–149. [Google Scholar] [CrossRef]
- Ricardo, I.A.; Alberto, E.A.; Júnior, A.H.S.; Macuvele, D.L.P.; Padoin, N.; Soares, C.; Riella, H.G.; Starling, M.C.V.; Trovo, A.G. A critical review on microplastics, interaction with organic and inorganic pollutants, impacts and effectiveness of advanced oxidation processes applied for their removal from aqueous matrices. Chem. Eng. J. 2021, 424, 130282. [Google Scholar] [CrossRef]
- Sathicq, M.B.; Sabatino, R.; di Cesare, A.; Eckert, E.M.; Fontaneto, D.; Rogora, M.; Corno, G. PET particles raise microbiological concerns for human health while tyre wear microplastic particles potentially affect ecosystem services in waters. J. Hazard. Mater. 2022, 429, 128397. [Google Scholar] [CrossRef]
- Eriksen, M.K.; Damgaard, A.; Boldrin, A.; Astrup, T.F. Quality assessment and circularity potential of recovery systems for household plastic waste. J. Ind. Ecol. 2019, 23, 156–168. [Google Scholar] [CrossRef]
- Gaustad, G.; Krystofik, M.; Bustamante, M.; Badami, K. Circular economy strategies for mitigating critical material supply issues. Resour. Conserv. Recycl. 2018, 135, 24–33. [Google Scholar] [CrossRef]
- Geisendorf, S.; Pietrulla, F. The circular economy and circular economic concepts—A literature analysis and redefinition. Thunderbird Int. Bus. Rev. 2018, 60, 771–782. [Google Scholar] [CrossRef]
- Ali, M.; Courtenay, P. Evaluating the progress of the UK’s Material Recycling Facilities: A mini review. Waste Manag. Res. 2014, 32, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, M.; Picuno, C.; van Velzen, E.U.T.; Kuchta, K.; de Meester, S.; Ragaert, K. The impact of collection portfolio expansion on key performance indicators of the Dutch recycling system for Post-Consumer Plastic Packaging Waste, a comparison between 2014 and 2017. Waste Manag. 2019, 100, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Huysveld, S.; Ragaert, K.; Demets, R.; Nhu, T.T.; Civancik-Uslu, D.; Kusenberg, M.; van Geem, K.; de Meester, S.; Dewulf, J. Technical and market substitutability of recycled materials: Calculating the environmental benefits of mechanical and chemical recycling of plastic packaging waste. Waste Manag. 2022, 152, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, S.; Hodzic, A.; Scelsi, L.; Hayes, S.; Soutis, C.; Alma’Adeed, M.; Kahraman, R. Plastics recycling: Insights into life cycle impact assessment methods. Plast. Rubber Compos. 2013, 42, 1–10. [Google Scholar] [CrossRef]
- Abdelraheem, E.M.; Busch, H.; Hanefeld, U.; Tonin, F. Biocatalysis explained: From pharmaceutical to bulk chemical production. React. Chem. Eng. 2019, 4, 1878–1894. [Google Scholar] [CrossRef]
- Corvellec, H.; Stowell, A.F.; Johansson, N. Critiques of the circular economy. J. Ind. Ecol. 2022, 26, 421–432. [Google Scholar] [CrossRef]
- European Commission. Directorate-General for Research and Innovation. In A Sustainable Bioeconomy for Europe Strengthening the Connection between Economy, Society and the Environment, Updated Bioeconomy Strategy; European Commission: Brussels, Belgium, 2018; Available online: https://data.europa.eu/doi/10.2777/792130 (accessed on 17 July 2022).
- Chamas, A.; Moon, H.; Zheng, J.; Qiu, Y.; Tabassum, T.; Jang, J.H.; Abu-Omar, M.; Scott, S.L.; Suh, S. Degradation rates of plastics in the environment. ACS Sustain. Chem. Eng. 2020, 8, 3494–3511. [Google Scholar] [CrossRef]
- Blank, L.M.; Narancic, T.; Mampel, J.; Tiso, T.; O’Connor, K. Biotechnological upcycling of plastic waste and other non-conventional feedstocks in a circular economy. Curr. Opin. Biotechnol. 2020, 62, 212–219. [Google Scholar] [CrossRef]
- Vollmer, I.; Jenks, M.J.; Roelands, M.C.; White, R.J.; van Harmelen, T.; de Wild, P.; van Der Laan, G.P.; Meirer, F.; Keurentjes, J.T.; Weckhuysen, B.M. Beyond mechanical recycling: Giving new life to plastic waste. Angew. Chem. Int. Ed. 2020, 59, 15402–15423. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Dai, L.; Ma, L.; Guo, R.-T. Enzymatic degradation of plant biomass and synthetic polymers. Nat. Rev. Chem. 2020, 4, 114–126. [Google Scholar] [CrossRef]
- Wei, R.; Tiso, T.; Bertling, J.; O’Connor, K.; Blank, L.M.; Bornscheuer, U.T. Possibilities and limitations of biotechnological plastic degradation and recycling. Nat. Catal. 2020, 3, 867–871. [Google Scholar] [CrossRef]
- Amobonye, A.; Bhagwat, P.; Singh, S.; Pillai, S. Plastic biodegradation: Frontline microbes and their enzymes. Sci. Total Environ. 2021, 759, 143536. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.D.; Rorrer, N.A.; Sullivan, K.P.; Otto, M.; McGeehan, J.E.; Román-Leshkov, Y.; Wierckx, N.; Beckham, G.T. Chemical and biological catalysis for plastics recycling and upcycling. Nat. Catal. 2021, 4, 539–556. [Google Scholar] [CrossRef]
- Inderthal, H.; Tai, S.L.; Harrison, S.T. Non-hydrolyzable plastics–An interdisciplinary look at plastic bio-oxidation. Trends Biotechnol. 2021, 39, 12–23. [Google Scholar] [CrossRef]
- Abdelmoez, W.; Dahab, I.; Ragab, E.M.; Abdelsalam, O.A.; Mustafa, A. Bio-and oxo-degradable plastics: Insights on facts and challenges. Polym. Adv. Technol. 2021, 32, 1981–1996. [Google Scholar] [CrossRef]
- Ali, S.S.; Elsamahy, T.; Al-Tohamy, R.; Zhu, D.; Mahmoud, Y.A.-G.; Koutra, E.; Metwally, M.A.; Kornaros, M.; Sun, J. Plastic wastes biodegradation: Mechanisms, challenges and future prospects. Sci. Total Environ. 2021, 780, 146590. [Google Scholar] [CrossRef]
- Qin, Z.H.; Mou, J.H.; Chao, C.Y.H.; Chopra, S.S.; Daoud, W.; Leu, S.Y.; Ning, Z.; Tso, C.Y.; Chan, C.K.; Tang, S.; et al. Biotechnology of plastic waste degradation, recycling, and valorization: Current advances and future perspectives. ChemSusChem 2021, 14, 4103–4114. [Google Scholar] [CrossRef]
- Zhu, B.; Wang, D.; Wei, N. Enzyme discovery and engineering for sustainable plastic recycling. Trends Biotechnol. 2022, 40, 22–37. [Google Scholar] [CrossRef]
- Zhao, X.; Boruah, B.; Chin, K.F.; Đokić, M.; Modak, J.M.; Soo, H.S. Upcycling to sustainably reuse plastics. Adv. Mater. 2022, 34, 2100843. [Google Scholar] [CrossRef]
- Andler, R.; Tiso, T.; Blank, L.; Andreeßen, C.; Zampolli, J.; D’Afonseca, V.; Guajardo, C.; Díaz-Barrera, A. Current progress on the biodegradation of synthetic plastics: From fundamentals to biotechnological applications. Rev. Environ. Sci. Bio/Technol. 2022, 21, 829–850. [Google Scholar] [CrossRef]
- Plastics—The Facts 2020. Available online: https://plasticseurope.org/wp-content/uploads/2021/09/Plastics_the_facts-WEB-2020_versionJun21_final.pdf (accessed on 24 December 2021).
- How Plastic Are Made. Available online: https://plasticseurope.org/plastics-explained/how-plastics-are-made/ (accessed on 24 December 2021).
- Grigore, M.E. Methods of recycling, properties and applications of recycled thermoplastic polymers. Recycling 2017, 2, 24. [Google Scholar] [CrossRef]
- Schyns, Z.O.; Shaver, M.P. Mechanical recycling of packaging plastics: A review. Macromol. Rapid Commun. 2021, 42, 2000415. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.; Amin, M.; Thermoplastic Elastomeric (TPE) Materials and Their Use in Outdoor Electrical Insulation. Reviews on Advanced Materials Science 29. 2011. Available online: https://ipme.ru/e-journals/RAMS/no_12911/02_amin.pdf (accessed on 20 October 2022).
- Zhao, X.; Liu, X.; Feng, K.; An, W.L.; Tian, F.; Du, R.; Xu, S.; Chen, L.; Wu, G.; Wang, Y.Z. Multicycling of Epoxy Thermoset Through a Two-Step Strategy of Alcoholysis and Hydrolysis using a Self-Separating Catalysis System. ChemSusChem 2022, 15, e202101607. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Y.; Chen, W.Q.; Zhu, B.; Qu, S.; Xu, M. Critical review of global plastics stock and flow data. J. Ind. Ecol. 2021, 25, 1300–1317. [Google Scholar] [CrossRef]
- Hsu, W.-T.; Domenech, T.; McDowall, W. How circular are plastics in the EU?: MFA of plastics in the EU and pathways to circularity. Clean. Environ. Syst. 2021, 2, 100004. [Google Scholar] [CrossRef]
- Bishop, G.; Styles, D.; Lens, P.N. Recycling of European plastic is a pathway for plastic debris in the ocean. Environ. Int. 2020, 142, 105893. [Google Scholar] [CrossRef]
- Ryberg, M.W.; Hauschild, M.Z.; Wang, F.; Averous-Monnery, S.; Laurent, A. Global environmental losses of plastics across their value chains. Resour. Conserv. Recycl. 2019, 151, 104459. [Google Scholar] [CrossRef]
- Datta, J.; Kopczyńska, P.; Simón, D.; Rodríguez, J.F. Thermo-chemical decomposition study of polyurethane elastomer through glycerolysis route with using crude and refined glycerine as a transesterification agent. J. Polym. Environ. 2018, 26, 166–174. [Google Scholar] [CrossRef]
- Kemona, A.; Piotrowska, M. Polyurethane recycling and disposal: Methods and prospects. Polymers 2020, 12, 1752. [Google Scholar] [CrossRef]
- Hahladakis, J.N.; Iacovidou, E. An overview of the challenges and trade-offs in closing the loop of post-consumer plastic waste (PCPW): Focus on recycling. J. Hazard. Mater. 2019, 380, 120887. [Google Scholar] [CrossRef] [PubMed]
- Larrain, M.; van Passel, S.; Thomassen, G.; van Gorp, B.; Nhu, T.T.; Huysveld, S.; van Geem, K.M.; de Meester, S.; Billen, P. Techno-economic assessment of mechanical recycling of challenging post-consumer plastic packaging waste. Resour. Conserv. Recycl. 2021, 170, 105607. [Google Scholar] [CrossRef]
- Lee, A.; Liew, M.S. Tertiary recycling of plastics waste: An analysis of feedstock, chemical and biological degradation methods. J. Mater. Cycles Waste Manag. 2021, 23, 32–43. [Google Scholar] [CrossRef]
- Schwarz, A.; Ligthart, T.; Bizarro, D.G.; de Wild, P.; Vreugdenhil, B.; van Harmelen, T. Plastic recycling in a circular economy; determining environmental performance through an LCA matrix model approach. Waste Manag. 2021, 121, 331–342. [Google Scholar] [CrossRef]
- Krahling, H.; Sartorius, I. Plastics after Use: Sustainable Management of Material and Energy Resources. Polym. Sci. A Compr. Ref. 2012, 10, 581–595. [Google Scholar]
- Ragaert, K.; Delva, L.; van Geem, K. Mechanical and chemical recycling of solid plastic waste. Waste Manag. 2017, 69, 24–58. [Google Scholar] [CrossRef]
- Antonopoulos, I.; Faraca, G.; Tonini, D. Recycling of post-consumer plastic packaging waste in the EU: Recovery rates, material flows, and barriers. Waste Manag. 2021, 126, 694–705. [Google Scholar] [CrossRef]
- Deloitte, 2017. Blueprint for PPW: Quality Sorting and Recycling. Final Report. Available online: https://www2.deloitte.com/content/dam/Deloitte/my/Documents/risk/my-risk-blueprint-plastics-packaging-waste-2017.pdf (accessed on 20 October 2022).
- Rozenstein, O.; Puckrin, E.; Adamowski, J. Development of a new approach based on midwave infrared spectroscopy for post-consumer black plastic waste sorting in the recycling industry. Waste Manag. 2017, 68, 38–44. [Google Scholar] [CrossRef]
- Faraca, G.; Astrup, T. Plastic waste from recycling centres: Characterisation and evaluation of plastic recyclability. Waste Manag. 2019, 95, 388–398. [Google Scholar] [CrossRef]
- Jönsson, C.; Wei, R.; Biundo, A.; Landberg, J.; Schwarz Bour, L.; Pezzotti, F.; Toca, A.; Jacques, L.M.; Bornscheuer, U.T.; Syrén, P.O. Biocatalysis in the recycling landscape for synthetic polymers and plastics towards circular textiles. ChemSusChem 2021, 14, 4028–4040. [Google Scholar] [CrossRef] [PubMed]
- LI, W.C.; Tse, H.; Fok, L. Plastic waste in the marine environment: A review of sources, occurrence and effects. Sci. Total Environ. 2016, 566, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.; Hamid, A.K.; Krebsbach, S.A.; He, J.; Wang, D. Critical review of microplastics removal from the environment. Chemosphere 2022, 293, 133557. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.G.; Moore, C.J.; van Franeker, J.A.; Moloney, C.L. Monitoring the abundance of plastic debris in the marine environment. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 1999–2012. [Google Scholar] [CrossRef]
- Sienkiewicz, M.; Janik, H.; Borzędowska-Labuda, K.; Kucińska-Lipka, J. Environmentally friendly polymer-rubber composites obtained from waste tyres: A review. J. Clean. Prod. 2017, 147, 560–571. [Google Scholar] [CrossRef]
- Ronkay, F.; Molnar, B.; Gere, D.; Czigany, T. Plastic waste from marine environment: Demonstration of possible routes for recycling by different manufacturing technologies. Waste Manag. 2021, 119, 101–110. [Google Scholar] [CrossRef]
- Sun, J.; Dai, X.; Wang, Q.; van Loosdrecht, M.C.; Ni, B.-J. Microplastics in wastewater treatment plants: Detection, occurrence and removal. Water Res. 2019, 152, 21–37. [Google Scholar] [CrossRef]
- Dey, T.K.; Uddin, M.; Jamal, M. Detection and removal of microplastics in wastewater: Evolution and impact. Environ. Sci. Pollut. Res. 2021, 28, 16925–16947. [Google Scholar] [CrossRef]
- Galafassi, S.; Sabatino, R.; Sathicq, M.B.; Eckert, E.M.; Fontaneto, D.; Dalla Fontana, G.; Mossotti, R.; Corno, G.; Volta, P.; di Cesare, A. Contribution of microplastic particles to the spread of resistances and pathogenic bacteria in treated wastewaters. Water Res. 2021, 201, 117368. [Google Scholar] [CrossRef]
- Galafassi, S.; di Cesare, A.; di Nardo, L.; Sabatino, R.; Valsesia, A.; Fumagalli, F.S.; Corno, G.; Volta, P. Microplastic retention in small and medium municipal wastewater treatment plants and the role of the disinfection. Environ. Sci. Pollut. Res. 2022, 29, 10535–10546. [Google Scholar] [CrossRef]
- Tiwari, N.; Santhiya, D.; Sharma, J.G. Microbial remediation of micro-nano plastics: Current knowledge and future trends. Environ. Pollut. 2020, 265, 115044. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ren, X.; Rene, E.R.; Wang, Z.; Zhou, L.; Zhang, Z.; Wang, Q. The degradation performance of different microplastics and their effect on microbial community during composting process. Bioresour. Technol. 2021, 332, 125133. [Google Scholar] [CrossRef] [PubMed]
- Gaur, V.K.; Gupta, S.; Sharma, P.; Gupta, P.; Varjani, S.; Srivastava, J.K.; Chang, J.-S.; Bui, X.-T. Metabolic Cascade for Remediation of Plastic Waste: A Case Study on Microplastic Degradation. Curr. Pollut. Rep. 2022, 8, 30–50. [Google Scholar] [CrossRef]
- Arpia, A.A.; Chen, W.-H.; Ubando, A.T.; Naqvi, S.R.; Culaba, A.B. Microplastic degradation as a sustainable concurrent approach for producing biofuel and obliterating hazardous environmental effects: A state-of-the-art review. J. Hazard. Mater. 2021, 418, 126381. [Google Scholar] [CrossRef]
- Miri, S.; Saini, R.; Davoodi, S.M.; Pulicharla, R.; Brar, S.K.; Magdouli, S. Biodegradation of microplastics: Better late than never. Chemosphere 2022, 286, 131670. [Google Scholar] [CrossRef]
- Zhou, Y.; Kumar, M.; Sarsaiya, S.; Sirohi, R.; Awasthi, S.K.; Sindhu, R.; Binod, P.; Pandey, A.; Bolan, N.S.; Zhang, Z.; et al. Challenges and opportunities in bioremediation of micro-nano plastics: A review. Sci. Total Environ. 2022, 802, 149823. [Google Scholar] [CrossRef]
- Yang, S.; Hai, F.I.; Nghiem, L.D.; Price, W.E.; Roddick, F.; Moreira, M.T.; Magram, S.F. Understanding the factors controlling the removal of trace organic contaminants by white-rot fungi and their lignin modifying enzymes: A critical review. Bioresour. Technol. 2013, 141, 97–108. [Google Scholar] [CrossRef]
- Park, G.W.; Gong, G.; Joo, J.C.; Song, J.; Lee, J.; Lee, J.-P.; Kim, H.T.; Ryu, M.H.; Sirohi, R.; Zhuang, X.; et al. Recent progress and challenges in biological degradation and biotechnological valorization of lignin as an emerging source of bioenergy: A state-of-the-art review. Renew. Sustain. Energy Rev. 2022, 157, 112025. [Google Scholar] [CrossRef]
- Antranikian, G.; Streit, W.R. Microorganisms harbor keys to a circular bioeconomy making them useful tools in fighting plastic pollution and rising CO2 levels. Extremophiles 2022, 26, 10. [Google Scholar] [CrossRef]
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019, 85, e01095-19. [Google Scholar] [CrossRef]
- Lee, S.; Lee, Y.R.; Kim, S.J.; Lee, J.-S.; Min, K. Recent advances and challenges in the biotechnological upcycling of plastic wastes for constructing a circular bioeconomy. Chem. Eng. J. 2022, 454, 140470. [Google Scholar] [CrossRef]
- Brott, S.; Pfaff, L.; Schuricht, J.; Schwarz, J.N.; Böttcher, D.; Badenhorst, C.P.; Wei, R.; Bornscheuer, U.T. Engineering and evaluation of thermostable IsPETase variants for PET degradation. Eng. Life Sci. 2022, 22, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Palm, G.J.; Reisky, L.; Böttcher, D.; Müller, H.; Michels, E.A.; Walczak, M.C.; Berndt, L.; Weiss, M.S.; Bornscheuer, U.T.; Weber, G.; et al. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nat. Commun. 2019, 10, 1717. [Google Scholar] [CrossRef]
- Tarazona, N.A.; Wei, R.; Brott, S.; Pfaff, L.; Bornscheuer, U.T.; Lendlein, A.; Machatschek, R. Rapid depolymerization of poly (ethylene terephthalate) thin films by a dual-enzyme system and its impact on material properties. Chem Catal. 2022, 2, 3573–3589. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.-L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Pfaff, L.; Gao, J.; Li, Z.; Jäckering, A.; Weber, G.; Mican, J.; Chen, Y.; Dong, W.; Han, X.; Feiler, C.G.; et al. Multiple substrate binding mode-guided engineering of a thermophilic PET hydrolase. ACS Catal. 2022, 12, 9790–9800. [Google Scholar] [CrossRef] [PubMed]
- Sonnendecker, C.; Oeser, J.; Richter, P.K.; Hille, P.; Zhao, Z.; Fischer, C.; Lippold, H.; Blázquez-Sánchez, P.; Engelberger, F.; Ramírez-Sarmiento, C.A.; et al. Low Carbon Footprint Recycling of Post-Consumer PET Plastic with a Metagenomic Polyester Hydrolase. ChemSusChem 2022, 15, e202101062. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Diaz, D.J.; Czarnecki, N.J.; Zhu, C.; Kim, W.; Shroff, R.; Acosta, D.J.; Alexander, B.R.; Cole, H.O.; Zhang, Y.; et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 2022, 604, 662–667. [Google Scholar] [CrossRef]
- Bell, E.L.; Smithson, R.; Kilbride, S.; Foster, J.; Hardy, F.J.; Ramachandran, S.; Tedstone, A.A.; Haigh, S.J.; Garforth, A.A.; Day, P.J.; et al. Directed evolution of an efficient and thermostable PET depolymerase. Nat. Catal. 2022, 5, 673–681. [Google Scholar] [CrossRef]
- Pirillo, V.; Orlando, M.; Tessaro, D.; Pollegioni, L.; Molla, G. An efficient protein evolution workflow for the improvement of bacterial PET hydrolyzing enzymes. Int. J. Mol. Sci. 2022, 23, 264. [Google Scholar] [CrossRef]
- Yamamoto, N.; Nakayama, A.; Oshima, M.; Kawasaki, N.; Aiba, S.-I. Enzymatic hydrolysis of lysine diisocyanate based polyurethanes and segmented polyurethane ureas by various proteases. React. Funct. Polym. 2007, 67, 1338–1345. [Google Scholar] [CrossRef]
- Peng, Y.-H.; Shih, Y.-H.; Lai, Y.-C.; Liu, Y.-Z.; Liu, Y.-T.; Lin, N.-C. Degradation of polyurethane by bacterium isolated from soil and assessment of polyurethanolytic activity of a Pseudomonas putida strain. Environ. Sci. Pollut. Res. 2014, 21, 9529–9537. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.R.; Huang, J.; Anand, P.; Kucera, K.; Sandoval, A.G.; Dantzler, K.W.; Hickman, D.; Jee, J.; Kimovec, F.M.; Koppstein, D.; et al. Biodegradation of polyester polyurethane by endophytic fungi. Appl. Environ. Microbiol. 2011, 77, 6076–6084. [Google Scholar] [CrossRef]
- Liu, J.; Liu, J.; Xu, B.; Xu, A.; Cao, S.; Wei, R.; Zhou, J.; Jiang, M.; Dong, W. Biodegradation of polyether-polyurethane foam in yellow mealworms (Tenebrio molitor) and effects on the gut microbiome. Chemosphere 2022, 304, 135263. [Google Scholar] [CrossRef] [PubMed]
- Magnin, A.; Pollet, E.; Perrin, R.; Ullmann, C.; Persillon, C.; Phalip, V.; Avérous, L. Enzymatic recycling of thermoplastic polyurethanes: Synergistic effect of an esterase and an amidase and recovery of building blocks. Waste Manag. 2019, 85, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Barragán, J.; Domínguez-Malfavón, L.; Vargas-Suárez, M.; González-Hernández, R.; Aguilar-Osorio, G.; Loza-Tavera, H. Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams. Appl. Environ. Microbiol. 2016, 82, 5225–5235. [Google Scholar] [CrossRef] [PubMed]
- Mathur, G.; Prasad, R. Degradation of polyurethane by Aspergillus flavus (ITCC 6051) isolated from soil. Appl. Biochem. Biotechnol. 2012, 167, 1595–1602. [Google Scholar] [CrossRef]
- Shah, Z.; Gulzar, M.; Hasan, F.; Shah, A.A. Degradation of polyester polyurethane by an indigenously developed consortium of Pseudomonas and Bacillus species isolated from soil. Polym. Degrad. Stab. 2016, 134, 349–356. [Google Scholar] [CrossRef]
- Matsumiya, Y.; Murata, N.; Tanabe, E.; Kubota, K.; Kubo, M. Isolation and characterization of an ether-type polyurethane-degrading micro-organism and analysis of degradation mechanism by Alternaria sp. J. Appl. Microbiol. 2010, 108, 1946–1953. [Google Scholar] [CrossRef]
- Bombelli, P.; Howe, C.J.; Bertocchini, F. Polyethylene bio-degradation by caterpillars of the wax moth Galleria mellonella. Curr. Biol. 2017, 27, R292–R293. [Google Scholar] [CrossRef]
- Barrionuevo, J.M.R.; Vilanova-Cuevas, B.; Alvarez, A.; Martín, E.; Malizia, A.; Galindo-Cardona, A.; de Cristóbal, R.E.; Occhionero, M.A.; Chalup, A.; Monmany-Garzia, A.C.; et al. The bacterial and fungal gut microbiota of the greater wax moth, Galleria mellonella L. consuming polyethylene and polystyrene. Front. Microbiol. 2022, 13, 918861. [Google Scholar] [CrossRef]
- Sanluis-Verdes, A.; Colomer-Vidal, P.; Rodriguez-Ventura, F.; Bello-Villarino, M.; Spinola-Amilibia, M.; Ruiz-Lopez, E.; Illanes-Vicioso, R.; Castroviejo, P.; Cigliano, R.A.; Montoya, M.; et al. Wax worm saliva and the enzymes therein are the key to polyethylene degradation by Galleria mellonella. Nat. Commun. 2022, 13, 5568. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, M.; Girdhar, A.; Tiwari, A.; Nayarisseri, A. Implications of a novel Pseudomonas species on low density polyethylene biodegradation: An in vitro to in silico approach. SpringerPlus 2014, 3, 497. [Google Scholar] [CrossRef] [PubMed]
- Montazer, Z.; Habibi Najafi, M.B.; Levin, D.B. Microbial degradation of low-density polyethylene and synthesis of polyhydroxyalkanoate polymers. Can. J. Microbiol. 2019, 65, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Brandon, A.M.; Gao, S.-H.; Tian, R.; Ning, D.; Yang, S.-S.; Zhou, J.; Wu, W.-M.; Criddle, C.S. Biodegradation of polyethylene and plastic mixtures in mealworms (larvae of Tenebrio molitor) and effects on the gut microbiome. Environ. Sci. Technol. 2018, 52, 6526–6533. [Google Scholar] [CrossRef]
- Sarmah, P.; Rout, J. Efficient biodegradation of low-density polyethylene by cyanobacteria isolated from submerged polyethylene surface in domestic sewage water. Environ. Sci. Pollut. Res. 2018, 25, 33508–33520. [Google Scholar] [CrossRef]
- Ward, P.G.; Goff, M.; Donner, M.; Kaminsky, W.; O’Connor, K.E. A two step chemo-biotechnological conversion of polystyrene to a biodegradable thermoplastic. Environ. Sci. Technol. 2006, 40, 2433–2437. [Google Scholar] [CrossRef]
- Mohan, A.J.; Sekhar, V.C.; Bhaskar, T.; Nampoothiri, K.M. Microbial assisted high impact polystyrene (HIPS) degradation. Bioresour. Technol. 2016, 213, 204–207. [Google Scholar] [CrossRef]
- Sekhar, V.C.; Nampoothiri, K.M.; Mohan, A.J.; Nair, N.R.; Bhaskar, T.; Pandey, A. Microbial degradation of high impact polystyrene (HIPS), an e-plastic with decabromodiphenyl oxide and antimony trioxide. J. Hazard. Mater. 2016, 318, 347–354. [Google Scholar] [CrossRef]
- Auta, H.; Emenike, C.; Fauziah, S. Screening of Bacillus strains isolated from mangrove ecosystems in Peninsular Malaysia for microplastic degradation. Environ. Pollut. 2017, 231, 1552–1559. [Google Scholar] [CrossRef]
- Cacciari, I.; Quatrini, P.; Zirletta, G.; Mincione, E.; Vinciguerra, V.; Lupattelli, P.; Giovannozzi Sermanni, G. Isotactic polypropylene biodegradation by a microbial community: Physicochemical characterization of metabolites produced. Appl. Environ. Microbiol. 1993, 59, 3695–3700. [Google Scholar] [CrossRef]
- Müller, R.J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.D. Enzymatic degradation of poly (ethylene terephthalate): Rapid hydrolyse using a hydrolase from T. fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405. [Google Scholar] [CrossRef]
- Webb, H.K.; Arnott, J.; Crawford, R.J.; Ivanova, E.P. Plastic degradation and its environmental implications with special reference to poly (ethylene terephthalate). Polymers 2012, 5, 1–18. [Google Scholar] [CrossRef]
- Oh, C.; Kim, T.D.; Kim, K.K. Carboxylic ester hydrolases in bacteria: Active site, structure, function and application. Crystals 2019, 9, 597. [Google Scholar] [CrossRef]
- Danso, D.; Schmeisser, C.; Chow, J.; Zimmermann, W.; Wei, R.; Leggewie, C.; Li, X.; Hazen, T.; Streit, W.R. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl. Environ. Microbiol. 2018, 84, e02773-17. [Google Scholar] [CrossRef] [PubMed]
- Carr, C.M.; Clarke, D.J.; Dobson, A.D. Microbial polyethylene terephthalate hydrolases: Current and future perspectives. Front. Microbiol. 2020, 11, 2825. [Google Scholar] [CrossRef]
- Kawai, F. The current state of research on PET hydrolyzing enzymes available for biorecycling. Catalysts 2021, 11, 206. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K.; et al. A bacterium that degrades and assimilates poly (ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Taniguchi, I.; Oda, K. Ideonella sakaiensis, PETase, and MHETase: From identification of microbial PET degradation to enzyme characterization. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 648, pp. 187–205. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, J.; Jiang, L. Comment on “A bacterium that degrades and assimilates poly (ethylene terephthalate)”. Science 2016, 353, 759. [Google Scholar] [CrossRef]
- Wallace, N.E.; Adams, M.C.; Chafin, A.C.; Jones, D.D.; Tsui, C.L.; Gruber, T.D. The highly crystalline PET found in plastic water bottles does not support the growth of the PETase-producing bacterium Ideonella sakaiensis. Environ. Microbiol. Rep. 2020, 12, 578–582. [Google Scholar] [CrossRef]
- Falkenstein, P.; Wei, R.; Matysik, J.; Song, C. Mechanistic investigation of enzymatic degradation of polyethylene terephthalate by nuclear magnetic resonance. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 648, pp. 231–252. [Google Scholar] [CrossRef]
- Brizendine, R.K.; Erickson, E.; Haugen, S.J.; Ramirez, K.J.; Miscall, J.; Salvachúa, D.; Pickford, A.R.; Sobkowicz, M.J.; McGeehan, J.E.; Beckham, G.T.; et al. Particle size reduction of poly (ethylene terephthalate) increases the rate of enzymatic depolymerization but does not increase the overall conversion extent. ACS Sustain. Chem. Eng. 2022, 10, 9131–9140. [Google Scholar] [CrossRef]
- Thomsen, T.B.; Hunt, C.J.; Meyer, A.S. Influence of substrate crystallinity and glass transition temperature on enzymatic degradation of polyethylene terephthalate (PET). New Biotechnol. 2022, 69, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Son, H.F.; Joo, S.; Seo, H.; Sagong, H.-Y.; Lee, S.H.; Hong, H.; Kim, K.-J. Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzym. Microb. Technol. 2020, 141, 109656. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, Y.; Liu, X.; Dong, S.; Tian, Y.E.; Qiao, Y.; Mitra, R.; Han, J.; Li, C.; Han, X. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy. ACS Catal. 2021, 11, 1340–1350. [Google Scholar] [CrossRef]
- Meyer-Cifuentes, I.E.; Werner, J.; Jehmlich, N.; Will, S.E.; Neumann-Schaal, M.; Öztürk, B. Synergistic biodegradation of aromatic-aliphatic copolyester plastic by a marine microbial consortium. Nat. Commun. 2020, 11, 5790. [Google Scholar] [CrossRef]
- Xi, X.; Ni, K.; Hao, H.; Shang, Y.; Zhao, B.; Qian, Z. Secretory expression in Bacillus subtilis and biochemical characterization of a highly thermostable polyethylene terephthalate hydrolase from bacterium HR29. Enzym. Microb. Technol. 2021, 143, 109715. [Google Scholar] [CrossRef] [PubMed]
- Eiamthong, B.; Meesawat, P.; Wongsatit, T.; Jitdee, J.; Sangsri, R.; Patchsung, M.; Aphicho, K.; Suraritdechachai, S.; Huguenin-Dezot, N.; Tang, S.; et al. Discovery and genetic code expansion of a polyethylene terephthalate (PET) hydrolase from the human saliva metagenome for the degradation and bio-functionalization of PET. Angew. Chem. 2022, 61, e202203061. [Google Scholar] [CrossRef]
- Sulaiman, S.; Yamato, S.; Kanaya, E.; Kim, J.-J.; Koga, Y.; Takano, K.; Kanaya, S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 2012, 78, 1556–1562. [Google Scholar] [CrossRef]
- Carbios—Enzymes Powering the Circular Economy. Available online: https://www.carbios.com/wp-content/uploads/2022/01/carbios_presentation-january-2022.pdf (accessed on 20 October 2022).
- García, J.L. Enzymatic recycling of polyethylene terephthalate through the lens of proprietary processes. Microb. Biotechnol. 2022, 15, 2699–2704. [Google Scholar] [CrossRef]
- Singh, A.; Rorrer, N.A.; Nicholson, S.R.; Erickson, E.; DesVeaux, J.S.; Avelino, A.F.; Lamers, P.; Bhatt, A.; Zhang, Y.; Avery, G.; et al. Techno-economic, life-cycle, and socioeconomic impact analysis of enzymatic recycling of poly (ethylene terephthalate). Joule 2021, 5, 2479–2503. [Google Scholar] [CrossRef]
- Pasula, R.R.; Lim, S.; Ghadessy, F.J.; Sana, B. The influences of substrates’ physical properties on enzymatic PET hydrolysis: Implications for PET hydrolase engineering. Eng. Biol. 2022, 6, 17–22. [Google Scholar] [CrossRef]
- Wei, R.; von Haugwitz, G.; Pfaff, L.; Mican, J.; Badenhorst, C.P.; Liu, W.; Weber, G.; Austin, H.P.; Bednar, D.; Damborsky, J.; et al. Mechanism-based design of efficient PET hydrolases. ACS Catal. 2022, 12, 3382–3396. [Google Scholar] [CrossRef] [PubMed]
- Erickson, E.; Shakespeare, T.J.; Bratti, F.; Buss, B.L.; Graham, R.; Hawkins, M.A.; König, G.; Michener, W.E.; Miscall, J.; Ramirez, K.J.; et al. Comparative performance of PETase as a function of reaction conditions, substrate properties, and product accumulation. ChemSusChem 2022, 15, e202101932. [Google Scholar] [CrossRef] [PubMed]
- Pirillo, V.; Orlando, M.; Battaglia, C.; Pollegioni, L.; Molla, G. Efficient polyethylene terephthalate degradation at moderate temperature: A protein engineering study of LC-cutinase highlights the key role of residue 243. FEBS J. 2023, accepted. [Google Scholar] [CrossRef] [PubMed]
- Bååth, J.A.; Jensen, K.; Borch, K.; Westh, P.; Kari, J. Sabatier principle for rationalizing enzymatic hydrolysis of a synthetic polyester. JACS Au 2022, 2, 1223–1231. [Google Scholar] [CrossRef]
- Schubert, S.W.; Schaller, K.; Bååth, J.A.; Hunt, C.; Borch, K.; Jensen, K.; Brask, J.; Westh, P. Reaction pathways for the enzymatic degradation of poly (ethylene terephthalate): What characterizes an efficient PET-hydrolase? ChemBioChem 2022, 24, e202200516. [Google Scholar] [CrossRef]
- Erickson, E.; Gado, J.E.; Avilán, L.; Bratti, F.; Brizendine, R.K.; Cox, P.A.; Gill, R.; Graham, R.; Kim, D.-J.; König, G.; et al. Sourcing thermotolerant poly (ethylene terephthalate) hydrolase scaffolds from natural diversity. Nat. Commun. 2022, 13, 7850. [Google Scholar] [CrossRef]
- Quartinello, F.; Vajnhandl, S.; Volmajer Valh, J.; Farmer, T.J.; Vončina, B.; Lobnik, A.; Herrero Acero, E.; Pellis, A.; Guebitz, G.M. Synergistic chemo-enzymatic hydrolysis of poly (ethylene terephthalate) from textile waste. Microb. Biotechnol. 2017, 10, 1376–1383. [Google Scholar] [CrossRef]
- Statista. Price of Polyethylene Terephthalate (PET) Worldwide from 2017 to 2020 with Estimated Figures for 2021 to 2022. 2022. Available online: https://www.statista.com/statistics/1171088/price-polyethylene-terephthalate-forecast-globally/ (accessed on 30 May 2022).
- Uekert, T.; DesVeaux, J.S.; Singh, A.; Nicholson, S.R.; Lamers, P.; Ghosh, T.; McGeehan, J.E.; Carpenter, A.C.; Beckham, G.T. Life cycle assessment of enzymatic poly (ethylene terephthalate) recycling. Green Chem. 2022, 24, 6531–6543. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, J.K.; Cha, H.G.; Kang, M.J.; Lee, H.S.; Khang, T.U.; Yun, E.J.; Lee, D.-H.; Song, B.K.; Park, S.J.; et al. Biological valorization of poly (ethylene terephthalate) monomers for upcycling waste PET. ACS Sustain. Chem. Eng. 2019, 7, 19396–19406. [Google Scholar] [CrossRef]
- Kim, H.T.; Hee Ryu, M.; Jung, Y.J.; Lim, S.; Song, H.M.; Park, J.; Hwang, S.Y.; Lee, H.S.; Yeon, Y.J.; Sung, B.H.; et al. Chemo-Biological Upcycling of Poly (ethylene terephthalate) to Multifunctional Coating Materials. ChemSusChem 2021, 14, 4251–4259. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.J.; Kim, H.T.; Lee, M.-W.; Kim, K.-A.; Khang, T.U.; Song, H.M.; Park, S.J.; Joo, J.C.; Cha, H.G. A chemo-microbial hybrid process for the production of 2-pyrone-4, 6-dicarboxylic acid as a promising bioplastic monomer from PET waste. Green Chem. 2020, 22, 3461–3469. [Google Scholar] [CrossRef]
- Werner, A.Z.; Clare, R.; Mand, T.D.; Pardo, I.; Ramirez, K.J.; Haugen, S.J.; Bratti, F.; Dexter, G.N.; Elmore, J.R.; Huenemann, J.D.; et al. Tandem chemical deconstruction and biological upcycling of poly (ethylene terephthalate) to β-ketoadipic acid by Pseudomonas putida KT2440. Metab. Eng. 2021, 67, 250–261. [Google Scholar] [CrossRef]
- Tiso, T.; Narancic, T.; Wei, R.; Pollet, E.; Beagan, N.; Schröder, K.; Honak, A.; Jiang, M.; Kenny, S.T.; Wierckx, N.; et al. Towards bio-upcycling of polyethylene terephthalate. Metab. Eng. 2021, 66, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Sadler, J.C.; Wallace, S. Microbial synthesis of vanillin from waste poly (ethylene terephthalate). Green Chem. 2021, 23, 4665–4672. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhang, T.; Zheng, Y.; Li, Q.; Su, T.; Qi, Q. Potential one-step strategy for PET degradation and PHB biosynthesis through co-cultivation of two engineered microorganisms. Eng. Microbiol. 2021, 1, 100003. [Google Scholar] [CrossRef]
- Liu, P.; Zheng, Y.; Yuan, Y.; Zhang, T.; Li, Q.; Liang, Q.; Su, T.; Qi, Q. Valorization of Polyethylene Terephthalate to Muconic Acid by Engineering Pseudomonas Putida. Int. J. Mol. Sci. 2022, 23, 10997. [Google Scholar] [CrossRef]
- Mückschel, B.; Simon, O.; Klebensberger, J.; Graf, N.; Rosche, B.; Altenbuchner, J.; Pfannstiel, J.; Huber, A.; Hauer, B. Ethylene glycol metabolism by Pseudomonas putida. Appl. Environ. Microbiol. 2012, 78, 8531–8539. [Google Scholar] [CrossRef]
- Utomo, R.N.C.; Li, W.-J.; Tiso, T.; Eberlein, C.; Doeker, M.; Heipieper, H.J.; Jupke, A.; Wierckx, N.; Blank, L.M. Defined microbial mixed culture for utilization of polyurethane monomers. ACS Sustain. Chem. Eng. 2020, 8, 17466–17474. [Google Scholar] [CrossRef]
- Son, H.F.; Cho, I.J.; Joo, S.; Seo, H.; Sagong, H.-Y.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for highly efficient PET degradation. ACS Catal. 2019, 9, 3519–3526. [Google Scholar] [CrossRef]
- Gautam, R.; Bassi, A.; Yanful, E. A review of biodegradation of synthetic plastic and foams. Appl. Biochem. Biotechnol. 2007, 141, 85–108. [Google Scholar] [CrossRef] [PubMed]
- Magnin, A.; Pollet, E.; Phalip, V.; Avérous, L. Evaluation of biological degradation of polyurethanes. Biotechnol. Adv. 2020, 39, 107457. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Dong, J.; Guo, X.; Ding, M.; Bao, R.; Luo, Y. Current advances in polyurethane biodegradation. Polym. Int. 2022, 71, 1384–1392. [Google Scholar] [CrossRef]
- Gamerith, C.; Acero, E.H.; Pellis, A.; Ortner, A.; Vielnascher, R.; Luschnig, D.; Zartl, B.; Haernvall, K.; Zitzenbacher, S.; Strohmeier, G.; et al. Improving enzymatic polyurethane hydrolysis by tuning enzyme sorption. Polym. Degrad. Stab. 2016, 132, 69–77. [Google Scholar] [CrossRef]
- Magnin, A.; Entzmann, L.; Pollet, E.; Avérous, L. Breakthrough in polyurethane bio-recycling: An efficient laccase-mediated system for the degradation of different types of polyurethanes. Waste Manag. 2021, 132, 23–30. [Google Scholar] [CrossRef]
- Branson, Y.; Söltl, S.; Buchmann, C.; Wei, R.; Schaffert, L.; Badenhorst, C.P.; Reisky, L.; Jäger, G.; Bornscheuer, U. Urethanases for the enzymatic hydrolysis of low molecular weight carbamates and the recycling of polyurethanes. Angew. Chem. 2023, e202216220. [Google Scholar] [CrossRef]
- Mi, H.-Y.; Jing, X.; Napiwocki, B.N.; Hagerty, B.S.; Chen, G.; Turng, L.-S. Biocompatible, degradable thermoplastic polyurethane based on polycaprolactone-block-polytetrahydrofuran-block-polycaprolactone copolymers for soft tissue engineering. J. Mater. Chem. B 2017, 5, 4137–4151. [Google Scholar] [CrossRef]
- Jeong, J.; Oh, D.; Goh, M. Synthesis, antibacterial activity, and enzymatic decomposition of bio-polyurethane foams containing propolis. J. Ind. Eng. Chem. 2022, 109, 182–188. [Google Scholar] [CrossRef]
- Liu, J.; He, J.; Xue, R.; Xu, B.; Qian, X.; Xin, F.; Blank, L.M.; Zhou, J.; Wei, R.; Dong, W.; et al. Biodegradation and up-cycling of polyurethanes: Progress, challenges, and prospects. Biotechnol. Adv. 2021, 48, 107730. [Google Scholar] [CrossRef]
- Liu, J.; Zeng, Q.; Lei, H.; Xin, K.; Xu, A.; Wei, R.; Li, D.; Zhou, J.; Dong, W.; Jiang, M.; et al. Biodegradation of polyester polyurethane by Cladosporium sp. P7: Evaluating its degradation capacity and metabolic pathways. J. Hazard. Mater. 2023, 448, 130776. [Google Scholar] [CrossRef]
- Chen, L.; Rong, M.; Yang, L.; Yu, J.; Qu, H.; Meng, Q.; Ni, S.; Xu, Z.; Zhu, X.; Wang, L.; et al. Construction of super-hydrophobic hypercrosslinked porous polymers for selectively removing aromatic diamines from the polyurethane bio-hydrolysate. Chem. Eng. J. 2022, 428, 132509. [Google Scholar] [CrossRef]
- Kotova, I.; Taktarova, Y.V.; Tsavkelova, E.; Egorova, M.; Bubnov, I.; Malakhova, D.; Shirinkina, L.; Sokolova, T.; Bonch-Osmolovskaya, E. Microbial Degradation of Plastics and Approaches to Make it More Efficient. Microbiology 2021, 90, 671–701. [Google Scholar] [CrossRef]
- Palmer, R.J.; Staff, U.B. Polyamides, plastics. In Kirk-Othmer Encyclopedia of Chemical Technology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Negoro, S. Biodegradation of nylon oligomers. Appl. Microbiol. Biotechnol. 2000, 54, 461–466. [Google Scholar] [CrossRef]
- Biundo, A.; Subagia, R.; Maurer, M.; Ribitsch, D.; Syrén, P.-O.; Guebitz, G.M. Switched reaction specificity in polyesterases towards amide bond hydrolysis by enzyme engineering. RSC Adv. 2019, 9, 36217–36226. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, T.; Kitaoka, Y.; Kakezawa, M.; Nishida, T. Purification and characterization of a nylon-degrading enzyme. Appl. Environ. Microbiol. 1998, 64, 1366–1371. [Google Scholar] [CrossRef] [PubMed]
- Santo, M.; Weitsman, R.; Sivan, A. The role of the copper-binding enzyme-laccase—In the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeterior. Biodegrad. 2013, 84, 204–210. [Google Scholar] [CrossRef]
- Zhang, J.; Pedersen, N.J.; Eser, B.E.; Guo, Z. Biodegradation of polyethylene and polystyrene: From microbial deterioration to enzyme discovery. Biotechnol. Adv. 2022, 60, 107991. [Google Scholar] [CrossRef]
- Mohanan, N.; Montazer, Z.; Sharma, P.K.; Levin, D.B. Microbial and enzymatic degradation of synthetic plastics. Front. Microbiol. 2020, 11, 580709. [Google Scholar] [CrossRef]
- Eyheraguibel, B.; Traikia, M.; Fontanella, S.; Sancelme, M.; Bonhomme, S.; Fromageot, D.; Lemaire, J.; Lauranson, G.; Lacoste, J.; Delort, A.; et al. Characterization of oxidized oligomers from polyethylene films by mass spectrometry and NMR spectroscopy before and after biodegradation by a Rhodococcus rhodochrous strain. Chemosphere 2017, 184, 366–374. [Google Scholar] [CrossRef]
- Sudhakar, M.; Doble, M.; Murthy, P.S.; Venkatesan, R. Marine microbe-mediated biodegradation of low-and high-density polyethylenes. Int. Biodeterior. Biodegrad. 2008, 61, 203–213. [Google Scholar] [CrossRef]
- Jeon, H.J.; Kim, M.N. Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int. Biodeterior. Biodegrad. 2015, 103, 141–146. [Google Scholar] [CrossRef]
- Yang, S.-S.; Wu, W.-M.; Brandon, A.M.; Fan, H.-Q.; Receveur, J.P.; Li, Y.; Wang, Z.-Y.; Fan, R.; McClellan, R.L.; Gao, S.-H.; et al. Ubiquity of polystyrene digestion and biodegradation within yellow mealworms, larvae of Tenebrio molitor Linnaeus (Coleoptera: Tenebrionidae). Chemosphere 2018, 212, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Harper, C.A. Handbook of Plastics, Elastomers, and Composites; McGraw-Hill: New York, NY, USA, 2002; Volume 4. [Google Scholar]
- Jain, K.; Bhunia, H.; Sudhakara Reddy, M. Degradation of polypropylene–poly-L-lactide blend by bacteria isolated from compost. Bioremediation J. 2018, 22, 73–90. [Google Scholar] [CrossRef]
- Braun, D. Poly (vinyl chloride) on the way from the 19th century to the 21st century. J. Polym. Sci. Part A Polym. Chem. 2004, 42, 578–586. [Google Scholar] [CrossRef]
- Zhang, Z.; Peng, H.; Yang, D.; Zhang, G.; Zhang, J.; Ju, F. Polyvinyl chloride degradation by a bacterium isolated from the gut of insect larvae. Nat. Commun. 2022, 13, 5360. [Google Scholar] [CrossRef]
- Shah, M.M.; Barr, D.P.; Chung, N.; Aust, S.D. Use of white rot fungi in the degradation of environmental chemicals. Toxicol. Lett. 1992, 64, 493–501. [Google Scholar] [CrossRef]
- Sullivan, K.P.; Werner, A.Z.; Ramirez, K.J.; Ellis, L.D.; Bussard, J.R.; Black, B.A.; Brandner, D.G.; Bratti, F.; Buss, B.L.; Dong, X.; et al. Mixed plastics waste valorization through tandem chemical oxidation and biological funneling. Science 2022, 378, 207–211. [Google Scholar] [CrossRef]
- Guzik, M.W.; Nitkiewicz, T.; Wojnarowska, M.; Sołtysik, M.; Kenny, S.T.; Babu, R.P.; Best, M.; O’Connor, K.E. Robust process for high yield conversion of non-degradable polyethylene to a biodegradable plastic using a chemo-biotechnological approach. Waste Manag. 2021, 135, 60–69. [Google Scholar] [CrossRef]
- Tzouvelekis, L.; Tzelepi, E.; Tassios, P.; Legakis, N. CTX-M-type β-lactamases: An emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 2000, 14, 137–142. [Google Scholar] [CrossRef]
- Wang, W.; Themelis, N.J.; Sun, K.; Bourtsalas, A.C.; Huang, Q.; Zhang, Y.; Wu, Z. Current influence of China’s ban on plastic waste imports. Waste Dispos. Sustain. Energy 2019, 1, 67–78. [Google Scholar] [CrossRef]
- Gruber, F.; Grählert, W.; Wollmann, P.; Kaskel, S. Classification of black plastics waste using fluorescence imaging and machine learning. Recycling 2019, 4, 40. [Google Scholar] [CrossRef]
- Erkinay Ozdemir, M.; Ali, Z.; Subeshan, B.; Asmatulu, E. Applying machine learning approach in recycling. J. Mater. Cycles Waste Manag. 2021, 23, 855–871. [Google Scholar] [CrossRef]
- Platonova, E.; Chechenov, I.; Pavlov, A.; Solodilov, V.; Afanasyev, E.; Shapagin, A.; Polezhaev, A. Thermally Remendable Polyurethane Network Cross-Linked via Reversible Diels-Alder Reaction. Polymers 2021, 13, 1935. [Google Scholar] [CrossRef] [PubMed]
- Zahedifar, P.; Pazdur, L.; Vande Velde, C.M.; Billen, P. Multistage chemical recycling of polyurethanes and dicarbamates: A glycolysis–hydrolysis demonstration. Sustainability 2021, 13, 3583. [Google Scholar] [CrossRef]
- Sivan, A. New perspectives in plastic biodegradation. Curr. Opin. Biotechnol. 2011, 22, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Carmen, S. Microbial capability for the degradation of chemical additives present in petroleum-based plastic products: A review on current status and perspectives. J. Hazard. Mater. 2021, 402, 123534. [Google Scholar] [CrossRef]
- Chu, M.; Liu, Y.; Lou, X.; Zhang, Q.; Chen, J. Rational Design of Chemical Catalysis for Plastic Recycling. ACS Catal. 2022, 12, 4659–4679. [Google Scholar] [CrossRef]
- He, J.; Han, L.; Wang, F.; Ma, C.; Cai, Y.; Ma, W.; Xu, E.G.; Xing, B.; Yang, Z. Photocatalytic strategy to mitigate microplastic pollution in aquatic environments: Promising catalysts, efficiencies, mechanisms, and ecological risks. Crit. Rev. Environ. Sci. Technol. 2022, 53, 504–526. [Google Scholar] [CrossRef]
- Jehanno, C.; Alty, J.W.; Roosen, M.; de Meester, S.; Dove, A.P.; Chen, E.Y.-X.; Leibfarth, F.A.; Sardon, H. Critical advances and future opportunities in upcycling commodity polymers. Nature 2022, 603, 803–814. [Google Scholar] [CrossRef]
- Tiso, T.; Winter, B.; Wei, R.; Hee, J.; de Witt, J.; Wierckx, N.; Quicker, P.; Bornscheuer, U.T.; Bardow, A.; Nogales, J.; et al. The metabolic potential of plastics as biotechnological carbon sources—Review and targets for the future. Metab. Eng. 2022, 71, 77–98. [Google Scholar] [CrossRef]
- Rosenboom, J.-G.; Langer, R.; Traverso, G. Bioplastics for a circular economy. Nat. Rev. Mater. 2022, 7, 117–137. [Google Scholar] [CrossRef] [PubMed]
- Vert, M.; Doi, Y.; Hellwich, K.-H.; Hess, M.; Hodge, P.; Kubisa, P.; Rinaudo, M.; Schué, F. Terminology for biorelated polymers and applications (IUPAC Recommendations 2012). Pure Appl. Chem. 2012, 84, 377–410. [Google Scholar] [CrossRef]
- Shaw, N.; Monahan, F.; O’Riordan, E.; O’Sullivan, M. Physical properties of WPI films plasticized with glycerol, xylitol, or sorbitol. J. Food Sci. 2002, 67, 164–167. [Google Scholar] [CrossRef]
- Sessa, D.J.; Selling, G.W.; Willett, J.; Palmquist, D.E. Viscosity control of zein processing with sodium dodecyl sulfate. Ind. Crops Prod. 2006, 23, 15–22. [Google Scholar] [CrossRef]
- Wihodo, M.; Moraru, C.I. Effect of Pulsed Light treatment on the functional properties of casein films. LWT Food Sci. Technol. 2015, 64, 837–844. [Google Scholar] [CrossRef]
- Nuvoli, D.; Montevecchi, G.; Lovato, F.; Masino, F.; van der Borght, M.; Messori, M.; Antonelli, A. Protein films from black soldier fly (Hermetia illucens, Diptera: Stratiomyidae) prepupae: Effect of protein solubility and mild crosslinking. J. Sci. Food Agric. 2021, 101, 4506–4513. [Google Scholar] [CrossRef]
- Li, H.; Kognou, A.L.M.; Jiang, Z.H.; Qin, W.; Xu, C.C. Production of bio-polyurethane (BPU) foams from greenhouse/agricultural wastes, and their biodegradability. Biofuels Bioprod. Biorefin. 2022, 16, 826–837. [Google Scholar] [CrossRef]
- Paul, U.C.; Bayer, G.; Grasselli, S.; Malchiodi, A.; Bayer, I.S. Biodegradable, stretchable and transparent plastic films from modified waterborne polyurethane dispersions. Polymers 2022, 14, 1199. [Google Scholar] [CrossRef]
- Pedersen, D.D.; Kim, S.; Wagner, W.R. Biodegradable polyurethane scaffolds in regenerative medicine: Clinical translation review. J. Biomed. Mater. Res. Part A 2022, 110, 1460–1487. [Google Scholar] [CrossRef]
- Lopes, R.V.; Loureiro, N.P.; Quirino, R.L.; Gomes, A.C.M.; Pezzin, A.P.T.; Manzur, L.P.; dos Santos, M.L.; Sales, M.J. Biodegradation Study of Polyurethanes from Linseed and Passion Fruit Oils. Coatings 2022, 12, 617. [Google Scholar] [CrossRef]
- Di Bartolo, A.; Infurna, G.; Dintcheva, N.T. A review of bioplastics and their adoption in the circular economy. Polymers 2021, 13, 1229. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Gao, Z.; Wang, Z.; Su, T.; Yang, L.; Li, P. Enzymatic degradation of poly (butylene succinate) by cutinase cloned from Fusarium solani. Polym. Degrad. Stab. 2016, 134, 211–219. [Google Scholar] [CrossRef]
- Wallace, P.W.; Haernvall, K.; Ribitsch, D.; Zitzenbacher, S.; Schittmayer, M.; Steinkellner, G.; Gruber, K.; Guebitz, G.M.; Birner-Gruenberger, R. PpEst is a novel PBAT degrading polyesterase identified by proteomic screening of Pseudomonas pseudoalcaligenes. Appl. Microbiol. Biotechnol. 2017, 101, 2291–2303. [Google Scholar] [CrossRef] [PubMed]
- Austin, H.P.; Allen, M.D.; Donohoe, B.S.; Rorrer, N.A.; Kearns, F.L.; Silveira, R.L.; Pollard, B.C.; Dominick, G.; Duman, R.; El Omari, K.; et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA 2018, 115, E4350–E4357. [Google Scholar] [CrossRef] [PubMed]
- Naser, A.Z.; Deiab, I.; Darras, B.M. Poly (lactic acid)(PLA) and polyhydroxyalkanoates (PHAs), green alternatives to petroleum-based plastics: A review. RSC Adv. 2021, 11, 17151–17196. [Google Scholar] [CrossRef]
- Gricajeva, A.; Nadda, A.K.; Gudiukaite, R. Insights into polyester plastic biodegradation by carboxyl ester hydrolases. J. Chem. Technol. Biotechnol. 2022, 97, 359–380. [Google Scholar] [CrossRef]
- Zhao, X.; Cornish, K.; Vodovotz, Y. Narrowing the gap for bioplastic use in food packaging: An update. Environ. Sci. Technol. 2020, 54, 4712–4732. [Google Scholar] [CrossRef]
- European Bioplastics. Bioplastic Facts and Figures. 2022. Available online: https://www.docs.european-bioplastics.org/publications/EUBP_Facts_and_figures.pdf (accessed on 20 October 2022).
- Zrimec, J.; Kokina, M.; Jonasson, S.; Zorrilla, F.; Zelezniak, A. Plastic-degrading potential across the global microbiome correlates with recent pollution trends. mBio 2021, 12, e02155-21. [Google Scholar] [CrossRef]
- Jiménez, D.J.; Öztürk, B.; Wei, R.; Bugg, T.D.; Amaya Gomez, C.V.; Salcedo Galan, F.; Castro-Mayorga, J.L.; Saldarriaga, J.F.; Tarazona, N.A. Merging plastics, microbes, and enzymes: Highlights from an international workshop. Appl. Environ. Microbiol. 2022, 88, e00721–e00722. [Google Scholar] [CrossRef]
- Kraemer, S.A.; Boynton, P.J. Evidence for microbial local adaptation in nature. Mol. Ecol. 2017, 26, 1860–1876. [Google Scholar] [CrossRef]
- Edwards, S.; León-Zayas, R.; Ditter, R.; Laster, H.; Sheehan, G.; Anderson, O.; Beattie, T.; Mellies, J.L. Microbial Consortia and Mixed Plastic Waste: Pangenomic Analysis Reveals Potential for Degradation of Multiple Plastic Types via Previously Identified PET Degrading Bacteria. Int. J. Mol. Sci. 2022, 23, 5612. [Google Scholar] [CrossRef] [PubMed]
- Bååth, J.A.; Borch, K.; Westh, P. A suspension-based assay and comparative detection methods for characterization of polyethylene terephthalate hydrolases. Anal. Biochem. 2020, 607, 113873. [Google Scholar] [CrossRef] [PubMed]
- Frank, R.; Krinke, D.; Sonnendecker, C.; Zimmermann, W.; Jahnke, H.-G. Real-Time Noninvasive Analysis of Biocatalytic PET Degradation. ACS Catal. 2021, 12, 25–35. [Google Scholar] [CrossRef]
- Qiao, Y.; Hu, R.; Chen, D.; Wang, L.; Wang, Z.; Yu, H.; Fu, Y.; Li, C.; Dong, Z.; Weng, Y.-X.; et al. Fluorescence-activated droplet sorting of PET degrading microorganisms. J. Hazard. Mater. 2022, 424, 127417. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; Liu, J.; Cao, S.; Xu, B.; Guo, C.; Yu, Z.; Chen, X.; Zhou, J.; Dong, W.; Jiang, M.; et al. Application of a novel fluorogenic polyurethane analogue probe in polyester-degrading microorganisms screening by microfluidic droplet. Microb. Biotechnol. 2022, 16, 474–480. [Google Scholar] [CrossRef]
- Kim, D.-W.; Ahn, J.-H.; Cha, C.-J. Biodegradation of plastics: Mining of plastic-degrading microorganisms and enzymes using metagenomics approaches. J. Microbiol. 2022, 60, 969–976. [Google Scholar] [CrossRef]
- National Library of Medicine. GenBank. 2021. Available online: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 14 October 2022).
- EBI. MGnify. 2022. Available online: https://www.ebi.ac.uk/metagenomics/ (accessed on 14 October 2022).
- Vasina, M.; Vanacek, P.; Hon, J.; Kovar, D.; Faldynova, H.; Kunka, A.; Buryska, T.; Badenhorst, C.P.; Mazurenko, S.; Bednar, D.; et al. Advanced database mining of efficient haloalkane dehalogenases by sequence and structure bioinformatics and microfluidics. Chem. Catal. 2022, 2, 2704–2725. [Google Scholar] [CrossRef]
- Gan, Z.; Zhang, H. PMBD: A comprehensive plastics microbial biodegradation database. Database 2019, 2019, baz119. [Google Scholar] [CrossRef]
- Gambarini, V.; Pantos, O.; Kingsbury, J.M.; Weaver, L.; Handley, K.M.; Lear, G. PlasticDB: A database of microorganisms and proteins linked to plastic biodegradation. Database 2022, 2022, baac008. [Google Scholar] [CrossRef]
- Buchholz, P.C.; Feuerriegel, G.; Zhang, H.; Perez-Garcia, P.; Nover, L.L.; Chow, J.; Streit, W.R.; Pleiss, J. Plastics degradation by hydrolytic enzymes: The plastics-active enzymes database—PAZy. Proteins Struct. Funct. Bioinform. 2022, 90, 1443–1456. [Google Scholar] [CrossRef]
- Chow, J.; Perez-Garcia, P.; Dierkes, R.; Streit, W.R. Microbial enzymes will offer limited solutions to the global plastic pollution crisis. Microb. Biotechnol. 2022, 16, 195–217. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Rives, A.; Meier, J.; Sercu, T.; Goyal, S.; Lin, Z.; Liu, J.; Guo, D.; Ott, M.; Zitnick, C.L.; Ma, J.; et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl. Acad. Sci. USA 2021, 118, e2016239118. [Google Scholar] [CrossRef]
- Zheng, L.; Baumann, U.; Reymond, J.-L. Molecular mechanism of enantioselective proton transfer to carbon in catalytic antibody 14D9. Proc. Natl. Acad. Sci. USA 2004, 101, 3387–3392. [Google Scholar] [CrossRef]
- Hie, B.L.; Yang, K.K. Adaptive machine learning for protein engineering. Curr. Opin. Struct. Biol. 2022, 72, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.; Erickson, E.; Brizendine, R.K.; Salvachúa, D.; Michener, W.E.; Li, Y.; Tan, Z.; Beckham, G.T.; McGeehan, J.E.; Pickford, A.R.; et al. The role of binding modules in enzymatic poly (ethylene terephthalate) hydrolysis at high-solids loadings. Chem. Catal. 2022, 2, 2644–2657. [Google Scholar] [CrossRef]
- Chen, K.; Dong, X.; Sun, Y. Sequentially co-immobilized PET and MHET hydrolases via Spy chemistry in calcium phosphate nanocrystals present high-performance PET degradation. J. Hazard. Mater. 2022, 438, 129517. [Google Scholar] [CrossRef]
- Chen, Z.; Duan, R.; Xiao, Y.; Wei, Y.; Zhang, H.; Sun, X.; Wang, S.; Cheng, Y.; Wang, X.; Tong, S.; et al. Biodegradation of highly crystallized poly (ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin. Nat. Commun. 2022, 13, 7138. [Google Scholar] [CrossRef]
- Burelo, M.; Gaytán, I.; Loza-Tavera, H.; Cruz-Morales, J.A.; Zárate-Saldaña, D.; Cruz-Gómez, M.J.; Gutiérrez, S. Synthesis, characterization and biodegradation studies of polyurethanes: Effect of unsaturation on biodegradability. Chemosphere 2022, 307, 136136. [Google Scholar] [CrossRef]

| Main Topics | Outlook | Reference |
|---|---|---|
| Biotechnological options and challenges for the valorization of plastic waste degradation products with a focus on microbial metabolic engineering. | Few industrially implemented examples until now. The challenges are understanding degradation pathways and measuring the efficiency of microbial conversion to target markets. | [31] |
| Chemical recycling routes of plastics with reference to life cycle analysis; a small parenthesis on biotechnology; evaluation of industrial systems of sorting and collection. | New or improved chemical catalysts are needed, targeting superior plastic contact and their stability over multiple uses and improving heat distribution. Technological developments will benefit from improvement of cleaning and sorting and from avoiding the commercialization of composite and multilayer materials. | [32] |
| Catalytic mechanisms and structural rationale of microbial enzymes able to decompose both non-starch plant biomass and synthetic plastics. | Enzymes that can degrade macromolecular polymers (either lignin or recalcitrant plastics) share common biochemical features. Novel plastic polymer-degrading enzymes may be discovered to allow for investigations of the mechanisms by which they operate. | [33] |
| Biotechnology-based plastic deconstruction; clarification of biodegradable materials and microplastic pollution; challenges and future directions. | Plastics should be made from biomass and CO2 and with degradation-on-demand features for products that potentially reach the environment. Consumers must be willing to pay an extra tax to compensate for higher biotech-based recycled plastic prices. | [34] |
| Microbial degradation of synthetic plastics and probable enzymatic mechanisms. | The biochemical and structural properties of enzymes degrading more recalcitrant plastics need further studies to allow for their modification towards better degradation efficiency. The inclusion of the appropriate pretreatment/additives might yield better results. | [35] |
| Comprehensive update on challenges and opportunities in chemical and biological catalysis for plastics deconstruction and recycling; suggestions to find standards to compare different mechanisms of plastic deconstruction and their relative performance. | Biological and chemical catalysis should be combined to depolymerize plastics and generate commodity chemicals. These efforts could be synergistic with the development of alternative materials with better end-of-life functionalities that increase their amenability to catalytic deconstruction. | [36] |
| Enzymatic mechanisms of plastic degradation and factors influencing their performance. | To unravel reaction mechanisms in recalcitrant C-C plastics, basic investigations of changes in substrate polydispersity and the resulting product molecules are required. A ‘bottom-up’ approach of structure-guided de novo enzyme system design is needed. | [37] |
| Literature survey and challenges of biodegradability of recalcitrant plastics in the presence of pro-oxidants. | There are concerns on microfragments of oxo-plastics reaching marine environments, as no previous study had reported a 100% complete biodegradation of oxo-biodegradable plastic. In the near future, bioplastics are expected to be favored. | [38] |
| Microbial biodegradation of various synthetic plastic types with a focus on algae and the gut microbial consortium of insects. | The mode of action and mechanism of microbial degradation calls for further studies to detect an effective enzymatic system that fits the tested polymeric material. A practical system for plastic biodegradation is still not available. | [39] |
| Recent advances in the biotechnology-based biodegradation of both traditional and bio-based plastics with a focus on known degradation mechanisms and valorization of plastic waste. | Studies of the recycling and valorization of plastic waste could offer solutions to plastic industries. Synthetic biology studies on the functioning of microbial cell factories are needed to further improve the adaptability of microbes to a circular economy of plastics; the degradation mechanisms of some types of plastics are still missing and must be studied. | [40] |
| Comprehensive update on strategies for the discovery and engineering of plastic-degrading enzymes. | New high-throughput screening methods are needed to identify plastic-degrading enzymes. A better connection between protein features and functions is needed to guide accurate protein engineering. The development of engineered thermophilic microorganisms can overcome the problem of short enzyme lifetimes in plastic degradation processes. | [41] |
| Comprehensive update on plastic depolymerization and upcycling routes, including the modification of plastics to make them more degradable and a mention of biotechnological systems | Academia and industry need to cooperate to create marketable solutions from different plastic recycling technologies with a business model in mind. There should be more international and local government efforts to promote recycling/upcycling and penalize disposal with enforcement. Policies should be introduced to offer more convenient and effective recycling choices for consumers, avoiding low recycling rates due to the collection of highly mixed recyclables. Governments, non-profit organizations, and academia should work together to inform and encourage consumers to choose upcycled and/or biodegradable materials. More interdisciplinary research is needed to create innovative and safe products from plastic upcycling. | [42] |
| The most promising biotechnological open-loop recycling processes for synthetic plastics with a focus on how to improve degradation with abiotic pretreatments, enzyme engineering, and novel bioreactor designs | Higher degradation activities on polyester and C–C backbone plastics will be fundamental. This can be achieved by engineering existing enzymes and microorganisms, by applying synergistic degradation strategies with multiple enzymes and pretreatments, by focusing on the optimization of the reaction conditions in the reactor, and by evaluating the economic feasibility of plastic monomer upcycling to high-value products | [43] |
| Resin Type | Acronym | Applications | 2019 EU Demand Distribution (Mass %) | Chemical Structure | |
|---|---|---|---|---|---|
| Thermoplastics | Polypropylene | PP | Food packaging, sweet and snack wrappers, hinged caps, microwave containers, pipes, automotive parts, bank notes, etc. | 19.4 | –[CH2-CH(CH3)]n– |
| Low-Density Polyethylene | LDPE | Reusable bags, trays and containers, agricultural film, food packaging film, etc. | 17.4 | –(CH2-CH2)n– | |
| High-Density Polyethylene | HDPE | Toys, milk bottles, shampoo bottles, pipes, houseware, etc. | 12.4 | –(CH2-CH2)n– | |
| Polyvinyl Chloride | PVC | Window frames, profiles, floors, wall covering, pipes, cable insulation, garden hoses, inflatable pools, etc. | 10 | –(CH2-CHCl)n– | |
| Polyethylene Terephthalate | PET | Bottles for water, soft drinks, juices, and cleaners; food packaging; textiles; etc. | 7.9 | –[CO(CH2)4CO-OCH2CH2O]n– | |
| Polystyrene | PS + Expanded polystyrene (EPS) | Food packaging (diary, fishery), building insulation, electrical and electronic equipment, inner liner for fridges, eyeglass frames, etc. | 6.2 | –[CH2-CH(C6H5)]n– | |
| Other Thermoplastics | ABS, PBT, PC, Polytetrafluoroethylene (PTFE), PMMA. | Hub caps (ABS), optical fibers (PBT), eyeglasses lenses, roofing sheets (PC), touch screens (PMMA), cable coating in telecommunications (PTFE), and many other applications in aerospace, as well as medical implants, surgical devices, membranes, valves and seals, protective coatings, etc. | 11.3 | Many formulations | |
| Thermosets | Polyurethane | Polyurethane (PUR) | Building insulation, pillows, mattresses, insulating foams for fridges, etc. | 7.9 | –[R-OCO-NH-R2-NH-CO-O]n– |
| Other Thermosets 1 | Phenol formaldehyde resins (PF), urea–formaldehyde (UF) | Decorative laminates, textiles, paper, foundry sand molds, foam insulation, paints, coatings, adhesives, etc. | 7.5 | - | |
| Polymer Type | PPW Composition | PPW Collection Rate | MRF Sorting Rate | REC Recycling Rate |
|---|---|---|---|---|
| PET | 18 | 62 | 85 | 81 |
| HDPE | 20 | 44 | 85 | 88 |
| PP | 20 | 32 | 64 | 66 |
| PS | 7 | 30 | 37 | 66 |
| PVC | 3 | 20 | 73 | 80 |
| LDPE (films) | 32 | 36 | 59 | 71 |
| Plastic Polymer | Material | Source | Reaction Condition | Degradation Rate | Reference |
|---|---|---|---|---|---|
| PET | Ultrathin PET film (2.5 to 7 nm) obtained from amorphous PET sheets of 2 mm thickness | IsPETaseTM [87] and IsMHETaseSM [88], two variants of I. sakaiensis PETase and METase | PBS buffer, pH 7.4, 50 °C | ≈70% in 1 h | [89] |
| PET | Amorphized and micronized PET (≈200–250 µm) from post-consumer bottle-grade PET | Cutinase from leaf–branch compost (ICCG variant) | 100 mM potassium phosphate buffer, pH 8.0, 72 °C | ≈90% in 10 h | [90] |
| PET | 2 × 1 cm2 amorphous Goodfellow PET film and low-crystallinity (13%) PET powder | PHL7/PES-H1, a cutinase from a compost site (L92F/Q94Y variant) | 1 M potassium phosphate buffer, pH 8.0, 72 °C, shaking at 1000 rpm | ≈100% in 24 h | [91] |
| PET | 3 × 0.5 cm2 flakes of an amorphous PET clamshell container | PHL7/PES-H1, a cutinase from a compost site | 1 M potassium phosphate buffer, pH 8.0, 70 °C, shaking at 1000 rpm | >95% in 24 h | [92] |
| PET | Low-crystallinity (1.2 to 6.2%) discs (6 mm) from 51 different post-consumer PET products | FAST-PETase, a variant of I. sakaiensis PETase | 100 mM potassium phosphate buffer, pH 8.0, 50 °C, shaking at 180 rpm | ≈100% in 1 to 7 days | [93] |
| PET | PET/PE composite packaging tray lid (4 mg, thickness of 325 μm PET and 40 μm PE) | HotPETase, a directed-evolved variant of I. sakaiensis PETase | pH 9.2, 50 mM gly-OH buffer with 4% BugBuster | ≈20% in 24 h | [94] |
| PET | ≈37% crystallinity PET microplastics (≈300 µm) | TS-ΔIsPET, a variant of PETase from Ideonella sakaiensis | 100 mM potassium phosphate buffer, pH 8.0, 40 °C, shaking | ≈26% in 2 days | [95] |
| Polyester-type PUR | Lab-prepared 10 µm thin PUR and segmented PUR urea films based on lysine diisocyanate | Papain, Bromelain, Ficin, Chymotrypsin, Proteinase K | PBS, pH 7.0, 37 °C | Up to ≈50% in 7 days | [96] |
| Polyester-type PUR | Impranil-DLN® | Pseudomonas putida A12 | pH 8.0, 25 °C | 92% in 4 days | [97] |
| Polyester-type PUR | Impranil-DLN® | Pestalotiopsis Microspora E2712A | 25 °C in a rotary incubator | 99% in 2 weeks | [98] |
| Polyether-type PUR | PUR foam | Tenebrio molitor | Guts of the larvae (probably microbiota-assisted) | 67% after 30 days | [99] |
| Polyester-type PUR | Lab-prepared 0.3 mm thin polycaprolactone thermoplastic PUR film (Capa 2302) | Amidase E4143 and esterase E3576 | PBS, pH 7.0, 37 °C | 33% in 51 days | [100] |
| Polyester-type PUR | Impranil-DLN® | Cladosporium pseudocladospo-rioides, Cladosporium tenuissimum, Cladosporium asperulatum, Cladosporium montecillanum, Aspergillus fumigatus, Penicillium chrysogenum | 25 °C, no shaking | 40–87% in 2 weeks | [101] |
| Polyester-type PUR | Commercial 1 mm thin PUR film | Aspergillus flavus ITCC 6051 | 28 °C under 120 rpm shaking | 60.6% in 30 days | [102] |
| Polyester-type PUR | Lab-prepared ~0.2 mm thin PUR | Bacillus subtilis MZA-75 and Pseudomonas aeruginosa MZA-85 | 37 °C under 150 rpm shaking | 40% in 30 days | [103] |
| Polyether-type PUR | PUR foam used for commercial production of mattress cushioning | Cladosporium pseudocladospo-rioides, Cladosporium tenuissimum, Cladosporium asperulatum, Cladosporium montecillanum, Aspergillus fumigatus, Penicillium chrysogenum | 25–30 °C, no shaking | 10–65% in 21 days | [101] |
| Polyether-type PUR | Cubical ether–PUR (1 cm3) | Alternaria sp. PURDK2 | 30 °C, no shaking | 27.5% in 70 days | [104] |
| LDPE | LDPE bag | Galleria mellonella | - | 13% in 14 h | [105,106,107] |
| LDPE | LDPE film | Pseudomonas citronellolis | 37 °C under 15 rpm shaking | 17.8% in 4 days | [108] |
| LDPE | LDPE powder | Cupriavidus necator H16 | 30 °C in a rotary incubator | 33.7% in 21 days | [109] |
| LDPE | LDPE foam | Tenebrio molitor | Gut of the larvae (probably microbiota-assisted) | 49.0% in 32 days | [110] |
| LDPE | 20 μm thin PE film | Oscillatoria subbrevis | - | 30% in 42 days | [111] |
| PS | PS pyrolysate oil | Pseudomonas putida CA-3 | Four consecutive treatments at 30 °C for 48 h | 10% in 8 days | [112] |
| PS | Brominated high-impact PS | Bacillus sp. | 30 °C, no shaking | 23.7% in 30 days | [113] |
| PS | Brominated high-impact PS | Exiguobacterium sp. strain YT2 | 30 °C under 150 rpm shaking | 12.4% in 30 days | [114] |
| PP | Max 250 µm microplastic PP | Bacillus cereus | 33 °C under 150 rpm shaking | 12% in 40 days | [115] |
| PP | Isotactic PP strips | Pseudomonas sp., Vibrio sp., Aspergillus niger | pH 7.0, 30 °C | 60% in 175 days | [116] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orlando, M.; Molla, G.; Castellani, P.; Pirillo, V.; Torretta, V.; Ferronato, N. Microbial Enzyme Biotechnology to Reach Plastic Waste Circularity: Current Status, Problems and Perspectives. Int. J. Mol. Sci. 2023, 24, 3877. https://doi.org/10.3390/ijms24043877
Orlando M, Molla G, Castellani P, Pirillo V, Torretta V, Ferronato N. Microbial Enzyme Biotechnology to Reach Plastic Waste Circularity: Current Status, Problems and Perspectives. International Journal of Molecular Sciences. 2023; 24(4):3877. https://doi.org/10.3390/ijms24043877
Chicago/Turabian StyleOrlando, Marco, Gianluca Molla, Pietro Castellani, Valentina Pirillo, Vincenzo Torretta, and Navarro Ferronato. 2023. "Microbial Enzyme Biotechnology to Reach Plastic Waste Circularity: Current Status, Problems and Perspectives" International Journal of Molecular Sciences 24, no. 4: 3877. https://doi.org/10.3390/ijms24043877
APA StyleOrlando, M., Molla, G., Castellani, P., Pirillo, V., Torretta, V., & Ferronato, N. (2023). Microbial Enzyme Biotechnology to Reach Plastic Waste Circularity: Current Status, Problems and Perspectives. International Journal of Molecular Sciences, 24(4), 3877. https://doi.org/10.3390/ijms24043877








