A Novel miRNA in Rice Associated with the Low Seed Setting Rate Symptom of Rice Stripe Virus
Abstract
1. Introduction
2. Results
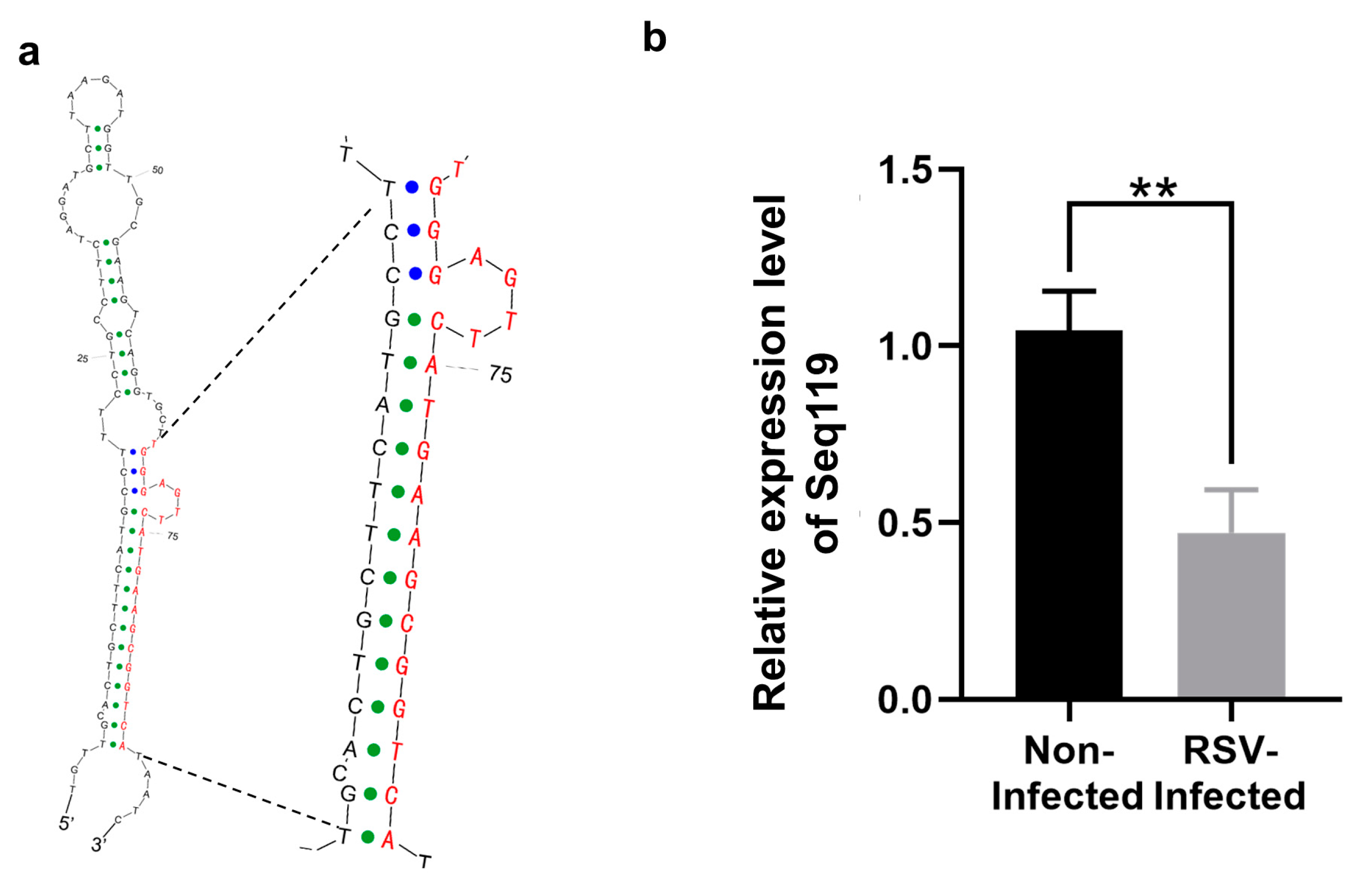
2.1. Identification of Seq119 in Rice
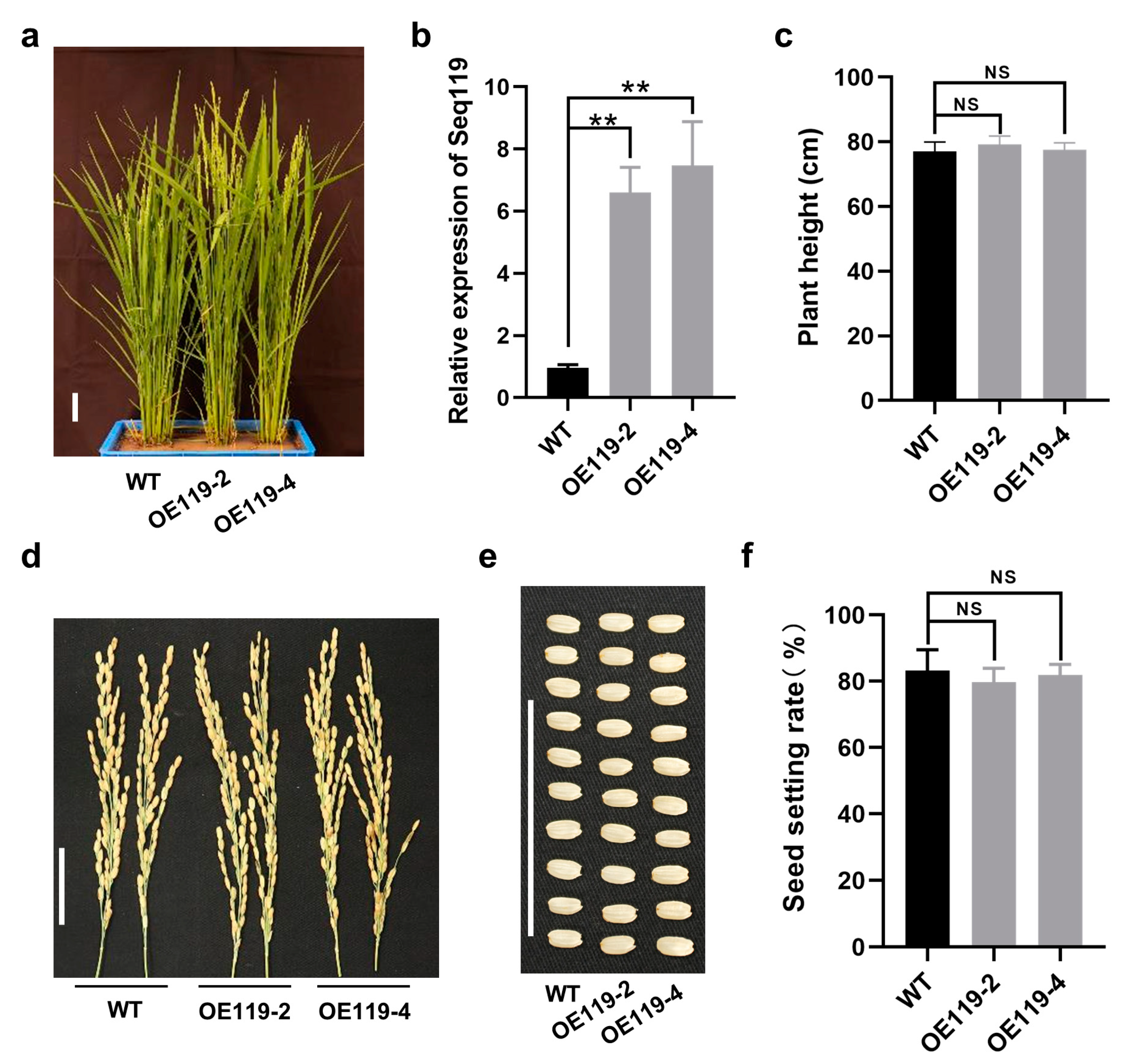
2.2. Overexpression of Seq119 in Rice Has No Obvious Effect on Rice Development
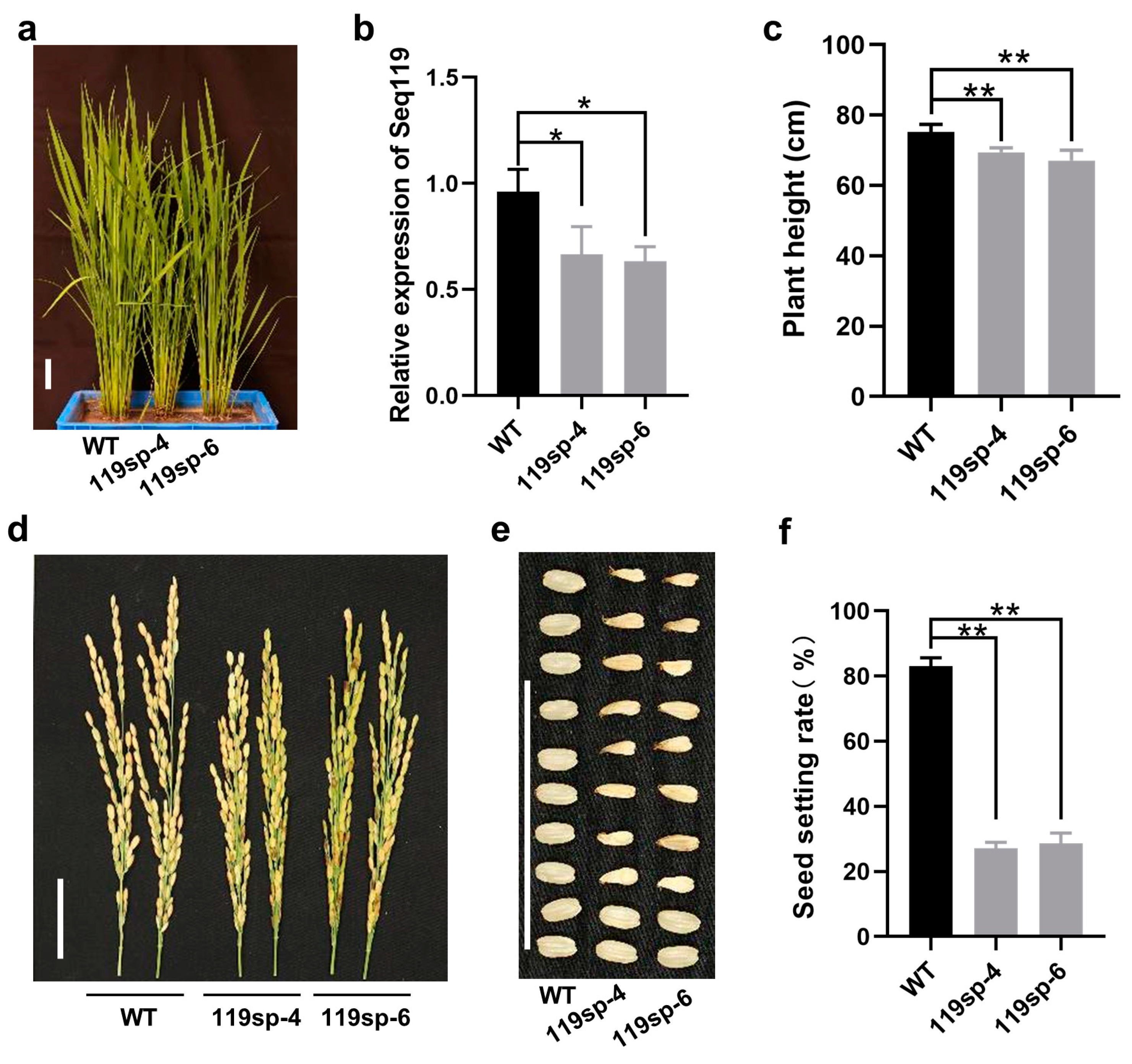
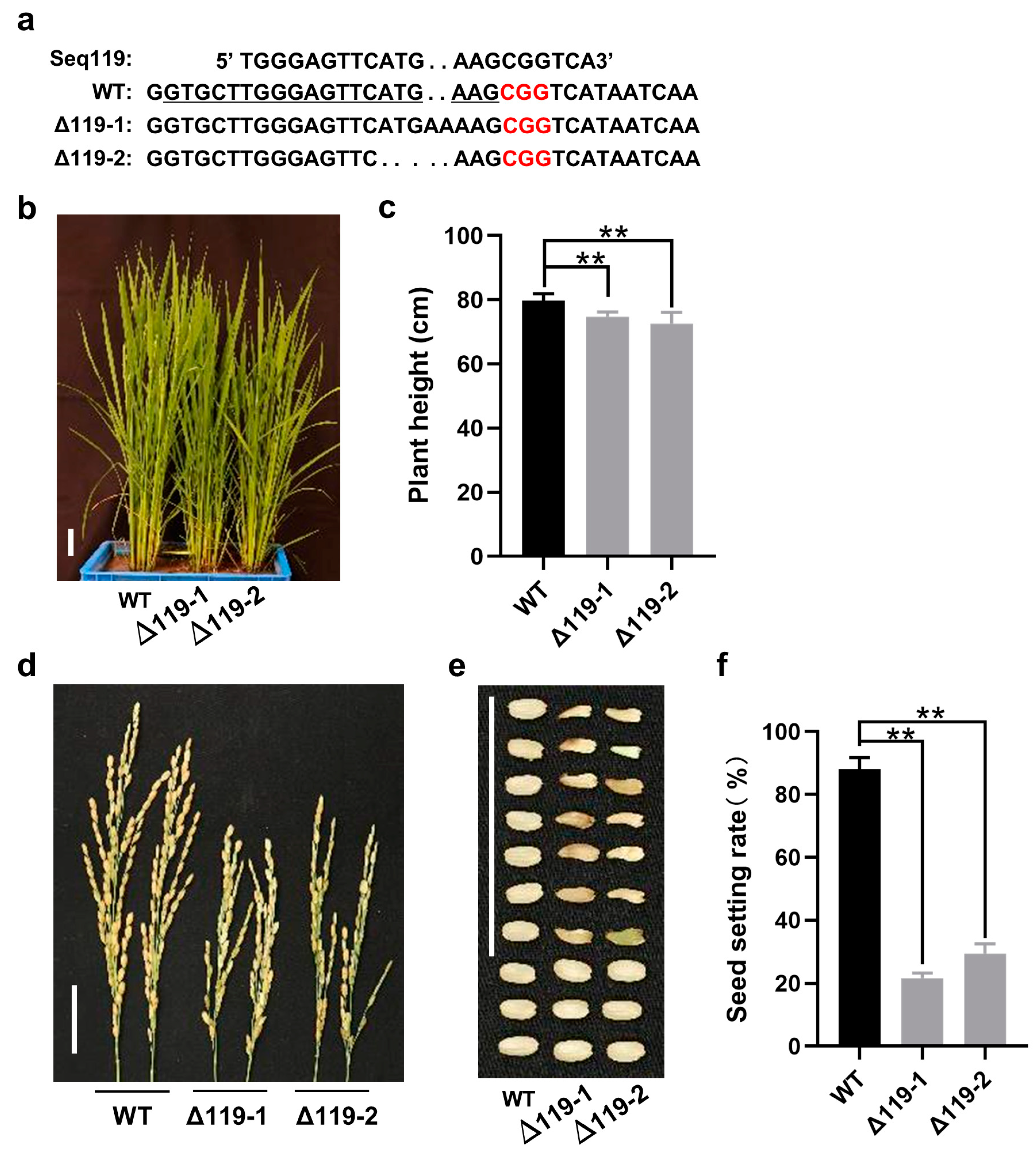
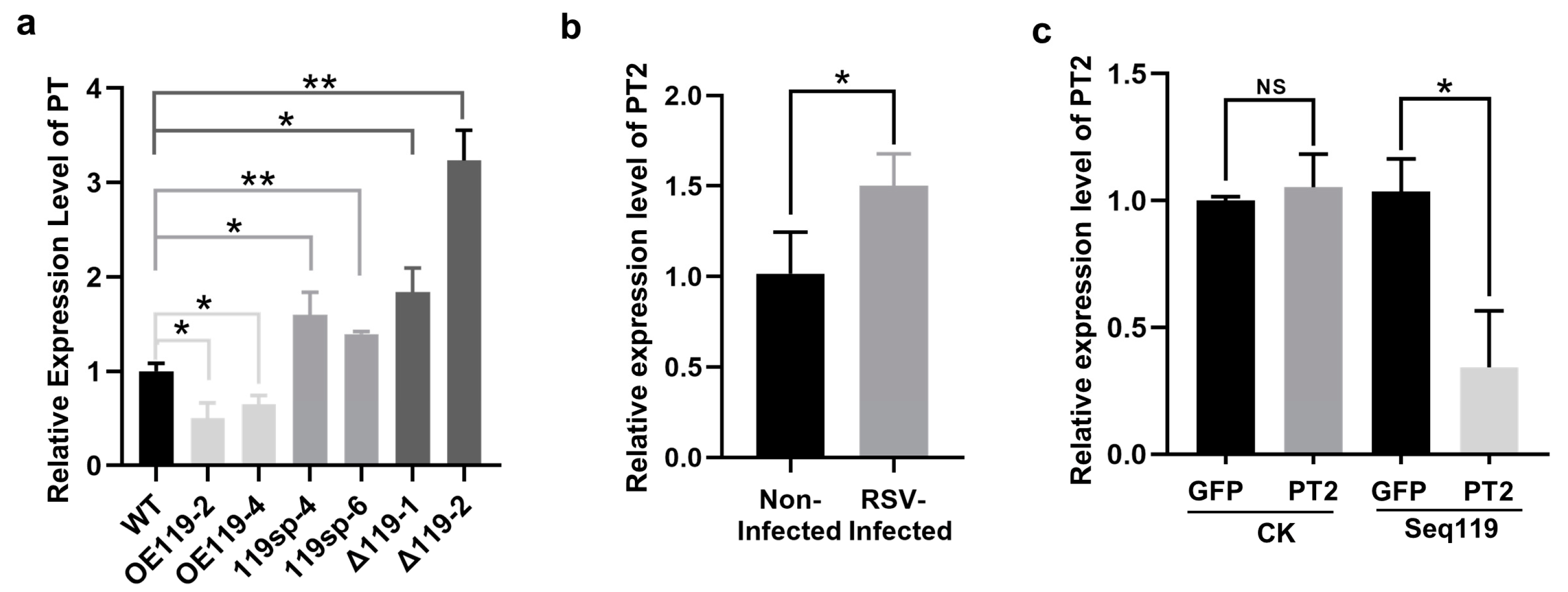
2.3. Loss of Function of Seq119 Reduces Seed Setting Rate in Rice
2.4. Identification of the Target Genes of Seq119
2.5. Overexpression of PT2 Causes a Low Seed Setting Rate in Rice
3. Discussion
4. Materials and Methods
4.1. Rice Transformation
4.2. Plant Materials and Growth Conditions
4.3. Virus Inoculation Assay
4.4. Expression in N. benthamiana Leaves
4.5. Total RNA Extraction and RNA Analysis
4.6. Fluorescence Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Carrington, J.C.; Ambros, V. Role of microRNAs in plant and animal development. Science 2003, 301, 336–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.; Xue, H.; Chen, X. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Weigel, D. Control of leaf morphogenesis by miRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef]
- Kidner, C.A.; Martienssen, R.A. The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 2005, 8, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.S.; Xie, Q.; Fei, J.F.; Chua, N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef]
- Chen, X.M. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Feng, J.; Liu, S.; Wang, M.; Lang, Q.; Jin, C. Identification of microRNAs and their targets in tomato infected with Cucumber mosaic virus based on deep sequencing. Planta 2014, 240, 1335–1352. [Google Scholar] [CrossRef]
- Yin, X.; Wang, J.; Cheng, H.; Wang, X.; Yu, D. Detection and evolutionary analysis of soybean miRNAs responsive to soybean mosaic virus. Planta 2013, 237, 1213–1225. [Google Scholar] [CrossRef]
- Naqvi, A.R.; Haq, Q.; Mukherjee, S.K. MicroRNA profiling of tomato leaf curl new delhi virus (tolcndv) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol. J. 2010, 7, 281. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Mou, G.; Wang, K.; Zhou, G. MicroRNAs responding to southern rice black-streaked dwarf virus infection and their target genes associated with symptom development in rice. Virus Res. 2014, 190, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Amin, I.; Patil, B.L.; Briddon, R.W.; Mansoor, S.; Fauquet, C.M. A common set of developmental miRNAs are upregulated in Nicotiana benthamiana by diverse begomoviruses. Virol. J. 2011, 8, 143. [Google Scholar] [CrossRef]
- Gao, H.; Zheng, X.M.; Fei, G.; Chen, J.; Jin, M.; Ren, Y.; Wu, W.; Zhou, K.; Sheng, P.; Feng, Z. Ehd4 Encodes a Novel and Oryza-Genus-Specific Regulator of Photoperiodic Flowering in Rice. PLoS Genet. 2013, 9, e1003281. [Google Scholar] [CrossRef]
- Bazzini, A.; Hopp, H.; Beachy, R.; Asurmendi, S. Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc. Natl. Acad. Sci. USA 2007, 104, 12157–12162. [Google Scholar] [CrossRef]
- Cillo, F.; Mascia, T.; Pasciuto, M.M.; Gallitelli, D. Differential Effects of Mild and Severe Cucumber mosaic virus Strains in the Perturbation of MicroRNA-Regulated Gene Expression in Tomato Map to the 3′ Sequence of RNA 2. Mol. Plant Microbe Interact 2009, 22, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, R.; García-Marcos, A.; Barajas, D.; Martiáñez, J.; Tenllado, F. PVX–potyvirus synergistic infections differentially alter microRNA accumulation in Nicotiana benthamiana. Virus Res. 2012, 165, 231–235. [Google Scholar] [CrossRef]
- Wang, K.; Wang, X.; Li, M.; Shi, T.; Yang, P. Low genetic diversity and functional constraint of miRNA genes participating pollen-pistil interaction in rice. Plant Mol. Biol. 2017, 95, 89–98. [Google Scholar] [CrossRef]
- Li, X.P.; Ma, X.C.; Wang, H.; Zhu, Y.; Liu, X.X.; Li, T.T.; Zheng, Y.P.; Zhao, J.Q.; Zhang, J.W.; Huang, Y.Y.; et al. Osa-miR162a fine-tunes rice resistance to Magnaporthe oryzae and Yield. Rice 2020, 13, 38. [Google Scholar] [CrossRef]
- Zhou, S.X.; Zhu, Y.; Wang, L.F.; Zheng, Y.P.; Chen, J.F.; Li, T.T.; Yang, X.M.; Wang, H.; Li, X.P.; Ma, X.C.; et al. Osa-miR1873 fine-tunes rice immunity against Magnaporthe oryzae and yield traits. J. Integr. Plant Biol. 2020, 62, 1213–1226. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, X.; Zhang, J. Alteration of osa-miR156e expression affects rice plant architecture and strigolactones (SLs) pathway. Plant Cell Rep 2015, 34, 767–781. [Google Scholar] [CrossRef]
- Shiba, T.; Hirae, M.; Hayano-Saito, Y.; Ohto, Y.; Uematsu, H.; Sugiyama, A.; Okuda, M. Spread and yield loss mechanisms of rice stripe disease in rice paddies. Field Crops Res. 2018, 217, 211–217. [Google Scholar] [CrossRef]
- Guo, W.X.; Wu, G.T.; Yan, F.; Lu, Y.W.; Zheng, H.Y.; Lin, L.; Chen, H.R.; Chen, J.P. Identification of novel Oryza sativa miRNAs in deep sequencing-based small RNA libraries of rice infected with Rice stripe virus. PLoS ONE 2012, 7, e46443. [Google Scholar] [CrossRef]
- Tong, A.; Yuan, Q.; Wang, S.; Peng, J.; Lu, Y.; Zheng, H.; Lin, L.; Chen, H.; Gong, Y.; Chen, J. Altered accumulation of osa-miR171b contributes to rice stripe virus infection by regulating disease symptoms. J. Exp. Bot. 2017, 68, 4357–4367. [Google Scholar] [CrossRef]
- Du, P.; Wu, J.; Zhang, J.; Zhao, S.; Zheng, H.; Gao, G.; Wei, L.; Li, Y. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 2011, 7, e1002176. [Google Scholar] [CrossRef] [PubMed]
- Warthmann, N.; Chen, H.; Ossowski, S.; Weigel, D.; Hervé, P. Highly Specific Gene Silencing by Artificial miRNAs in Rice. PLoS ONE 2008, 3, e1829. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Lu, Y.W.; Wu, G.T.; Peng, J.J.; Zheng, H.Y.; Lin, L.; Chen, J.P. A simplified method for constructing artificial microRNAs based on the osa-MIR528 precursor. J. Biotechnol. 2012, 160, 146–150. [Google Scholar] [CrossRef]
- Todesco, M.; Rubio-Somoza, I.; Paz-Ares, J.; Weigel, D. A Collection of Target Mimics for Comprehensive Analysis of MicroRNA Function in Arabidopsis thaliana. PLoS Genet. 2010, 6, e1001031. [Google Scholar] [CrossRef]
- Yan, J.; Gu, Y.; Jia, X.; Kang, W.; Pan, S.; Tang, X.; Chen, X.; Tang, G. Effective Small RNA Destruction by the Expression of a Short Tandem Target Mimic in Arabidopsis. Plant Cell 2012, 24, 415–427. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.L.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.B.; Zhuang, Z.H.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2018, 46, W49. [Google Scholar] [CrossRef]
- Singh, A.; Gautam, V.; Singh, S.; Das, S.; Sarkar; Verma, S.; Mishra, V.; Mukherjee, S.; Sarkar, A.K. Plant small RNAs: Advancement in the understanding of biogenesis and role in plant development. Planta 2018, 248, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Y.; Chern, M.; Zhu, Y.; Zhang, L.L.; Lu, J.H.; Li, X.P.; Dang, W.Q.; Ma, X.C.; Yang, Z.R.; et al. Suppression of rice miR168 improves yield, flowering time and immunity. Nat. Plants 2021, 7, 129–136. [Google Scholar] [CrossRef]
- Tao, H.; Jia, Z.; Gao, X.; Gui, M.; Li, Y.; Liu, Y. Analysis of the miRNA expression profile involved in the tomato spotted wilt orthotospovirus-pepper interaction. Virus Res. 2022, 312, 198710. [Google Scholar] [CrossRef] [PubMed]
- Bizabani, C.; Rogans, S.J.; Rey, M.E.C. Differential miRNA profiles in South African cassava mosaic virus-infected cassava landraces reveal clues to susceptibility and tolerance to cassava mosaic disease. Virus Res. 2021, 303, 198400. [Google Scholar] [CrossRef]
- Liu, J.; Fan, H.; Wang, Y.; Han, C.; Wang, X.; Yu, J.; Li, D.; Zhang, Y. Genome-Wide microRNA Profiling Using Oligonucleotide Microarray Reveals Regulatory Networks of microRNAs in Nicotiana benthamiana During Beet Necrotic Yellow Vein Virus Infection. Viruses 2020, 12, 310. [Google Scholar] [CrossRef]
- Prigigallo, M.I.; Kriznik, M.; Paola, D.; Catalano, D.; Gruden, K.; Finetti-Sialer, M.M.; Cillo, F. Potato Virus Y Infection Alters Small RNA Metabolism and Immune Response in Tomato. Viruses 2019, 11, 1100. [Google Scholar] [CrossRef]
- Marmisolle, F.E.; Arizmendi, A.; Ribone, A.; Rivarola, M.; Garcia, M.L.; Reyes, C.A. Up-regulation of microRNA targets correlates with symptom severity in Citrus sinensis plants infected with two different isolates of citrus psorosis virus. Planta 2019, 251, 7. [Google Scholar] [CrossRef]
- Yin, Z.; Murawska, Z.; Xie, F.; Pawelkowicz, M.; Michalak, K.; Zhang, B.; Lebecka, R. microRNA response in potato virus Y infected tobacco shows strain-specificity depending on host and symptom severity. Virus Res. 2019, 260, 20–32. [Google Scholar] [CrossRef]
- Simon-Mateo, C.; Garcia, J.A. MicroRNA-Guided Processing Impairs Plum Pox Virus Replication, but the Virus Readily Evolves To Escape This Silencing Mechanism. J. Virol. 2006, 80, 2429–2436. [Google Scholar] [CrossRef]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, C.; Zhang, X.; Jin, H. Host small RNAs are big contributors to plant innate immunity. Curr. Opin. Plant Biol. 2009, 12, 465–472. [Google Scholar] [CrossRef]
- Lu, Y.D.; Gan, Q.H.; Chi, X.Y.; Qin, S. Roles of microRNA in plant defense and virus offense interaction. Plant Cell Rep. 2008, 27, 1571–1579. [Google Scholar] [CrossRef]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N. A Plant miRNA Contributes to Antibacterial Resistance by Repressing Auxin Signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, P.V.; Chen, H.M.; Patel, K.; Bond, D.M.; Baulcombe, S.D. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lii, Y.F.; Liu, Y.; Jin, H.L. Contribution of Small RNA Pathway Components in Plant Immunity. Mol. Plant-Microbe Interact. 2013, 26, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 2015, 4, e05733. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, C.; Shi, C.; Wang, Y.; Li, Y. Rice stripe virus NS3 protein regulates primary miRNA processing through association with the miRNA biogenesis factor OsDRB1 and facilitates virus infection in rice. PLoS Pathog. 2017, 13, e1006662. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, R.; Yang, Z.; Yao, S.; Zhao, S.; Wang, Y.; Li, P.; Song, X.; Jin, L.; Zhou, T.; et al. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants 2017, 3, 16203. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Yang, Z.; Yang, R.; Huang, Y.; Guo, G.; Kong, X.; Lan, Y.; Zhou, T.; Wang, H.; Wang, W.; et al. Transcriptional Regulation of miR528 by OsSPL9 Orchestrates Antiviral Response in Rice. Mol. Plant 2019, 12, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wei, M.; Li, Y.; Tao, H.; Wu, H.; Chen, Z.; Li, C.; Xu, J.H. MiR529a controls plant height, tiller number, panicle architecture and grain size by regulating SPL target genes in rice (Oryza sativa L.). Plant Sci. 2021, 302, 110728. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Shen, Y.; Li, H.; Yang, J.; Cai, X.; Zheng, G.; Zhu, Y.; Jia, B.; Sun, X. The multiple roles of OsmiR535 in modulating plant height, panicle branching and grain shape. Plant Sci. 2019, 283, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; He, W.; Peng, Z.; Zhang, H.; Li, F.; Yao, J. A putative AGO protein, OsAGO17, positively regulates grain size and grain weight through OsmiR397b in rice. Plant Biotechnol. J. 2020, 18, 916–928. [Google Scholar] [CrossRef]
- Sun, W.; Xu, X.H.; Li, Y.; Xie, L.; He, Y.; Li, W.; Lu, X.; Sun, H.; Xie, X. OsmiR530 acts downstream of OsPIL15 to regulate grain yield in rice. New Phytol. 2020, 226, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.F.; Peng, T.; Sun, H.Z.; Teotia, S.; Wen, H.L.; Du, Y.X.; Zhang, J.; Li, J.Z.; Tang, G.L.; Xue, H.W.; et al. miR1432-OsACOT (Acyl-CoA thioesterase) module determines grain yield via enhancing grain filling rate in rice. Plant Biotechnol. J. 2019, 17, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
- Li, J.; Meng, X.B.; Zong, Y.; Chen, K.L.; Zhang, H.W.; Liu, J.X.; Li, J.Y.; Gao, C.X. Gene replacements and insertions in rice by intron targeting using CRISPR–Cas9. Nat. Plants 2016, 2, 16139. [Google Scholar] [CrossRef]

| Targets | Target Acc. | Expectation | Target Description |
|---|---|---|---|
| PT1 | LOC4352810 | 1 | uncharacterized protein |
| PT2 | LOC4348735 | 2.5 | uncharacterized protein |
| PT3 | LOC4348324 | 3 | uncharacterized protein |
| PT4 | LOC4325059 | 3 | TRANSPARENT TESTA 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Q.; Zhai, Y.; Zhou, L.; Ai, X.; Chen, J.; Yan, F. A Novel miRNA in Rice Associated with the Low Seed Setting Rate Symptom of Rice Stripe Virus. Int. J. Mol. Sci. 2023, 24, 3675. https://doi.org/10.3390/ijms24043675
Yuan Q, Zhai Y, Zhou L, Ai X, Chen J, Yan F. A Novel miRNA in Rice Associated with the Low Seed Setting Rate Symptom of Rice Stripe Virus. International Journal of Molecular Sciences. 2023; 24(4):3675. https://doi.org/10.3390/ijms24043675
Chicago/Turabian StyleYuan, Quan, Yushan Zhai, Liya Zhou, Xuhong Ai, Jianping Chen, and Fei Yan. 2023. "A Novel miRNA in Rice Associated with the Low Seed Setting Rate Symptom of Rice Stripe Virus" International Journal of Molecular Sciences 24, no. 4: 3675. https://doi.org/10.3390/ijms24043675
APA StyleYuan, Q., Zhai, Y., Zhou, L., Ai, X., Chen, J., & Yan, F. (2023). A Novel miRNA in Rice Associated with the Low Seed Setting Rate Symptom of Rice Stripe Virus. International Journal of Molecular Sciences, 24(4), 3675. https://doi.org/10.3390/ijms24043675






