A Systematic Review of the Human Accelerated Regions in Schizophrenia and Related Disorders: Where the Evolutionary and Neurodevelopmental Hypotheses Converge
Abstract
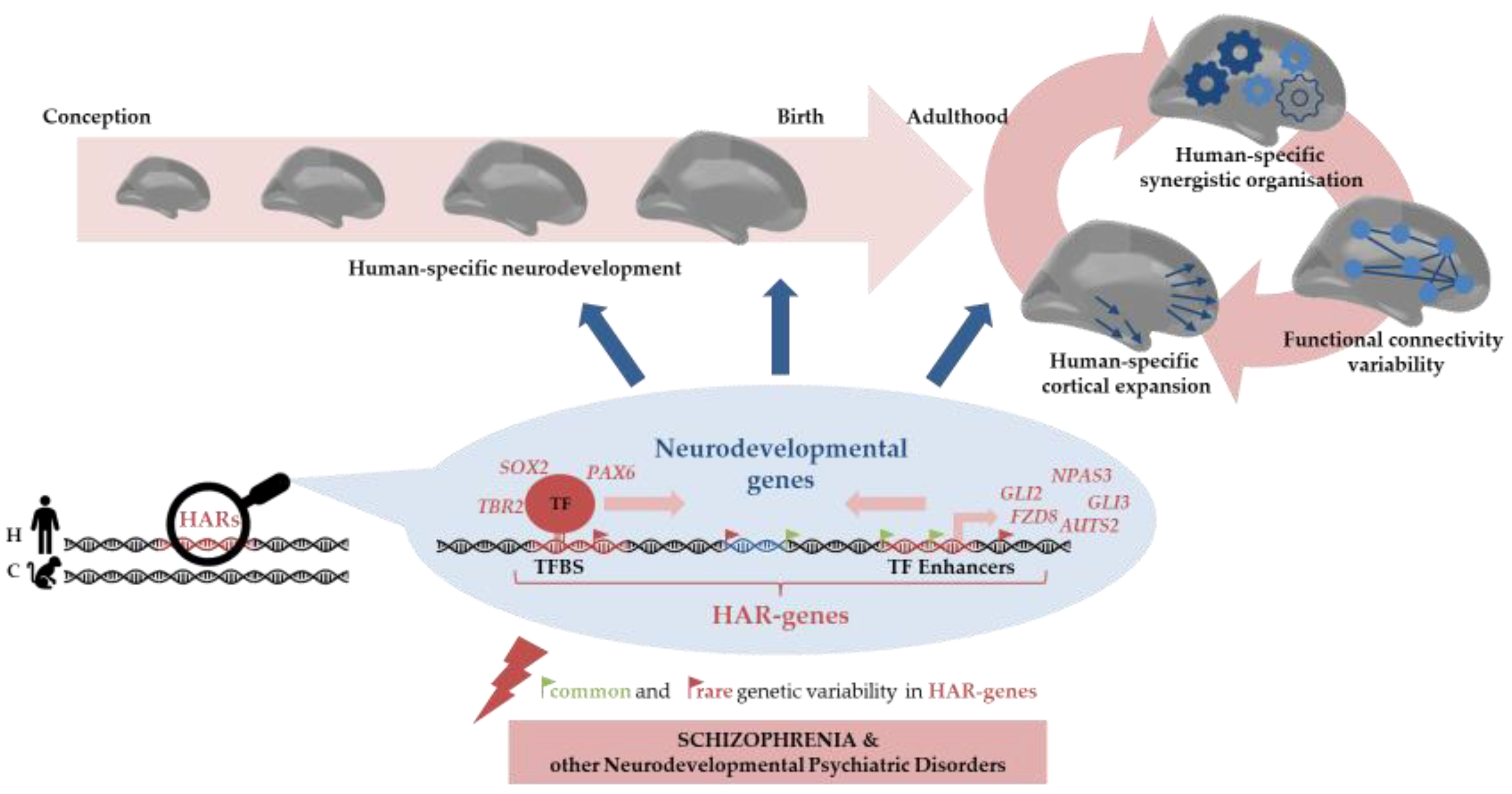
1. Introduction
2. Methods
3. Results
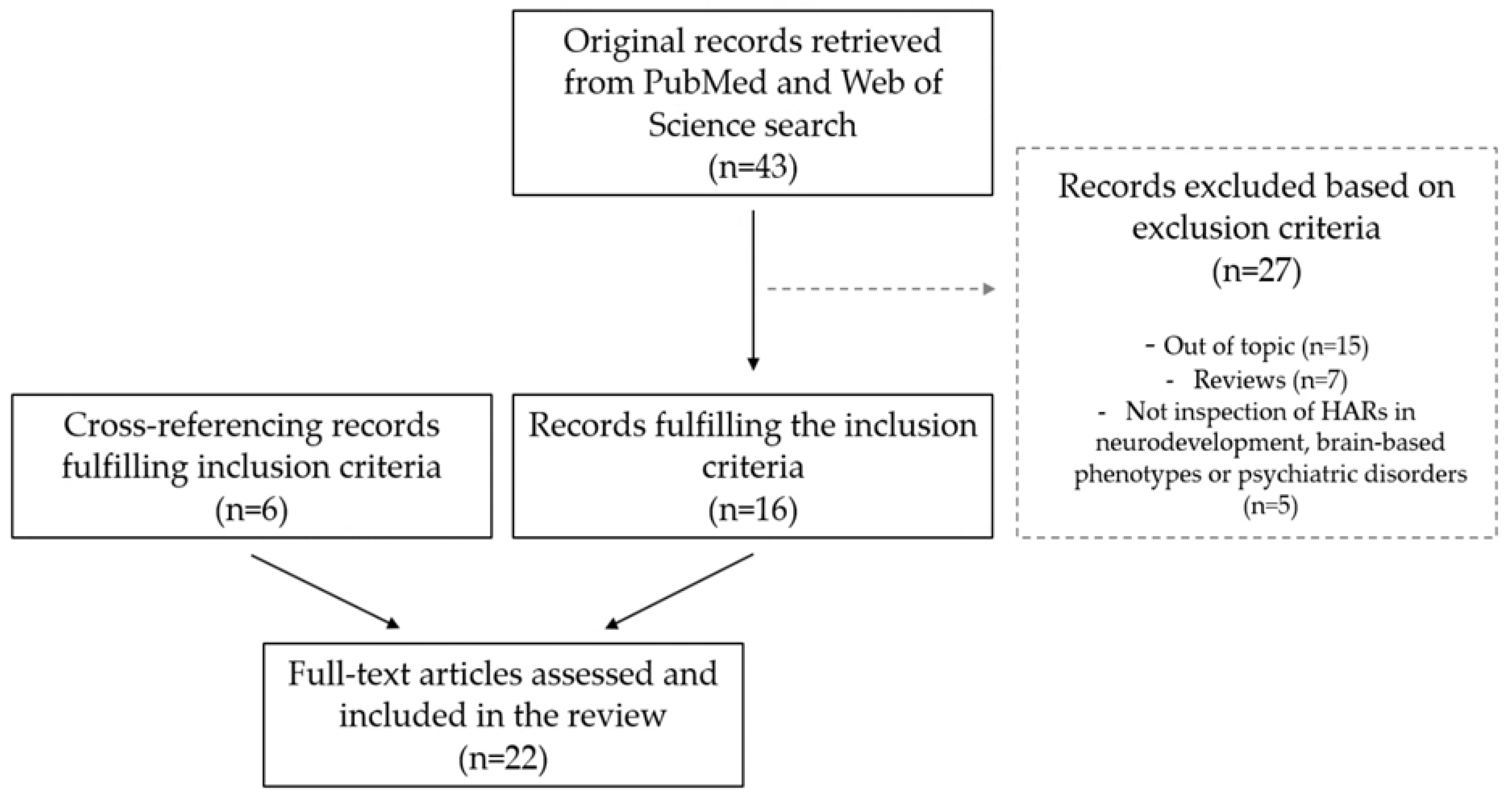
3.1. Literature Search and Study Selection
3.2. HARs and Neurodevelopment
3.2.1. HARs’ Function in Neurodevelopment
3.2.2. Genes Associated with HARs and Their Expression and Functional Patterns
3.2.3. Regulatory Effect of Candidate HARs on Their Proximal Genes
3.3. HARs and Brain and Cognitive Phenotypes
3.4. HARs and Psychiatric Disorders
3.4.1. HARs in Schizophrenia
3.4.2. HARs in Other Neurodevelopment-Related Psychiatric Disorders and Syndromes
4. Discussion
5. Future Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hilker, R.; Helenius, D.; Fagerlund, B.; Skytthe, A.; Christensen, K.; Werge, T.M.; Nordentoft, M.; Glenthøj, B. Heritability of Schizophrenia and Schizophrenia Spectrum Based on the Nationwide Danish Twin Register. Biol. Psychiatry 2018, 83, 492–498. [Google Scholar] [CrossRef]
- Trubetskoy, V.; Pardiñas, A.F.; Qi, T.; Panagiotaropoulou, G.; Awasthi, S.; Bigdeli, T.B.; Bryois, J.; Chen, C.-Y.; Dennison, C.A.; Hall, L.S.; et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 2022, 604, 502–508. [Google Scholar] [CrossRef]
- Singh, T.; Walters, J.T.R.; Johnstone, M.; Curtis, D.; Suvisaari, J.; Torniainen, M.; Rees, E.; Iyegbe, C.; Blackwood, D.; McIntosh, A.M.; et al. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat. Genet. 2017, 49, 1167–1173. [Google Scholar] [CrossRef]
- Rees, E.; GROUP Investigators; Han, J.; Morgan, J.; Carrera, N.; Escott-Price, V.; Pocklington, A.J.; Duffield, M.; Hall, L.S.; Legge, S.E.; et al. De novo mutations identified by exome sequencing implicate rare missense variants in SLC6A1 in schizophrenia. Nat. Neurosci. 2020, 23, 179–184. [Google Scholar] [CrossRef]
- Gulsuner, S.; Walsh, T.; Watts, A.C.; Lee, M.K.; Thornton, A.M.; Casadei, S.; Rippey, C.; Shahin, H.; Nimgaonkar, V.L.; Go, R.C.P.; et al. Spatial and Temporal Mapping of de Novo Mutations in Schizophrenia to a Fetal Prefrontal Cortical Network. Cell 2013, 154, 518–529. [Google Scholar] [CrossRef]
- O’dushlaine, C.; Rossin, L.; Lee, P.H.; Duncan, L.; Parikshak, N.N.; Newhouse, S.; Ripke, S.; Neale, B.M.; Purcell, S.M.; Posthuma, D.; et al. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 2015, 18, 199–209. [Google Scholar] [CrossRef]
- Stilo, S.A.; Murray, R.M. Non-Genetic Factors in Schizophrenia. Curr. Psychiatry Rep. 2019, 21, 100. [Google Scholar] [CrossRef]
- Weinberger, D.R. From neuropathology to neurodevelopment. Lancet 1995, 346, 552–557. [Google Scholar] [CrossRef]
- Birnbaum, R.; Weinberger, D.R. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat. Rev. Neurosci. 2017, 18, 727–740. [Google Scholar] [CrossRef]
- Jaaro-Peled, H.; Sawa, A. Neurodevelopmental Factors in Schizophrenia. Psychiatry Clin. North Am. 2020, 43, 263–274. [Google Scholar] [CrossRef]
- Schmitt, A.; Falkai, P.; Papiol, S. Neurodevelopmental disturbances in schizophrenia: Evidence from genetic and environmental factors. J. Neural Transm. 2022, 12, 1–11. [Google Scholar] [CrossRef]
- Owen, M.J.; O’Donovan, M.C. Schizophrenia and the neurodevelopmental continuum: Evidence from genomics. World Psychiatry 2017, 16, 227–235. [Google Scholar] [CrossRef]
- Anttila, V.; Bulik-Sullivan, B.; Finucane, H.K.; Walters, R.K.; Bras, J.; Duncan, L.; Escott-Price, V.; Falcone, G.J.; Gormley, P.; Malik, R.; et al. Analysis of shared heritability in common disorders of the brain. Science (1979) 2018, 360, eaap8757. [Google Scholar] [CrossRef]
- Rees, E.; Creeth, H.D.J.; Hwu, H.-G.; Chen, W.J.; Tsuang, M.; Glatt, S.J.; Rey, R.; Kirov, G.; Walters, J.T.R.; Holmans, P.; et al. Schizophrenia, autism spectrum disorders and developmental disorders share specific disruptive coding mutations. Nat. Commun. 2021, 12, 5353. [Google Scholar] [CrossRef]
- Kushima, I.; Nakatochi, M.; Aleksic, B.; Okada, T.; Kimura, H.; Kato, H.; Morikawa, M.; Inada, T.; Ishizuka, K.; Torii, Y.; et al. Cross-Disorder Analysis of Genic and Regulatory Copy Number Variations in Bipolar Disorder, Schizophrenia, and Autism Spectrum Disorder. Biol. Psychiatry 2022, 92, 362–374. [Google Scholar] [CrossRef]
- Hindley, G.; Frei, O.; Shadrin, A.A.; Cheng, W.; O’Connell, K.S.; Icick, R.; Parker, N.; Bahrami, S.; Karadag, N.; Roelfs, D.; et al. Charting the Landscape of Genetic Overlap Between Mental Disorders and Related Traits Beyond Genetic Correlation. Am. J. Psychiatry 2022, 179, 833–843. [Google Scholar] [CrossRef]
- Haukka, J.; Suvisaari, J.; Lönnqvist, J. Fertility of Patients with Schizophrenia, Their Siblings, and the General Population: A Cohort Study From 1950 to 1959 in Finland. Am. J. Psychiatry 2003, 160, 460–463. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.J.; Hearle, J.; Jenner, L.; Plant, K.; Drummond, A.; Barkla, J.M. The fertility and fecundity of patients with psychoses. Acta Psychiatr. Scand. 1999, 99, 441–446. [Google Scholar] [CrossRef]
- Huxley, J.; Mayr, E.; Osmond, H.; Hoffer, A. Schizophrenia as a Genetic Morphism. Nature 1964, 204, 220–221. [Google Scholar] [CrossRef]
- Kondrashov, F.A. Bioinformatical assay of human gene morbidity. Nucleic Acids Res. 2004, 32, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Charlson, F.J.; Ferrari, A.J.; Santomauro, D.F.; Diminic, S.; Stockings, E.; Scott, J.G.; McGrath, J.J.; Whiteford, H.A. Global Epidemiology and Burden of Schizophrenia: Findings from the Global Burden of Disease Study 2016. Schizophr. Bull. 2018, 44, 1195–1203. [Google Scholar] [CrossRef]
- Gulsuner, S.; Stein, D.J.; Susser, E.S.; Sibeko, G.; Pretorius, A.; Walsh, T.; Majara, L.; Mndini, M.M.; Mqulwana, S.G.; Ntola, O.A.; et al. Genetics of Schizophrenia in the South African Xhosa. Science 2020, 367, 569–573. [Google Scholar] [CrossRef]
- Lam, M.; Chen, C.-Y.; Li, Z.; Martin, A.R.; Bryois, J.; Ma, X.; Gaspar, H.; Ikeda, M.; Benyamin, B.; Brown, B.C.; et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 2019, 51, 1670–1678. [Google Scholar] [CrossRef]
- Escott-Price, V.; Pardiñas, A.F.; Santiago, E.; Walters, J.; Kirov, G.; Owen, M.J.; O’Donovan, M.C. The Relationship between Common Variant Schizophrenia Liability and Number of Offspring in the UK Biobank. Am. J. Psychiatry 2019, 176, 661–666. [Google Scholar] [CrossRef]
- Pardiñas, A.F.; Holmans, P.; Pocklington, A.J.; Escott-Price, V.; Ripke, S.; Carrera, N.; Legge, S.E.; Bishop, S.; Cameron, D.; Hamshere, M.L.; et al. Common Schizophrenia Alleles Are Enriched in Mutation-Intolerant Genes and in Regions under Strong Background Selection. Nat. Genet. 2018, 50, 381–389. [Google Scholar] [CrossRef]
- van Dongen, J.; Boomsma, D.I. The Evolutionary Paradox and the Missing Heritability of Schizophrenia. Am. J. Med. Genet. Part B: Neuropsychiatr. Genet. 2013, 162, 122–136. [Google Scholar] [CrossRef]
- Keller, M.C. Evolutionary Perspectives on Genetic and Environmental Risk Factors for Psychiatric Disorders. Annu. Rev. Clin. Psychol. 2018, 14, 471–493. [Google Scholar] [CrossRef]
- Crow, T.J. Schizophrenia as the price that Homo sapiens pays for language: A resolution of the central paradox in the origin of the species. Brain Res. Brain Res. Rev. 2000, 31, 118–129. [Google Scholar] [CrossRef]
- Burns, J.K. An Evolutionary Theory of Schizophrenia: Cortical Connectivity, Metarepresentation, and the Social Brain. Behav. Brain Sci. 2004, 27, 831–885. [Google Scholar] [CrossRef]
- Wynn, T.; Coolidge, F.L. The Implications of the Working Memory Model for the Evolution of Modern Cognition. Int. J. Evol. Biol. 2011, 2011, 1–12. [Google Scholar] [CrossRef]
- Rakic, P. Evolution of the neocortex: A perspective from developmental biology. Nat. Rev. Neurosci. 2009, 10, 724–735. [Google Scholar] [CrossRef]
- Geschwind, D.H.; Rakic, P. Cortical Evolution: Judge the Brain by Its Cover. Neuron 2013, 80, 633–647. [Google Scholar] [CrossRef]
- Rakic, P. Radial Unit Hypothesis of Neocortical Expansion. Novartis. Found. Symp. 2000, 228, 30–45. [Google Scholar] [CrossRef]
- Herculano-Houzel, S. The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Proc. Natl. Acad. Sci. USA 2012, 109, 10661–10668. [Google Scholar] [CrossRef]
- Otani, T.; Marchetto, M.C.; Gage, F.H.; Simons, B.D.; Livesey, F.J. 2D and 3D Stem Cell Models of Primate Cortical Development Identify Species-Specific Differences in Progenitor Behavior Contributing to Brain Size. Cell Stem Cell 2016, 18, 467–480. [Google Scholar] [CrossRef]
- Zhu, Y.; Sousa, A.M.M.; Gao, T.; Skarica, M.; Li, M.; Santpere, G.; Esteller-Cucala, P.; Juan, D.; Ferrández-Peral, L.; Gulden, F.O.; et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 2018, 362, eaat8077. [Google Scholar] [CrossRef]
- Mora-Bermúdez, F.; Badsha, F.; Kanton, S.; Camp, J.G.; Vernot, B.; Köhler, K.; Voigt, B.; Okita, K.; Maricic, T.; He, Z.; et al. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife 2016, 5, e18683. [Google Scholar] [CrossRef]
- O’Bleness, M.; Searles, V.B.; Varki, A.; Gagneux, P.; Sikela, J.M. Evolution of genetic and genomic features unique to the human lineage. Nat. Rev. Genet. 2012, 13, 853–866. [Google Scholar] [CrossRef]
- Levine, M.; Tjian, R. Transcription regulation and animal diversity. Nature 2003, 424, 147–151. [Google Scholar] [CrossRef]
- Liu, X.; Somel, M.; Tang, L.; Yan, Z.; Jiang, X.; Guo, S.; Yuan, Y.; He, L.; Oleksiak, A.; Zhang, Y.; et al. Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 2012, 22, 611–622. [Google Scholar] [CrossRef]
- Pollard, K.S.; Salama, S.R.; King, B.; Kern, A.D.; Dreszer, T.; Katzman, S.; Siepel, A.; Pedersen, J.S.; Bejerano, G.; Baertsch, R.; et al. Forces Shaping the Fastest Evolving Regions in the Human Genome. PLOS Genet. 2006, 2, 1599–1611. [Google Scholar] [CrossRef]
- Prabhakar, S.; Noonan, J.P.; Pääbo, S.; Rubin, E.M. Accelerated Evolution of Conserved Noncoding Sequences in Humans. Science 2006, 314, 786. [Google Scholar] [CrossRef]
- Bird, C.P.; Stranger, B.E.; Liu, M.; Thomas, D.J.; Ingle, C.E.; Beazley, C.; Miller, W.; Hurles, M.E.; Dermitzakis, E.T. Fast-evolving noncoding sequences in the human genome. Genome Biol. 2007, 8, R118. [Google Scholar] [CrossRef]
- Bush, E.C.; Lahn, B.T. A genome-wide screen for noncoding elements important in primate evolution. BMC Evol. Biol. 2008, 8, 17. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Garber, M.; Zuk, O.; Lin, M.F.; Parker, B.J.; Washietl, S.; Kheradpour, P.; Ernst, J.; Jordan, G.; Mauceli, E.; et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 2011, 478, 476–482. [Google Scholar] [CrossRef]
- Gittelman, R.M.; Hun, E.; Ay, F.; Madeoy, J.; Pennacchio, L.; Noble, W.S.; Hawkins, R.D.; Akey, J.M. Comprehensive identification and analysis of human accelerated regulatory DNA. Genome Res. 2015, 25, 1245–1255. [Google Scholar] [CrossRef]
- Pollard, K.S.; Salama, S.R.; Lambert, N.; Lambot, M.-A.; Coppens, S.; Pedersen, J.S.; Katzman, S.; King, B.; Onodera, C.; Siepel, A.; et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 2006, 443, 167–172. [Google Scholar] [CrossRef]
- McLean, C.Y.; Reno, P.L.; Pollen, A.A.; Bassan, A.I.; Capellini, T.D.; Guenther, C.; Indjeian, V.B.; Lim, X.; Menke, D.B.; Schaar, B.T.; et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 2011, 471, 216–219. [Google Scholar] [CrossRef]
- Somel, M.; Liu, X.; Khaitovich, P. Human brain evolution: Transcripts, metabolites and their regulators. Nat. Rev. Neurosci. 2013, 14, 112–127. [Google Scholar] [CrossRef]
- Capra, J.A.; Erwin, G.D.; McKinsey, G.; Rubenstein, J.L.R.; Pollard, K.S. Many human accelerated regions are developmental enhancers. Philos. Trans. R. Soc. B: Biol. Sci. 2013, 368, 20130025. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Pollard, K.S. Exploring the genesis and functions of Human Accelerated Regions sheds light on their role in human evolution. Curr. Opin. Genet. Dev. 2014, 29, 15–21. [Google Scholar] [CrossRef]
- Doan, R.N.; Bae, B.-I.; Cubelos, B.; Chang, C.; Hossain, A.A.; Al-Saad, S.; Mukaddes, N.M.; Oner, O.; Al-Saffar, M.; Balkhy, S.; et al. Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 2016, 167, 341–354.e12. [Google Scholar] [CrossRef]
- Burbano, H.A.; Green, R.E.; Maricic, T.; Lalueza-Fox, C.; de la Rasilla, M.; Rosas, A.; Kelso, J.; Pollard, K.S.; Lachmann, M.; Pääbo, S. Analysis of Human Accelerated DNA Regions Using Archaic Hominin Genomes. PLoS ONE 2012, 7, e32877. [Google Scholar] [CrossRef]
- Whalen, S.; Pollard, K.S. Enhancer Function and Evolutionary Roles of Human Accelerated Regions. Annu. Rev. Genet. 2022, 56, 423–439c. [Google Scholar] [CrossRef]
- Levchenko, A.; Kanapin, A.; Samsonova, A.; Gainetdinov, R.R. Human Accelerated Regions and Other Human-Specific Sequence Variations in the Context of Evolution and Their Relevance for Brain Development. Genome Biol. Evol. 2018, 10, 166–188. [Google Scholar] [CrossRef]
- Silver, D.L. Genomic divergence and brain evolution: How regulatory DNA influences development of the cerebral cortex. Bioessays 2016, 38, 162–171. [Google Scholar] [CrossRef]
- Doan, R.N.; Shin, T.; Walsh, C.A. Evolutionary Changes in Transcriptional Regulation: Insights into Human Behavior and Neurological Conditions. Annu. Rev. Neurosci. 2018, 41, 185–206. [Google Scholar] [CrossRef]
- Won, H.; Huang, J.; Opland, C.K.; Hartl, C.L.; Geschwind, D.H. Human evolved regulatory elements modulate genes involved in cortical expansion and neurodevelopmental disease susceptibility. Nat. Commun. 2019, 10, 2396. [Google Scholar] [CrossRef]
- Uebbing, S.; Gockley, J.; Reilly, S.K.; Kocher, A.A.; Geller, E.; Gandotra, N.; Scharfe, C.; Cotney, J.; Noonan, J.P. Massively parallel discovery of human-specific substitutions that alter enhancer activity. Proc. Natl. Acad. Sci. USA 2021, 118, e2102856118. [Google Scholar] [CrossRef]
- Girskis, K.M.; Stergachis, A.B.; DeGennaro, E.M.; Doan, R.N.; Qian, X.; Johnson, M.B.; Wang, P.P.; Sejourne, G.M.; Nagy, M.A.; Pollina, E.A.; et al. Rewiring of human neurodevelopmental gene regulatory programs by human accelerated regions. Neuron 2021, 109, 3239–3251.e7. [Google Scholar] [CrossRef]
- Suzuki, S.; Miyabe, E.; Inagaki, S. Novel brain-expressed noncoding RNA, HSTR1, identified at a human-specific variable number tandem repeat locus with a human accelerated region. Biochem. Biophys. Res. Commun. 2018, 503, 1478–1483. [Google Scholar] [CrossRef]
- Kamm, G.B.; Pisciottano, F.; Kliger, R.; Franchini, L.F. The Developmental Brain Gene NPAS3 Contains the Largest Number of Accelerated Regulatory Sequences in the Human Genome. Mol. Biol. Evol. 2013, 30, 1088–1102. [Google Scholar] [CrossRef]
- Kamm, G.B.; López-Leal, R.; Lorenzo, J.R.; Franchini, L.F. A fast-evolving human NPAS3 enhancer gained reporter expression in the developing forebrain of transgenic mice. Philos. Trans. R. Soc. B: Biol. Sci. 2013, 368, 20130019. [Google Scholar] [CrossRef]
- Oksenberg, N.; Stevison, L.; Wall, J.D.; Ahituv, N. Function and Regulation of AUTS2, a Gene Implicated in Autism and Human Evolution. PLOS Genet. 2013, 9, e1003221. [Google Scholar] [CrossRef]
- Boyd, J.L.; Skove, S.L.; Rouanet, J.P.; Pilaz, L.-J.; Bepler, T.; Gordân, R.; Wray, G.A.; Silver, D.L. Human-Chimpanzee Differences in a FZD8 Enhancer Alter Cell-Cycle Dynamics in the Developing Neocortex. Curr. Biol. 2015, 25, 772–779. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; de Lange, S.C.; Scholtens, L.H.; Watanabe, K.; Ardesch, D.J.; Jansen, P.R.; Savage, J.E.; Li, L.; Preuss, T.M.; Rilling, J.K.; et al. Genetic mapping and evolutionary analysis of human-expanded cognitive networks. Nat. Commun. 2019, 10, 4839. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wei, Y.; Zhang, J.; Ma, J.; Yi, Y.; Gu, Y.; Li, L.M.W.; Lin, Y.; Dai, Z. Gene expression associated with individual variability in intrinsic functional connectivity. Neuroimage 2021, 245, 118743. [Google Scholar] [CrossRef] [PubMed]
- Luppi, A.I.; Mediano, P.A.M.; Rosas, F.E.; Holland, N.; Fryer, T.D.; O’Brien, J.T.; Rowe, J.B.; Menon, D.K.; Bor, D.; Stamatakis, E.A. A synergistic core for human brain evolution and cognition. Nat. Neurosci. 2022, 25, 771–782. [Google Scholar] [CrossRef]
- Cheung, J.P.; Tubbs, J.D.; Sham, P.C. Extended gene set analysis of human neuro-psychiatric traits shows enrichment in brain-expressed human accelerated regions across development. Schizophr. Res. 2022, 246, 148–155. [Google Scholar] [CrossRef]
- Tolosa, A.; Sanjuán, J.; Leal, C.; Costas, J.; Moltó, M.D.; de Frutos, R. Rapid evolving RNA gene HAR1A and Schizophrenia. Schizophr. Res. 2008, 99, 370–372. [Google Scholar] [CrossRef]
- Xu, K.; Schadt, E.E.; Pollard, K.S.; Roussos, P.; Dudley, J.T. Genomic and Network Patterns of Schizophrenia Genetic Variation in Human Evolutionary Accelerated Regions. Mol. Biol. Evol. 2015, 32, 1148–1160. [Google Scholar] [CrossRef]
- González-Peñas, J.; Arrojo, M.; Paz, E.; Brenlla, J.; Páramo, M.; Costas, J. Cumulative role of rare and common putative functional genetic variants at NPAS3 in schizophrenia susceptibility. Am. J. Med. Genet. Part B: Neuropsychiatr. Genet. 2015, 168, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Bettella, F.; Hassani, S.; Wang, Y.; Witoelar, A.; Schork, A.J.; Thompson, W.K.; Collier, D.A.; Desikan, R.S.; Melle, I.; et al. Probing the Association between Early Evolutionary Markers and Schizophrenia. PLoS ONE 2017, 12, e0169227. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, U.; Bhatia, T.; Deshpande, S.N.; Thelma, B. Genetic variations in evolutionary accelerated regions disrupt cognition in schizophrenia. Psychiatry Res. 2022, 314, 114586. [Google Scholar] [CrossRef]
- Bhattacharyya, U.; Deshpande, S.N.; Bhatia, T.; Thelma, B.K. Revisiting Schizophrenia from an Evolutionary Perspective: An Association Study of Recent Evolutionary Markers and Schizophrenia. Schizophr. Bull. 2021, 47, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Erady, C.; Amin, K.; Onilogbo, T.O.A.E.; Tomasik, J.; Jukes-Jones, R.; Umrania, Y.; Bahn, S.; Prabakaran, S. Novel open reading frames in human accelerated regions and transposable elements reveal new leads to understand schizophrenia and bipolar disorder. Mol. Psychiatry 2022, 27, 1455–1468. [Google Scholar] [CrossRef]
- Takahashi, Y.; Terada, T.; Muto, Y. Systems Level Analysis and Identification of Pathways and Key Genes Associated with Delirium. Genes 2020, 11, 1225. [Google Scholar] [CrossRef]
- Schmidt, M.J.; Mirnics, K. Neurodevelopment, GABA System Dysfunction, and Schizophrenia. Neuropsychopharmacology 2015, 40, 190–206. [Google Scholar] [CrossRef]
- Han, J.H.; Suyama, J. Delirium and Dementia. Clin. Geriatr. Med. 2018, 34, 327–354. [Google Scholar] [CrossRef]
- Inouye, S. Delirium in Older Persons. N. Engl. J. Med. 2006, 354, 1157–1165. [Google Scholar] [CrossRef]
- Feng, Z.; Duren, Z.; Xiong, Z.; Wang, S.; Liu, F.; Wong, W.H.; Wang, Y. hReg-CNCC reconstructs a regulatory network in human cranial neural crest cells and annotates variants in a developmental context. Commun. Biol. 2021, 4, 442. [Google Scholar] [CrossRef] [PubMed]
- Cristino, A.S.; Williams, S.M.; Hawi, Z.; An, J.Y.; Bellgrove, M.A.; Schwartz, C.E.; Costa, L.D.F.; Claudianos, C. Neurodevelopmental and neuropsychiatric disorders represent an interconnected molecular system. Mol. Psychiatry 2014, 19, 294–301. [Google Scholar] [CrossRef]
- Padmanabhan, A.; Lynch, C.J.; Schaer, M.; Menon, V.; 401 Quarry, R. The Default Mode Network in Autism HHS Public Access. Biol Psychiatr. Cogn. Neurosci. Neuroimag. 2017, 2, 476–486. [Google Scholar] [CrossRef]
- Hu, M.-L.; Zong, X.-F.; Mann, J.J.; Zheng, J.-J.; Liao, Y.-H.; Li, Z.-C.; He, Y.; Chen, X.-G.; Tang, J.-S. A Review of the Functional and Anatomical Default Mode Network in Schizophrenia. Neurosci. Bull. 2017, 33, 73–84. [Google Scholar] [CrossRef]
- Chang, X.; Shen, H.; Wang, L.; Liu, Z.; Xin, W.; Hu, D.; Miao, D. Altered default mode and fronto-parietal network subsystems in patients with schizophrenia and their unaffected siblings. Brain Res. 2014, 1562, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Haghighat, H.; Mirzarezaee, M.; Araabi, B.N.; Khadem, A. Functional Networks Abnormalities in Autism Spectrum Disorder: Age-Related Hypo and Hyper Connectivity. Brain Topogr. 2021, 34, 306–322. [Google Scholar] [CrossRef] [PubMed]
- Mars, R.B.; Neubert, F.-X.; Noonan, M.A.P.; Sallet, J.; Toni, I.; Rushworth, M.F.S. On the relationship between the “default mode network” and the “social brain”. Front. Hum. Neurosci. 2012, 6, 189. [Google Scholar] [CrossRef]
- Marek, S.; Dosenbach, N.U.F. The frontoparietal network: Function, electrophysiology, and importance of individual precision mapping. Dialog- Clin. Neurosci. 2018, 20, 133–141. [Google Scholar] [CrossRef]
- Roth, G.; Dicke, U. Origin and Evolution of Human Cognition. In Progress in Brain Research; Hofman, M.A., Ed.; Elsevier, B.V.: Amsterdam, The Netherlands, 2019; Volume 250, pp. 285–316. ISBN 9780444643179. [Google Scholar]
- Lesh, T.A.; Niendam, T.A.; Minzenberg, M.J.; Carter, C.S. Cognitive Control Deficits in Schizophrenia: Mechanisms and Meaning. Neuropsychopharmacology 2011, 36, 316–338. [Google Scholar] [CrossRef]
- Barlati, S.; Minelli, A.; Ceraso, A.; Nibbio, G.; Carvalho Silva, R.; Deste, G.; Turrina, C.; Vita, A. Social Cognition in a Research Domain Criteria Perspective: A Bridge Between Schizophrenia and Autism Spectra Disorders. Front. Psychiatry 2020, 11, 806. [Google Scholar] [CrossRef]
- Van den Heuvel, M.P.; Scholtens, L.H.; de Lange, S.C.; Pijnenburg, R.; Cahn, W.; van Haren, N.E.M.; Sommer, I.E.; Bozzali, M.; Koch, K.; Boks, M.P.; et al. Evolutionary modifications in human brain connectivity associated with schizophrenia. Brain 2019, 142, 3991–4002. [Google Scholar] [CrossRef]
- Pickard, B.S.; Christoforou, A.; Thomson, P.A.; Fawkes, A.; Evans, K.L.; Morris, S.W.; Porteous, D.J.; Blackwood, D.H.; Muir, W.J. Interacting haplotypes at the NPAS3 locus alter risk of schizophrenia and bipolar disorder. Mol. Psychiatry 2009, 14, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Perlis, R.H.; Lee, P.H.; Rush, A.J.; Fava, M.; Sachs, G.S.; Lieberman, J.; Hamilton, S.P.; Sullivan, P.; Sklar, P.; et al. Cross-Disorder Genomewide Analysis of Schizophrenia, Bipolar Disorder, and Depression. Am. J. Psychiatry 2010, 167, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.F.; McGrath, J.A.; Thornquist, M.H.; Wolyniec, P.S.; Nestadt, G.; Swartz, K.L.; Lasseter, V.K.; Liang, K.Y.; Pulver, A.E. Genetic heterogeneity in schizophrenia II: Conditional analyses of affected schizophrenia sibling pairs provide evidence for an interaction between markers on chromosome 8p and 14q. Mol. Psychiatry 2002, 7, 658–664. [Google Scholar] [CrossRef]
- Elia, J.; Gai, X.; Xie, H.M.; Perin, J.C.; Geiger, E.; Glessner, J.T.; D’Arcy, M.; DeBerardinis, R.; Frackelton, E.; Kim, C.; et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry 2009, 15, 637–646. [Google Scholar] [CrossRef]
- Girirajan, S.; Brkanac, Z.; Coe, B.P.; Baker, C.; Vives, L.; Vu, T.H.; Shafer, N.; Bernier, R.; Ferrero, G.B.; Silengo, M.; et al. Relative Burden of Large CNVs on a Range of Neurodevelopmental Phenotypes. PLOS Genet. 2011, 7, e1002334. [Google Scholar] [CrossRef] [PubMed]
- Talkowski, M.E.; Rosenfeld, J.A.; Blumenthal, I.; Pillalamarri, V.; Chiang, C.; Heilbut, A.; Ernst, C.; Hanscom, C.; Rossin, E.; Lindgren, A.M.; et al. Sequencing Chromosomal Abnormalities Reveals Neurodevelopmental Loci that Confer Risk across Diagnostic Boundaries. Cell 2012, 149, 525–537. [Google Scholar] [CrossRef]
- Oksenberg, N.; Ahituv, N. The role of AUTS2 in neurodevelopment and human evolution. Trends Genet. 2013, 29, 600–608. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, Y.-H.; Wei, S.-G.; Zhang, H.-B.; Fu, D.-K.; Feng, Z.-F.; Guan, F.-L.; Zhu, Y.-S.; Li, S.-B. Association Study Identifying a New Susceptibility Gene (AUTS2) for Schizophrenia. Int. J. Mol. Sci. 2014, 15, 19406–19416. [Google Scholar] [CrossRef]
- Greenbaum, L.; Alkelai, A.; Rigbi, A.; Kohn, Y.; Lerer, B. Evidence for association of the GLI2 gene with tardive dyskinesia in patients with chronic schizophrenia. Mov. Disord. 2010, 25, 2809–2817. [Google Scholar] [CrossRef]
- Liu, Y.; Qu, H.-Q.; Chang, X.; Tian, L.; Glessner, J.; Sleiman, P.A.M.; Hakonarson, H. Expansion of Schizophrenia Gene Network Knowledge Using Machine Learning Selected Signals from Dorsolateral Prefrontal Cortex and Amygdala RNA-seq Data. Front. Psychiatry 2022, 13, 797329. [Google Scholar] [CrossRef]
- Rosato, M.; Stringer, S.; Gebuis, T.; Paliukhovich, I.; Li, K.W.; Posthuma, D.; Sullivan, P.F.; Smit, A.B.; van Kesteren, R.E. Combined cellomics and proteomics analysis reveals shared neuronal morphology and molecular pathway phenotypes for multiple schizophrenia risk genes. Mol. Psychiatry 2021, 26, 784–799. [Google Scholar] [CrossRef]
- Chu, X.-Y.; Quan, Y.; Zhang, H.-Y. Human accelerated genome regions with value in medical genetics and drug discovery. Drug Discov. Today 2020, 25, 821–827. [Google Scholar] [CrossRef]
- Florio, M.; Huttner, W.B. Neural progenitors, neurogenesis and the evolution of the neocortex. Development 2014, 141, 2182–2194. [Google Scholar] [CrossRef]
- Borrell, V.; Reillo, I. Emerging roles of neural stem cells in cerebral cortex development and evolution. Dev. Neurobiol. 2012, 72, 955–971. [Google Scholar] [CrossRef] [PubMed]
- Lewitus, E.; Kelava, I.; Huttner, W.B. Conical expansion of the outer subventricular zone and the role of neocortical folding in evolution and development. Front. Hum. Neurosci. 2013, 7, 424. [Google Scholar] [CrossRef]
- Faissner, A.; Reinhard, J. The extracellular matrix compartment of neural stem and glial progenitor cells. Glia 2015, 63, 1330–1349. [Google Scholar] [CrossRef]
- Kyrousi, C.; Cappello, S. Using brain organoids to study human neurodevelopment, evolution and disease. Wiley Interdiscip. Rev. Dev. Biol. 2020, 9, e347. [Google Scholar] [CrossRef]
- Rakic, P. Specification of Cerebral Cortical Areas. Science 1988, 241, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Grasby, K.L.; Jahanshad, N.; Painter, J.N.; Colodro-Conde, L.; Bralten, J.; Hibar, D.P.; Lind, P.A.; Pizzagalli, F.; Ching, C.R.K.; McMahon, M.A.B.; et al. The genetic architecture of the human cerebral cortex. Science 2020, 367, eaay6690. [Google Scholar] [CrossRef] [PubMed]
- Papiol, S.; Keeser, D.; Hasan, A.; Schneider-Axmann, T.; Raabe, F.; Degenhardt, F.; Rossner, M.J.; Bickeböller, H.; Cantuti-Castelvetri, L.; Simons, M.; et al. Polygenic burden associated to oligodendrocyte precursor cells and radial glia influences the hippocampal volume changes induced by aerobic exercise in schizophrenia patients. Transl. Psychiatry 2019, 9, 284. [Google Scholar] [CrossRef]
- Lisman, J.; Buzsáki, G.; Eichenbaum, H.; Nadel, L.; Ranganath, C.; Redish, A.D. Viewpoints: How the hippocampus contributes to memory, navigation and cognition. Nat. Neurosci. 2017, 20, 1434–1447. [Google Scholar] [CrossRef]
- Opitz, B. Memory Function and the Hippocampus. In The Hippocampus in Clinical Neuroscience; Szabo, K., Hennerici, M., Eds.; S. Karger AG: Berlin, Germany, 2014; Volume 34, pp. 51–59. ISBN 9783318025682. [Google Scholar]
- Franchini, L.F.; Pollard, K.S. Human evolution: The non-coding revolution. BMC Biol. 2017, 15, 89. [Google Scholar] [CrossRef]
- Huo, Y.; Li, S.; Liu, J.; Li, X.; Luo, X.-J. Functional genomics reveal gene regulatory mechanisms underlying schizophrenia risk. Nat. Commun. 2019, 10, 670. [Google Scholar] [CrossRef] [PubMed]
- Gusev, A.; Mancuso, N.; Won, H.; Kousi, M.; Finucane, H.K.; Reshef, Y.; Song, L.; Safi, A.; McCarroll, S.; Neale, B.M.; et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018, 50, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Gandal, M.J.; Zhang, P.; Hadjimichael, E.; Walker, R.L.; Chen, C.; Liu, S.; Won, H.; Van Bakel, H.; Varghese, M.; Wang, Y.; et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018, 362, eaat8127. [Google Scholar] [CrossRef] [PubMed]
- Parikshak, N.N.; Swarup, V.; Belgard, T.G.; Irimia, M.; Ramaswami, G.; Gandal, M.J.; Hartl, C.; Leppa, V.; Ubieta, L.D.L.T.; Huang, J.; et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 2016, 540, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Bettella, F.; Mattingsdal, M.; Wang, Y.; Witoelar, A.; Schork, A.J.; Thompson, W.K.; Zuber, V.; Winsvold, B.S.; Zwart, J.-A.; et al. Genetic Markers of Human Evolution Are Enriched in Schizophrenia. Biol. Psychiatry 2016, 80, 284–292. [Google Scholar] [CrossRef]
- Srinivasan, S.; Bettella, F.; Frei, O.; Hill, W.D.; Wang, Y.; Witoelar, A.; Schork, A.J.; Thompson, W.K.; Davies, G.; Desikan, R.S.; et al. Enrichment of genetic markers of recent human evolution in educational and cognitive traits. Sci. Rep. 2018, 8, 12585. [Google Scholar] [CrossRef]
- Pinson, A.; Xing, L.; Namba, T.; Kalebic, N.; Peters, J.; Oegema, C.E.; Traikov, S.; Reppe, K.; Riesenberg, S.; Maricic, T.; et al. Human TKTL1 implies greater neurogenesis in frontal neocortex of modern humans than Neanderthals. Science 2022, 377, eabl6422. [Google Scholar] [CrossRef]
- Banerjee, N.; Polushina, T.; Bettella, F.; Giddaluru, S.; Steen, V.M.; Andreassen, O.A.; Le Hellard, S. Recently evolved human-specific methylated regions are enriched in schizophrenia signals. BMC Evol. Biol. 2018, 18, 63. [Google Scholar] [CrossRef] [PubMed]
| Main Objective (Methodology) | Main Results |
|---|---|
| Doan et al. [52] To characterise the functions of HARs in neurodevelopment (in silico analyses) |
|
| Won et al. [58] To map and characterise HAR expression patterns, tissue, and cell specificity (in silico analyses) |
|
| Uerbbing et al. [59] To study the effect of HAR variability in human neurogenesis (massively parallel reporter assay (MPRA) in human neural stem cells) |
|
| Girskis et al. [60] To study HARs’ effect on the recent evolution of the human cerebral cortex (Capture MPRAs in human neural stem cells and neurospheres) |
|
| HAR | Gene—Function | Validation Methodology | Main Results |
|---|---|---|---|
| HAR1 [47] | HAR1F and HAR1R (Highly Accelerated Region 1A and 1B, 20q13.33) Non-Protein-Coding RNAs. Unknown function. | Expression assay on human embryonic and adult brain. Comparative expression analysis in embryonic macaque, mouse, and human brains |
|
| HACN96, HAR202, 2xHAR142, HAR89, 2xHAR223, 2xHAR157, 2xHAR122, HAR96, HACNS658, HAR189, HACNS553, HAR21, HACNS221, HAR173 [62,63] | NPAS3 (Neuronal PAS Domain Protein 3, 14q13.1) Transcription factor involved in the control of neurosignalling pathways during neurogenesis. | Expression assays in transgenic zebrafish and mice. Comparative expression of human, chimpanzee, and mouse HAR orthologs. Hybridisation in transgenic mice |
|
| HAR31, HACNS174, HACNS369 [64] | AUTS2 (Autism Susceptibility Candidate 2, 7q11.22) Transcription factor involved in neurodevelopmental regulation, axon and dendrite elongation, and neuronal migration. | Targeted expression assays in transgenic zebrafish and mice |
|
| HARE5 [65] | FZD8 (Frizzled Class Receptor 8, 10p11.21) Receptor in the WNT pathway implicated in cortical development. | Comparative expression of human and chimpanzee HAR orthologs in transgenic mice |
|
| HAR-HSTR1 [61] | HSTR1 (Human-Specific Tandem Repeat 1, 20p) Non-Protein-Coding RNA. Unknown function. Non-annotated gene. | Targeted expression assays in HEK293T cells. Comparative expression of human, chimpanzee, gorilla, and orangutan HAR orthologs through luciferase reporter assays |
|
| HAR426 [52] | CUX1 (Cut Like Homeobox 1, 7q22.1) Transcription factor involved in the control of neuronal differentiation. | Mutant and wild-type HAR mutation effect through luciferase reporter assays in mouse neural-precursor-like cells |
|
| HAR169 [52] | PTBP2 (Polypyrimidine Tract Binding Protein 2, 1p21.3) RNA-binding protein and brain-specific splicing regulator essential for neuronal differentiation. | Mutant and wild-type HAR mutation effect through luciferase reporter assays in mouse neural-precursor-like cells. Massively parallel reporter assays in mouse neurospheres |
|
| HAR1325 [52] | GPC4 (Glypican Proteoglycan 4, Xq26.2) Protein essential for excitatory synapse development in mice and dosage-sensitive gene in adult human brain. | Mutant and wild-type HAR mutation effect through luciferase reporter assays in mouse neural-precursor-like cells. Massively parallel reporter assays in mouse neurospheres |
|
| HAR4 [58] | GLI2 (GLI family zinc finger 2, 2q14.2) Transcription factor in the Sonic Hedgehog (Shh) pathway critical for neural tube formation. Involved in cell growth and specialisation. | Targeted expression in primary human neural progenitor cells |
|
| HAR1225 [58] | GLI3 (GLI family zinc finger 3, 7p14.1) Transcription factor in the Shh pathway critical for neural tube formation. Essential for dorsal–ventral patterning of telencephalon and cortex formation in humans. | Targeted expression in primary human neural progenitor cells |
|
| HAR342 [58] | TBR1 (T-Box Brain Transcription Factor 1, 2q24.2) Transcriptional factor repressor involved in neuronal migration, laminar and areal identity, and axonal projection. | Targeted expression in primary human neural progenitor cells |
|
| HAR2635, HAR2636 [60] | PPP1R17 (Protein Phosphatase 1 Regulatory Subunit 17, 7p14.3) Phosphatase inhibitor involved in neural progenitor cell proliferation and expression regulation in the developing human cortex. | Targeted chromatin conformation capture 3C interaction analysis |
|
| Main Objective | Main Methodology | Main Results |
|---|---|---|
| Wei et al. [66] To study the evolutionary genetics of cortical expansion using HAR gene expression | Correlation analyses of HAR genes expression with cortical expansion differences from human vs. chimpanzee (sMRI data) and human vs. primates comparative gene expression. Association analyses of HAR and HAR brain genes with DMN variability (resting state fMRI data), intelligence, and sociability (based on GWAS data) |
|
| Li et al. [67] To study the evolutionary genetics of brain connectivity using HAR brain gene expression | Correlation analyses of HAR brain gene expression with functional connectivity data (resting state fMRI data) |
|
| Luppi et al. [68] To study the evolutionary genetics of redundant and synergistic information organization using HAR brain gene expression | Correlation analyses of HAR brain gene expression with the spatial distribution of synergistic and redundant brain interactions (resting state fMRI data) |
|
| Cheung et al. [69] To test whether genes associated with intelligence are enriched in HARs | Enrichment analyses on HARs, brain expression, and their interaction on intelligence (based on GWAS data) |
|
| Main Objective | Main Methodology | Main Results |
|---|---|---|
| Schizophrenia | ||
| Xu et al. [71] To study HAR enrichment on common variability associated with SCZ | HAR enrichment analysis in SCZ (based on GWAS data) and gene co-expression network analyses |
|
| Srinivassan et al. [73] To study HAR enrichment of common variability associated with SCZ | HAR enrichment analysis in SCZ (based on GWAS data) |
|
| Wei et al. [66] To study the association of HAR genes and HAR brain genes with SCZ | Examination of potential associations of HAR and HAR brain genes with SCZ variability (based on GWAS data) |
|
| Cheung et al. [69] To study HAR gene enrichment on genes associated with neuropsychiatric disorders conditional to developmental gene-expression patterns | HAR gene enrichment analyses in five neuropsychiatric disorders (SCZ, BPD, ASD, MDD, and ADHD, based on GWAS data) conditional to gene expression in five developmental stages |
|
| Erady et al. [76] To investigate nORF associated with HARs in the genetic architecture of SCZ and BPD | Assess the overlap between nORF and nORF differentially expressed in SCZ and BPD and HARs. nORD and HARs overlap enrichment analyses in SCZ and BPD (based on GWAS data) |
|
| Tolosa et al. [70] To study the association of common variants in HAR1F gene with SCZ risk and AH in SCZ | Case–control association study (285 SCZ-spectrum disorders [221 AH and 64 no AH] and 337 HC) of HAR1F gene (six variants genotyped) with SCZ risk |
|
| González-Peñas et al. [72] To study the association of common and rare variants in NPAS3 HARs with SCZ risk | Case–control association study (538 SCZ and 539 HC) of NPAS3 gene (26 variants genotyped) with SCZ risk |
|
| Bhattacharyya et al. [74,75] To assess the association of variants in HARs with SCZ and cognitive performance | Case–control association study (Discovery: 494 patients and 436 healthy controls (HC); Replication: 552 patients and 551 HC) of HARs (49 variants genotyped) with SCZ risk. Case–control association study in a subsample (331 patients and 235 HC) of HARs (49 variants genotyped) with cognition variability |
|
| Other neurodevelopmental psychiatric disorders and related syndromes | ||
| Doan et al. [52] To evaluate the mutational landscape of HARs and their contribution to ASD | HAR gene mapping through in silico chromatin interaction data. Assessment of copy number variants (CNVs) in 2100 ASD-sibs sample. Assessment of rare mutations in HARs through whole-genome sequencing in 218 ASD families |
|
| Won et al. [58] To study the role of HAR genes in the susceptibility for neurodevelopmental disorders | HAR enrichment analysis with genes harbouring loss-of-function variants in ASD, SCZ, and DD data |
|
| Wei et al. [66] To study the association of HAR genes and HAR brain genes with ASD variability and brain structural changes found in psychiatric disorders | Examination of potential associations of HAR and HAR brain genes with genes associated with ASD (based on rare variants of brain disorders). Correlation analyses of HAR brain gene expression with structural alterations across psychiatric disorders (sMRI data on SCZ, BPD, ASD, MDD, OCD) |
|
| Takahashi et al. [77] To identify the molecular pathways associated with delirium and test the enrichment of HAR genes | Functional enrichment analysis of HAR genes in delirium-associated genes (obtained from the toxicogenomics database) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guardiola-Ripoll, M.; Fatjó-Vilas, M. A Systematic Review of the Human Accelerated Regions in Schizophrenia and Related Disorders: Where the Evolutionary and Neurodevelopmental Hypotheses Converge. Int. J. Mol. Sci. 2023, 24, 3597. https://doi.org/10.3390/ijms24043597
Guardiola-Ripoll M, Fatjó-Vilas M. A Systematic Review of the Human Accelerated Regions in Schizophrenia and Related Disorders: Where the Evolutionary and Neurodevelopmental Hypotheses Converge. International Journal of Molecular Sciences. 2023; 24(4):3597. https://doi.org/10.3390/ijms24043597
Chicago/Turabian StyleGuardiola-Ripoll, Maria, and Mar Fatjó-Vilas. 2023. "A Systematic Review of the Human Accelerated Regions in Schizophrenia and Related Disorders: Where the Evolutionary and Neurodevelopmental Hypotheses Converge" International Journal of Molecular Sciences 24, no. 4: 3597. https://doi.org/10.3390/ijms24043597
APA StyleGuardiola-Ripoll, M., & Fatjó-Vilas, M. (2023). A Systematic Review of the Human Accelerated Regions in Schizophrenia and Related Disorders: Where the Evolutionary and Neurodevelopmental Hypotheses Converge. International Journal of Molecular Sciences, 24(4), 3597. https://doi.org/10.3390/ijms24043597





