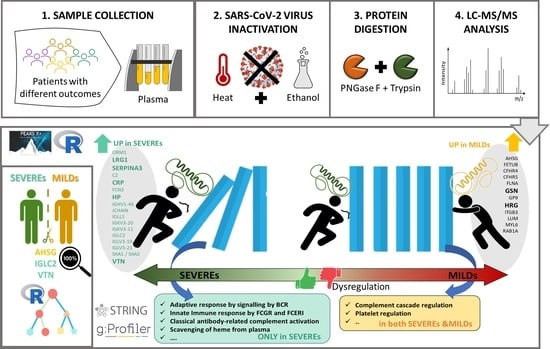
Plasma Proteomic Variables Related to COVID-19 Severity: An Untargeted nLC-MS/MS Investigation
Abstract
1. Introduction
2. Results
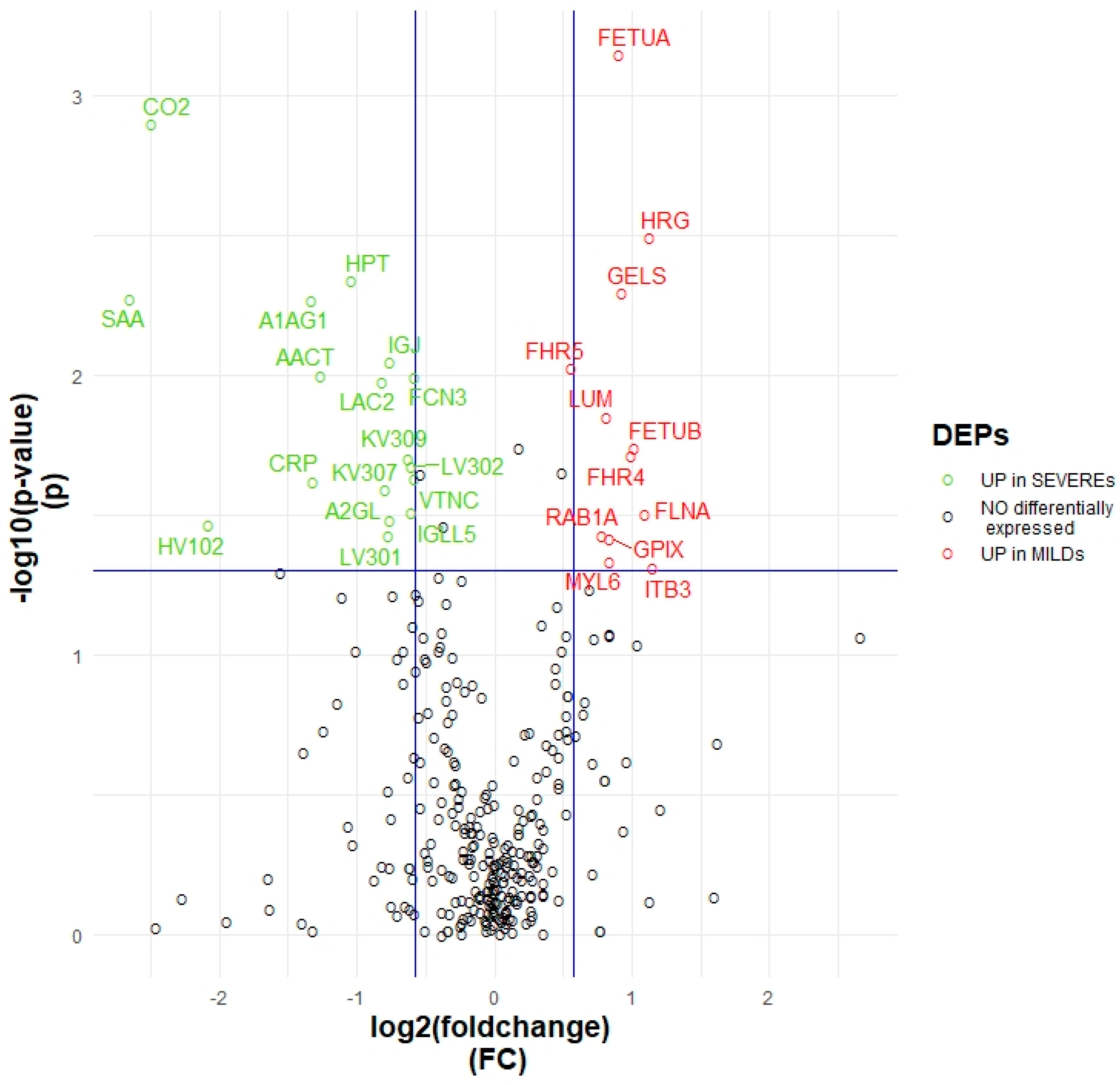
2.1. Proteomic Variables Related to Worse Outcome
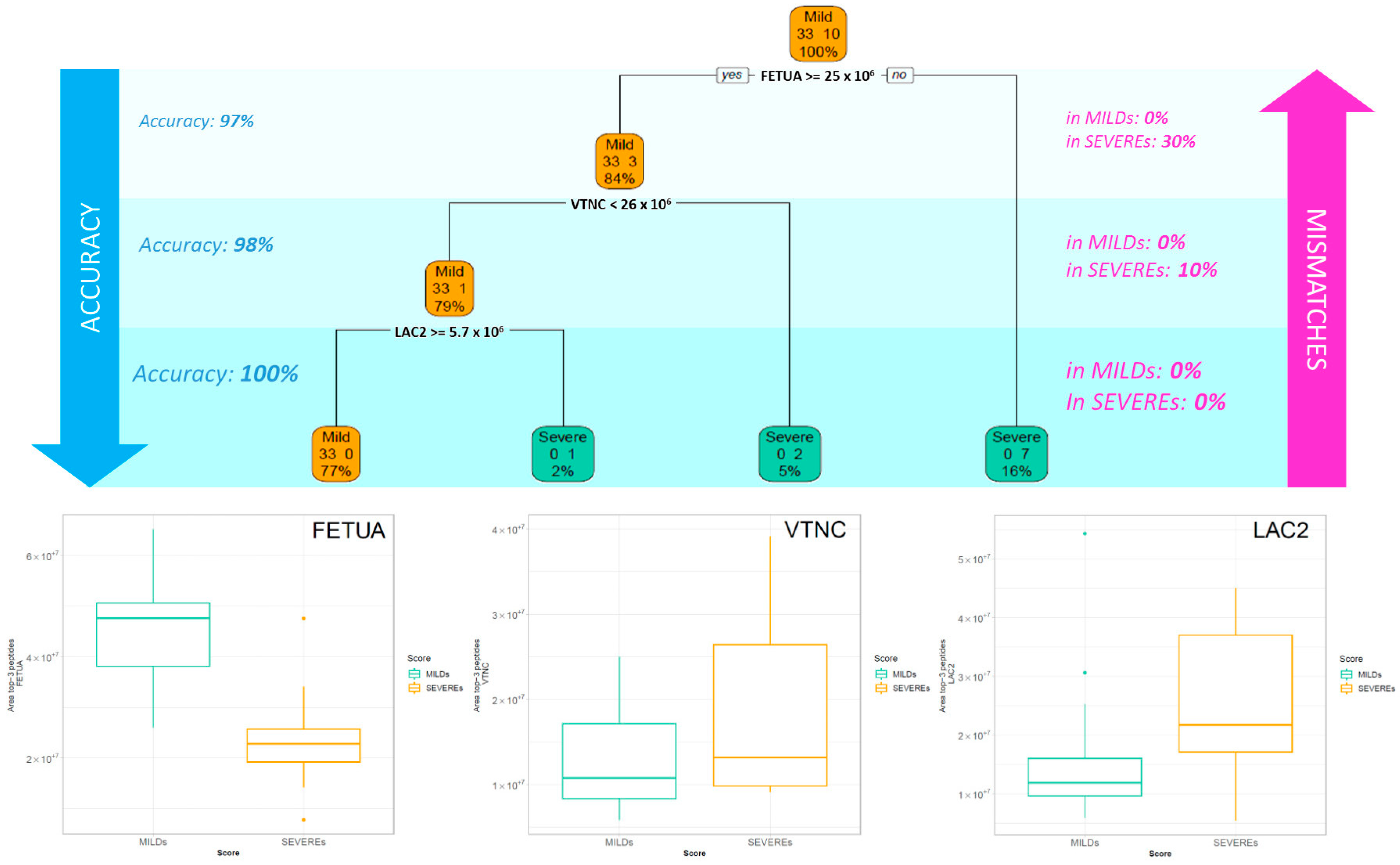
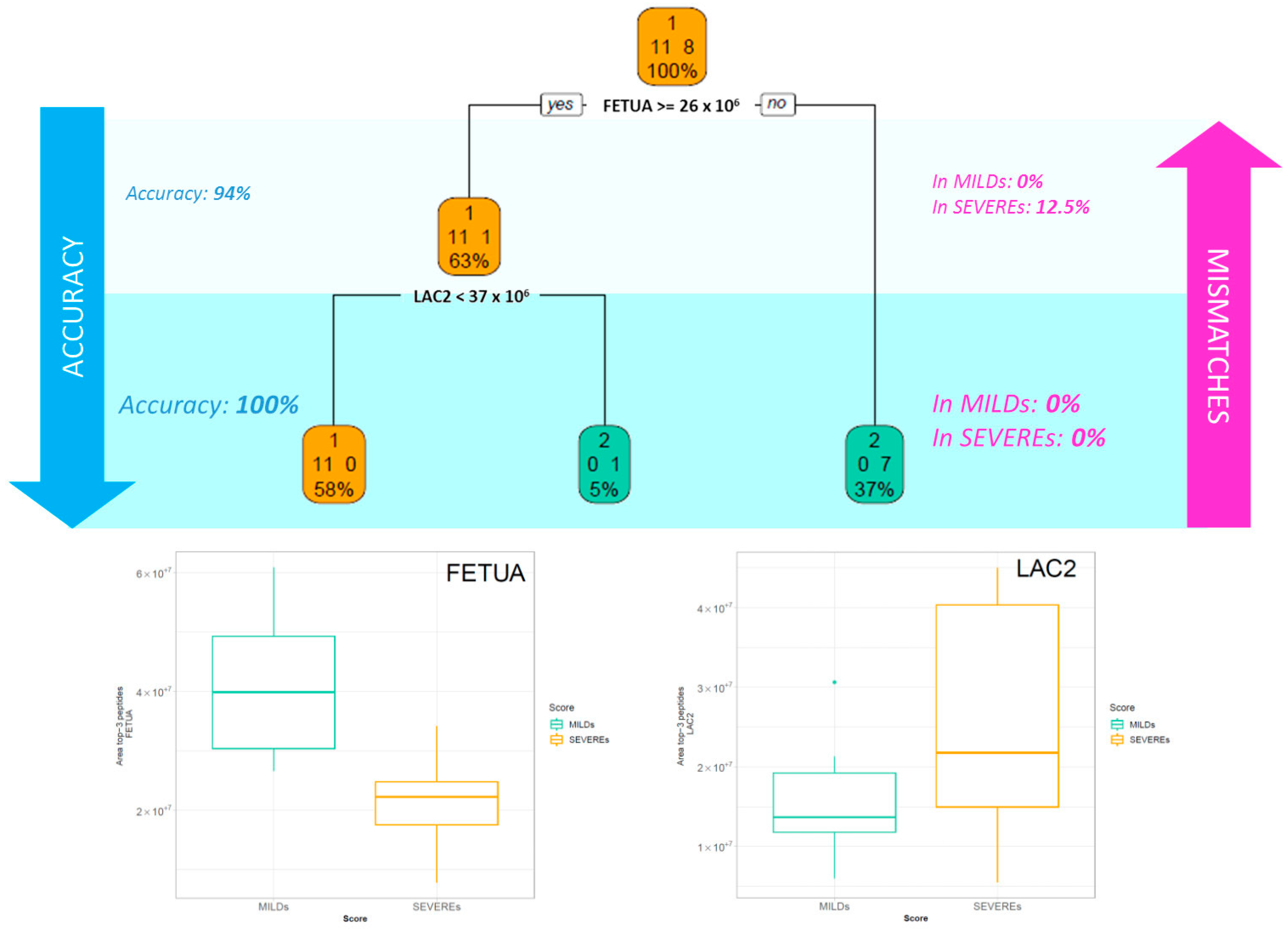
2.2. Classification Tree Related to Severity
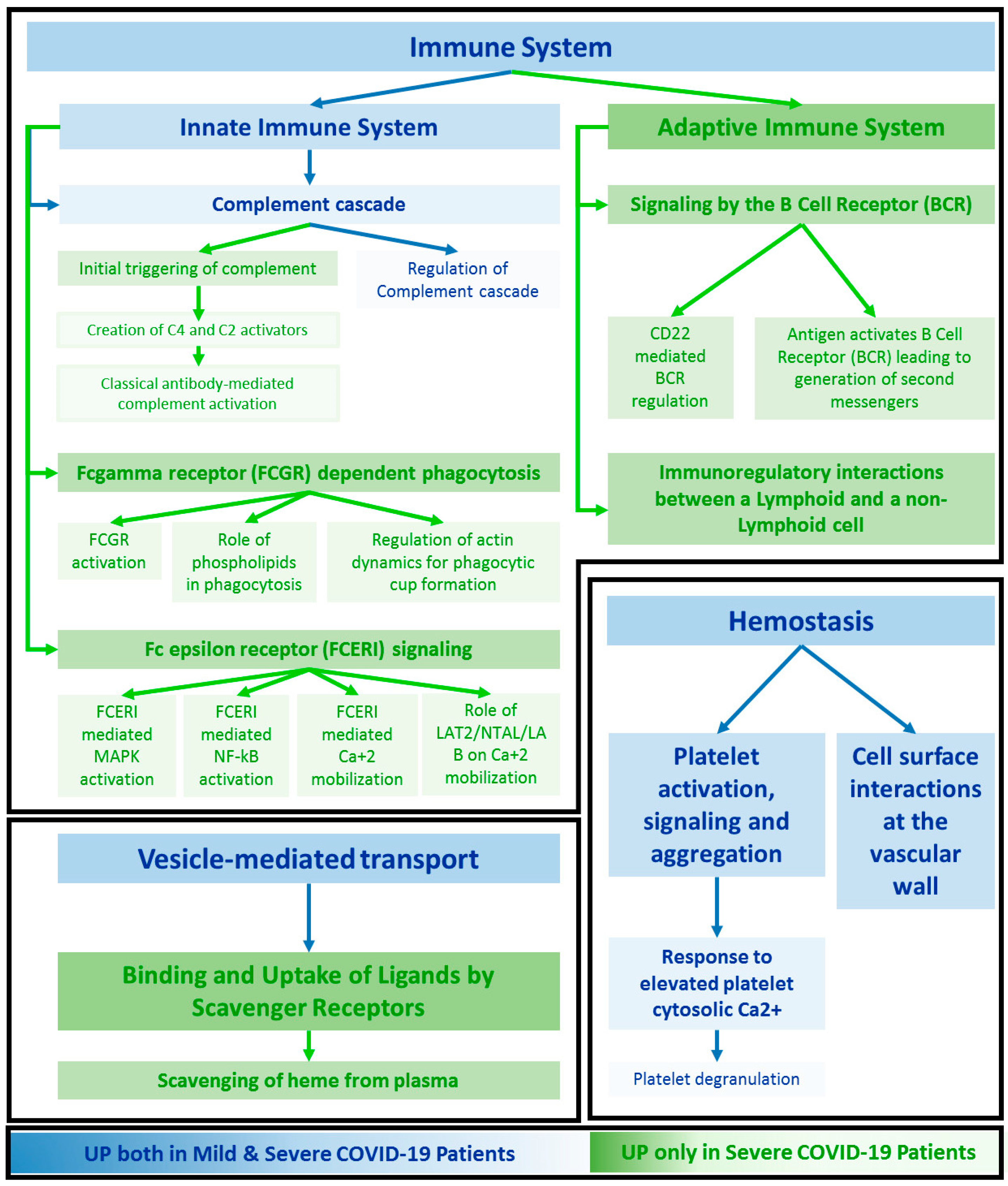
2.3. Functional Annotation
3. Discussion
3.1. Key proteins Characterising Patients Based on the Intensity of the Symptoms
3.2. Functional Signatures of Severity
3.3. Functional Patterns Associated to Both SEVEREs and MILDs
3.4. Specific Enrichment in SARS-CoV-2 Signalling Pathway
3.5. Limitations
4. Materials and Methods
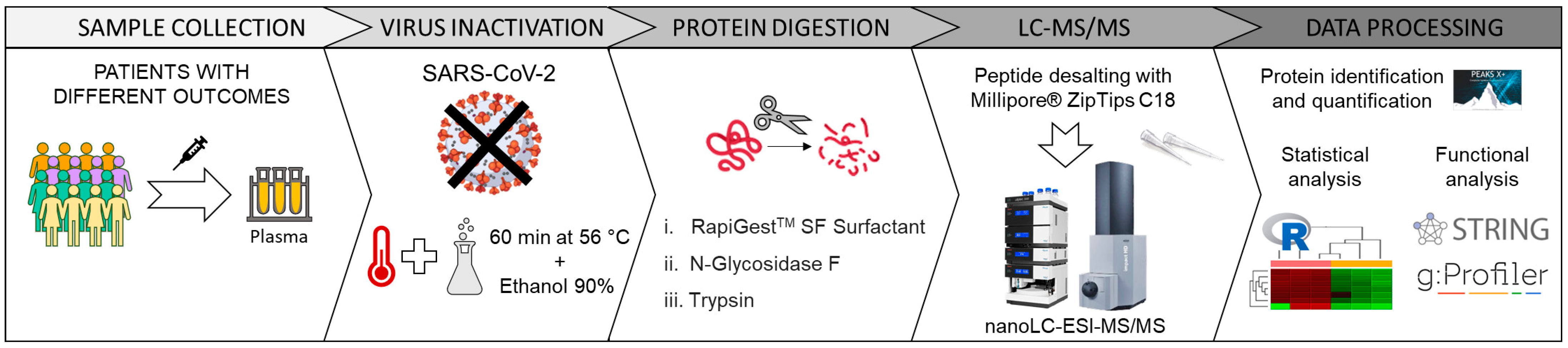
4.1. Plasma Sample Collection
4.2. Sample Inactivation and Deglycosylation
4.3. Protein Digestion
4.4. Mass Spectrometry Analysis
4.5. Data Processing
4.6. Statistical Analysis on Quantified Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Samadizadeh, S.; Masoudi, M.; Rastegar, M.; Salimi, V.; Shahbaz, M.B.; Tahamtan, A. COVID-19: Why does disease severity vary among individuals? Respir. Med. 2021, 180, 106356. [Google Scholar] [CrossRef] [PubMed]
- Merino, J.; Joshi, A.D.; Nguyen, L.H.; Leeming, E.R.; Mazidi, M.; Drew, D.; Gibson, R.; Graham, M.S.; Lo, C.-H.; Capdevila, J.; et al. Diet quality and risk and severity of COVID-19: A prospective cohort study. Gut 2021, 70, 2096–2104. [Google Scholar] [CrossRef] [PubMed]
- Monticelli, M.; Mele, B.H.; Andreotti, G.; Cubellis, M.V.; Riccio, G. Why does SARS-CoV-2 hit in different ways? Host genetic factors can influence the acquisition or the course of COVID-19. Eur. J. Med. Genet. 2021, 64, 104227. [Google Scholar] [CrossRef] [PubMed]
- Ciccosanti, F.; Antonioli, M.; Sacchi, A.; Notari, S.; Farina, A.; Beccacece, A.; Fusto, M.; Vergori, A.; D’Offizi, G.; Taglietti, F.; et al. Proteomic analysis identifies a signature of disease severity in the plasma of COVID-19 pneumonia patients associated to neutrophil, platelet and complement activation. Clin. Proteom. 2022, 19, 38. [Google Scholar] [CrossRef] [PubMed]
- Cosgriff, C.V.; Miano, T.A.; Mathew, D.; Huang, A.C.; Giannini, H.M.; Kuri-Cervantes, L.; Pampena, M.B.; Ittner, C.A.G.; Weisman, A.R.; Agyekum, R.S.; et al. Validating a Proteomic Signature of Severe COVID-19. Crit. Care Explor. 2022, 4, e0800. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Su, J.; Yang, K.; Wei, J.; Wan, H.; Cao, X.; Tan, W.; Wang, H. Elevations of serum cancer biomarkers correlate with severity of COVID-19. J. Med. Virol. 2020, 92, 2036–2041. [Google Scholar] [CrossRef]
- Costanzo, M.; Caterino, M.; Fedele, R.; Cevenini, A.; Pontillo, M.; Barra, L.; Ruoppolo, M. COVIDomics: The Proteomic and Metabolomic Signatures of COVID-19. Int. J. Mol. Sci. 2022, 23, 2414. [Google Scholar] [CrossRef]
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.-S.; Kreidl, M.; et al. Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection. Cell Syst. 2020, 11, 11–24.e4. [Google Scholar] [CrossRef]
- Csősz, É.; Kalló, G.; Márkus, B.; Deák, E.; Csutak, A.; Tőzsér, J. Quantitative body fluid proteomics in medicine—A focus on minimal invasiveness. J. Proteom. 2017, 153, 30–43. [Google Scholar] [CrossRef]
- Gisby, J.S.; Buang, N.B.; Papadaki, A.; Clarke, C.L.; Malik, T.H.; Medjeral-Thomas, N.; Pinheiro, D.; Mortimer, P.M.; Lewis, S.; Sandhu, E.; et al. Multi-omics identify falling LRRC15 as a COVID-19 severity marker and persistent pro-thrombotic signals in convalescence. Nat. Commun. 2022, 13, 7775. [Google Scholar] [CrossRef]
- Captur, G.; Moon, J.C.; Topriceanu, C.-C.; Joy, G.; Swadling, L.; Hallqvist, J.; Doykov, I.; Patel, N.; Spiewak, J.; Baldwin, T.; et al. Plasma proteomic signature predicts who will get persistent symptoms following SARS-CoV-2 infection. Ebiomedicine 2022, 85, 104293. [Google Scholar] [CrossRef]
- Sahin, A.T.; Yurtseven, A.; Dadmand, S.; Ozcan, G.; Akarlar, B.A.; Kucuk, N.E.O.; Senturk, A.; Ergonul, O.; Can, F.; Tuncbag, N.; et al. Plasma proteomics identify potential severity biomarkers from COVID-19 associated network. Proteom. Clin. Appl. 2022, e2200070. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, H.; Kim, S.Y.; Kim, Y.; Lee, J.-S.; Dan, K.; Seong, M.-W.; Han, D. In-depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients. Sci. Rep. 2020, 10, 22418. [Google Scholar] [CrossRef] [PubMed]
- Wynants, L.; Van Calster, B.; Collins, G.S.; Riley, R.D.; Heinze, G.; Schuit, E.; Bonten, M.M.J.; Dahly, D.L.; Damen, J.A.; Debray, T.P.A.; et al. Prediction models for diagnosis and prognosis of covid-19: Systematic review and critical appraisal. BMJ 2020, 369, m1328. [Google Scholar] [CrossRef]
- Patel, H.; Ashton, N.J.; Dobson, R.J.B.; Andersson, L.-M.; Yilmaz, A.; Blennow, K.; Gisslen, M.; Zetterberg, H. Proteomic blood profiling in mild, severe and critical COVID-19 patients. Sci. Rep. 2021, 11, 6357. [Google Scholar] [CrossRef]
- Gordon, A.D.; Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees, Wadsworth Statistics. Biometrics 1984, 40, 874. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Lebreton, J.P.; Joisel, F.; Raoult, J.P.; Lannuzel, B.; Rogez, J.P.; Humbert, G. Serum concentration of human alpha 2 HS glycoprotein during the inflammatory process: Evidence that alpha 2 HS glycoprotein is a negative acute-phase reactant. J. Clin. Investig. 1979, 64, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Przepiera-Będzak, H.; Fischer, K.; Brzosko, M. Axial spondyloarthritis and inflammatory bowel disease: Association between disease activity and endothelial dysfunction markers. Rheumatol. Int. 2021, 42, 273–277. [Google Scholar] [CrossRef]
- Sato, H.; Kazama, J.J.; Wada, Y.; Kuroda, T.; Narita, I.; Gejyo, F.; Gao, P.; Yamashita, H. Decreased Levels of Circulating α2-Heremans-Schmid Glycoprotein/Fetuin-A (AHSG) in Patients with Rheumatoid Arthritis. Intern. Med. 2007, 46, 1685–1692. [Google Scholar] [CrossRef] [PubMed]
- Ricken, F.; Can, A.D.; Gräber, S.; Häusler, M.; Jahnen-Dechent, W. Post-translational modifications glycosylation and phosphorylation of the major hepatic plasma protein fetuin-A are associated with CNS inflammation in children. PLoS ONE 2022, 17, e0268592. [Google Scholar] [CrossRef]
- Minas, M.; Mystridou, P.; Georgoulias, P.; Pournaras, S.; Kostikas, K.; Gourgoulianis, K.I. Fetuin-A is Associated with Disease Severity and Exacerbation Frequency in Patients with COPD. COPD 2012, 10, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Kukla, M.; Menżyk, T.; Dembiński, M.; Winiarski, M.; Garlicki, A.; Bociąga-Jasik, M.; Skonieczna, M.; Hudy, D.; Maziarz, B.; Kuśnierz-Cabala, B.; et al. Fetuin-A Deficiency but Not Pentraxin 3, FGF-21, or Irisin, Predisposes to More Serious COVID-19 Course. Biomolecules 2021, 11, 1422. [Google Scholar] [CrossRef] [PubMed]
- Reverté, L.; Yeregui, E.; Olona, M.; Gutiérrez-Valencia, A.; Buzón, M.J.; Martí, A.; Gómez-Bertomeu, F.; Auguet, T.; López-Cortés, L.F.; Burgos, J.; et al. Fetuin-A, inter-α-trypsin inhibitor, glutamic acid and ChoE (18:0) are key biomarkers in a panel distinguishing mild from critical coronavirus disease 2019 outcomes. Clin. Transl. Med. 2022, 12, e704. [Google Scholar] [CrossRef] [PubMed]
- de Boer, H.C.; Preissner, K.T.; Bouma, B.N.; de Groot, P.G. Binding of vitronectin-thrombin-antithrombin III complex to human endothelial cells is mediated by the heparin binding site of vitronectin. J. Biol. Chem. 1992, 267, 2264–2268. [Google Scholar] [CrossRef]
- Fox, C.R.; Parks, G.D. Complement Inhibitors Vitronectin and Clusterin Are Recruited from Human Serum to the Surface of Coronavirus OC43-Infected Lung Cells through Antibody-Dependent Mechanisms. Viruses 2021, 14, 29. [Google Scholar] [CrossRef]
- Välikangas, T.; Junttila, S.; Rytkönen, K.T.; Kukkonen-Macchi, A.; Suomi, T.; Elo, L.L. COVID-19-specific transcriptomic signature detectable in blood across multiple cohorts. Front. Genet. 2022, 13, 929887. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Park, S.; Jeong, H.W.; Ahn, J.Y.; Choi, S.J.; Lee, H.; Choi, B.; Nam, S.K.; Sa, M.; Kwon, J.-S.; et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020, 5, eabd1554. [Google Scholar] [CrossRef]
- McClain, M.T.; Constantine, F.J.; Henao, R.; Liu, Y.; Tsalik, E.L.; Burke, T.W.; Steinbrink, J.M.; Petzold, E.; Nicholson, B.P.; Rolfe, R.; et al. Dysregulated transcriptional responses to SARS-CoV-2 in the periphery. Nat. Commun. 2021, 12, 1079. [Google Scholar] [CrossRef]
- Mukund, K.; Nayak, P.; Ashokkumar, C.; Rao, S.; Almeda, J.; Betancourt-Garcia, M.M.; Sindhi, R.; Subramaniam, S. Immune Response in Severe and Non-Severe Coronavirus Disease 2019 (COVID-19) Infection: A Mechanistic Landscape. Front. Immunol. 2021, 12, 738073. [Google Scholar] [CrossRef]
- Leng, L.; Li, M.; Li, W.; Mou, D.; Liu, G.; Ma, J.; Zhang, S.; Li, H.; Cao, R. Sera proteomic features of active and recovered COVID-19 patients: Potential diagnostic and prognostic biomarkers. Signal Transduct. Target. Ther. 2021, 6, 216. [Google Scholar] [CrossRef]
- Lee, J.; Han, D.; Kim, S.Y.; Hong, K.H.; Jang, M.; Kim, M.J.; Kim, Y.; Park, J.H.; Cho, S.I.; Park, W.B.; et al. Longitudinal proteomic profiling provides insights into host response and proteome dynamics in COVID-19 progression. Proteomics 2021, 21, 2000278. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Beimdiek, J.; Janciauskiene, S.; Wrenger, S.; Volland, S.; Rozy, A.; Fuge, J.; Olejnicka, B.; Pink, I.; Illig, T.; Popov, A.; et al. Plasma markers of COVID-19 severity: A pilot study. Respir. Res. 2022, 23, 343. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, L.; Chen, J.; Gao, Y. Targeted capture enrichment and sequencing identifies HLA variants associated with the severity of COVID-19. Genes Genom. 2022, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Liu, X.; Shen, C. A systemic review of T-cell epitopes defined from the proteome of SARS-CoV-2. Virus Res. 2023, 324, 199024. [Google Scholar] [CrossRef]
- Wu, P.; Chen, D.; Ding, W.; Wu, P.; Hou, H.; Bai, Y.; Zhou, Y.; Li, K.; Xiang, S.; Liu, P.; et al. The trans-omics landscape of COVID-19. Nat. Commun. 2021, 12, 4543. [Google Scholar] [CrossRef]
- Ai, Z. Revealing key regulators of neutrophil function during inflammation by re-analysing single-cell RNA-seq. PLoS ONE 2022, 17, e0276460. [Google Scholar] [CrossRef]
- Java, A.; Apicelli, A.J.; Liszewski, M.K.; Coler-Reilly, A.; Atkinson, J.P.; Kim, A.H.; Kulkarni, H.S. The complement system in COVID-19: Friend and foe? J. Clin. Investig. 2020, 5, e140711. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Jing, Y.; Luo, L.; Chen, Y.; Westerberg, L.S.; Zhou, P.; Xu, Z.; Herrada, A.A.; Park, C.-S.; Kubo, M.; Mei, H.; et al. SARS-CoV-2 infection causes immunodeficiency in recovered patients by downregulating CD19 expression in B cells via enhancing B-cell metabolism. Signal Transduct. Target. Ther. 2021, 6, 345. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Gan, R.; Zhen, Z.; Hu, X.; Li, X.; Zhou, F.; Liu, Y.; Chen, C.; Xie, S.; Zhang, B.; et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct. Target. Ther. 2020, 5, 156. [Google Scholar] [CrossRef] [PubMed]
- Adeniji, O.S.; Giron, L.B.; Purwar, M.; Zilberstein, N.F.; Kulkarni, A.J.; Shaikh, M.W.; Balk, R.A.; Moy, J.N.; Forsyth, C.B.; Liu, Q.; et al. COVID-19 Severity Is Associated with Differential Antibody Fc-Mediated Innate Immune Functions. Mbio 2021, 12, e00281-21. [Google Scholar] [CrossRef]
- Nishibori, M. Novel aspects of sepsis pathophysiology: NETs, plasma glycoproteins, endotheliopathy and COVID-19. J. Pharmacol. Sci. 2022, 150, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Ousaka, D.; Nishibori, M. A new approach to combat the sepsis including COVID-19 by accelerating detoxification of hemolysis-related DAMPs. Nihon Yakurigaku Zasshi 2022, 157, 422–425. [Google Scholar] [CrossRef]
- Ulanowska, M.; Olas, B. Modulation of Hemostasis in COVID-19; Blood Platelets May Be Important Pieces in the COVID-19 Puzzle. Pathogens 2021, 10, 370. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, T.; Krammer, F.; Iwasaki, A. The first 12 months of COVID-19: A timeline of immunological insights. Nat. Rev. Immunol. 2021, 21, 245–256. [Google Scholar] [CrossRef]
- Pius-Sadowska, E.; Niedźwiedź, A.; Kulig, P.; Baumert, B.; Sobuś, A.; Rogińska, D.; Łuczkowska, K.; Ulańczyk, Z.; Wnęk, S.; Karolak, I.; et al. CXCL8, CCL2, and CMV Seropositivity as New Prognostic Factors for a Severe COVID-19 Course. Int. J. Mol. Sci. 2022, 23, 11338. [Google Scholar] [CrossRef] [PubMed]
- Breville, G.; Zamberg, I.; Sadallah, S.; Stephan, C.; Ponte, B.; Seebach, J.D. Case Report: Severe Complement-Mediated Thrombotic Microangiopathy in IgG4-Related Disease Secondary to Anti-Factor H IgG4 Autoantibodies. Front. Immunol. 2021, 11, 604759. [Google Scholar] [CrossRef]
- Cen, G.; Liu, L.; Wang, J.; Wang, X.; Chen, S.; Song, Y.; Liang, Z. Weighted Gene Co-Expression Network Analysis to Identify Potential Biological Processes and Key Genes in COVID-19-Related Stroke. Oxidative Med. Cell. Longev. 2022, 2022, 4526022. [Google Scholar] [CrossRef]
- Aramburu, I.V.; Hoving, D.; Vernardis, S.I.; Tin, M.C.; Ioannou, M.; Temkin, M.I.; De Vasconcelos, N.M.; Demichev, V.; Helbig, E.T.; Lippert, L.; et al. Functional proteomic profiling links deficient DNA clearance with increased mortality in individuals with severe COVID-19 pneumonia. Immunity 2022, 55, 2436–2453.e5. [Google Scholar] [CrossRef]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A.; et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2020, 12, 23–40.e7. [Google Scholar] [CrossRef]
- Asare-Werehene, M.; McGuinty, M.; Vranjkovic, A.; Galipeau, Y.; Cowan, J.; Cameron, B.; Cooper, C.L.; Langlois, M.-A.; Crawley, A.M.; Tsang, B.K. Longitudinal profiles of plasma gelsolin, cytokines and antibody expression predict COVID-19 severity and hospitalization outcomes. Front. Immunol. 2022, 13, 1011084. [Google Scholar] [CrossRef]
- Yang, Z.; Bedugnis, A.; Levinson, S.; DiNubile, M.; Stossel, T.; Lu, Q.; Kobzik, L. Delayed Administration of Recombinant Plasma Gelsolin Improves Survival in a Murine Model of Penicillin-Susceptible and Penicillin-Resistant Pneumococcal Pneumonia. J. Infect. Dis. 2019, 220, 1498–1502. [Google Scholar] [CrossRef] [PubMed]
- Oiwa, M.; Kuroda, K.; Kawanoue, N.; Morimatsu, H. Histidine-rich glycoprotein as a novel predictive biomarker of postoperative complications in intensive care unit patients: A prospective observational study. BMC Anesthesiol. 2022, 22, 232. [Google Scholar] [CrossRef] [PubMed]
- Wake, H. Histidine-rich Glycoprotein Modulates the Blood-vascular System in Septic Condition. Acta Med. Okayama 2019, 73, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Völlmy, F.; Toorn, H.V.D.; Chiozzi, R.Z.; Zucchetti, O.; Papi, A.; Volta, C.A.; Marracino, L.; Sega, F.V.D.; Fortini, F.; Demichev, V.; et al. A serum proteome signature to predict mortality in severe COVID-19 patients. Life Sci. Alliance 2021, 4, e202101099. [Google Scholar] [CrossRef]
- Pagani, L.; Chinello, C.; Mahajneh, A.; Clerici, F.; Criscuolo, L.; Favalli, A.; Gruarin, P.; Grifantini, R.; Bandera, A.; Lombardi, A.; et al. Untargeted Mass Spectrometry Approach to Study SARS-CoV-2 Proteins in Human Plasma and Saliva Proteome. BioChem 2022, 2, 64–82. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]

| Overall | Mild | Severe | |||||
|---|---|---|---|---|---|---|---|
| WHO Score 1 | WHO Score 4 | WHO Score 5 | |||||
| Participants (n) | 43 | 33 | 5 | 5 | p | Test | |
| AGE (median [IQR]) | 56.00 [39.50, 63.00] | 47.00 [35.00, 60.00] | 65.50 [58.75, 67.75] | 0.014 | Wilcoxon | ||
| SEX (%) | Female | 20 (46.5) | 16 (48.5) | 4 (40.0) | 0.728 | Exact Fisher | |
| Male | 23 (53.5) | 17 (51.5) | 6 (60.0) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pagani, L.; Chinello, C.; Risca, G.; Capitoli, G.; Criscuolo, L.; Lombardi, A.; Ungaro, R.; Mangioni, D.; Piga, I.; Muscatello, A.; et al. Plasma Proteomic Variables Related to COVID-19 Severity: An Untargeted nLC-MS/MS Investigation. Int. J. Mol. Sci. 2023, 24, 3570. https://doi.org/10.3390/ijms24043570
Pagani L, Chinello C, Risca G, Capitoli G, Criscuolo L, Lombardi A, Ungaro R, Mangioni D, Piga I, Muscatello A, et al. Plasma Proteomic Variables Related to COVID-19 Severity: An Untargeted nLC-MS/MS Investigation. International Journal of Molecular Sciences. 2023; 24(4):3570. https://doi.org/10.3390/ijms24043570
Chicago/Turabian StylePagani, Lisa, Clizia Chinello, Giulia Risca, Giulia Capitoli, Lucrezia Criscuolo, Andrea Lombardi, Riccardo Ungaro, Davide Mangioni, Isabella Piga, Antonio Muscatello, and et al. 2023. "Plasma Proteomic Variables Related to COVID-19 Severity: An Untargeted nLC-MS/MS Investigation" International Journal of Molecular Sciences 24, no. 4: 3570. https://doi.org/10.3390/ijms24043570
APA StylePagani, L., Chinello, C., Risca, G., Capitoli, G., Criscuolo, L., Lombardi, A., Ungaro, R., Mangioni, D., Piga, I., Muscatello, A., Blasi, F., Favalli, A., Martinovic, M., Gori, A., Bandera, A., Grifantini, R., & Magni, F. (2023). Plasma Proteomic Variables Related to COVID-19 Severity: An Untargeted nLC-MS/MS Investigation. International Journal of Molecular Sciences, 24(4), 3570. https://doi.org/10.3390/ijms24043570