Molecular Cloning and Functional Characterization of Galectin-1 in Yellow Drum (Nibea albiflora)
Abstract
:1. Introduction
2. Results
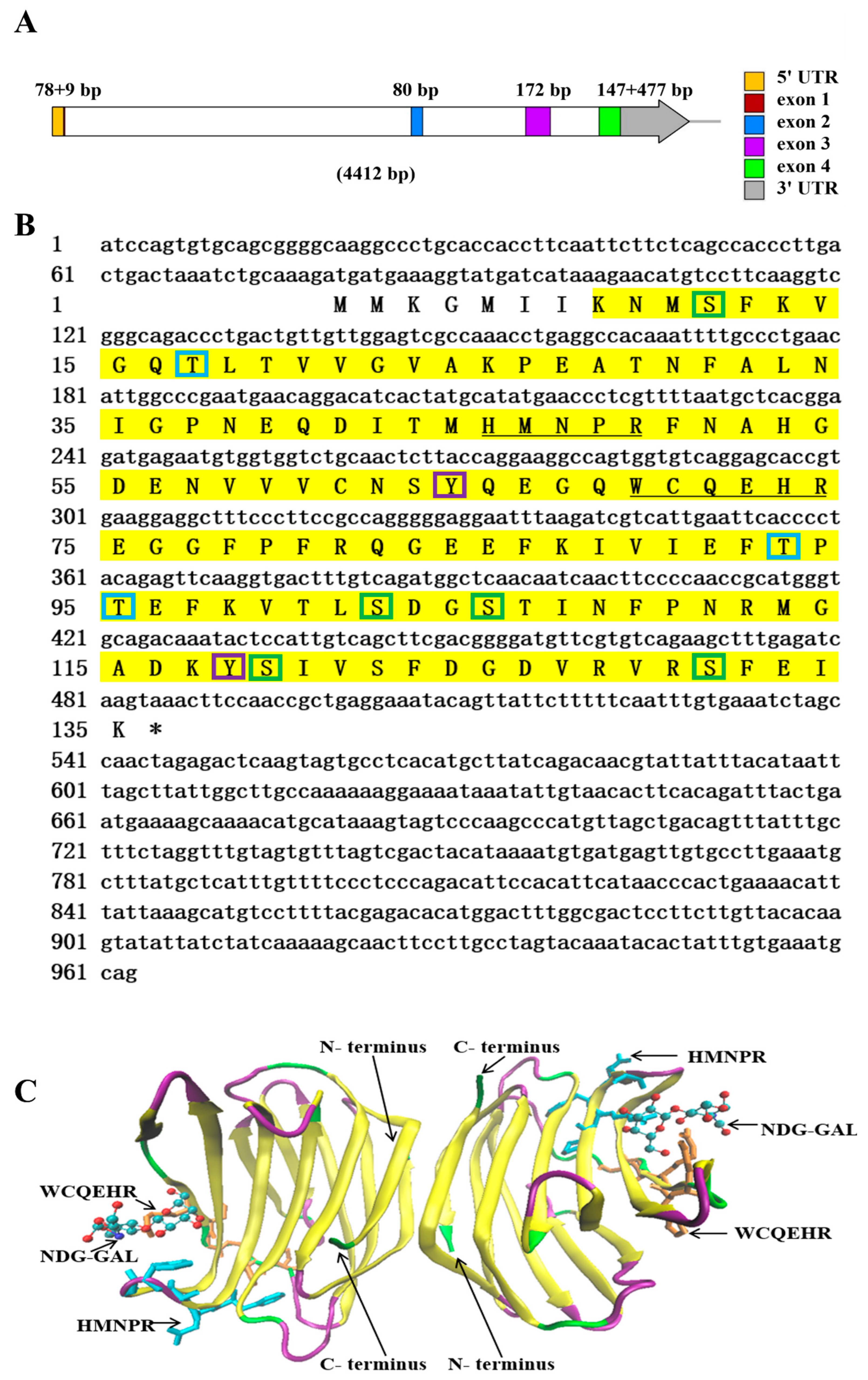
2.1. Sequence Characteristic Analysis of NaGal-1
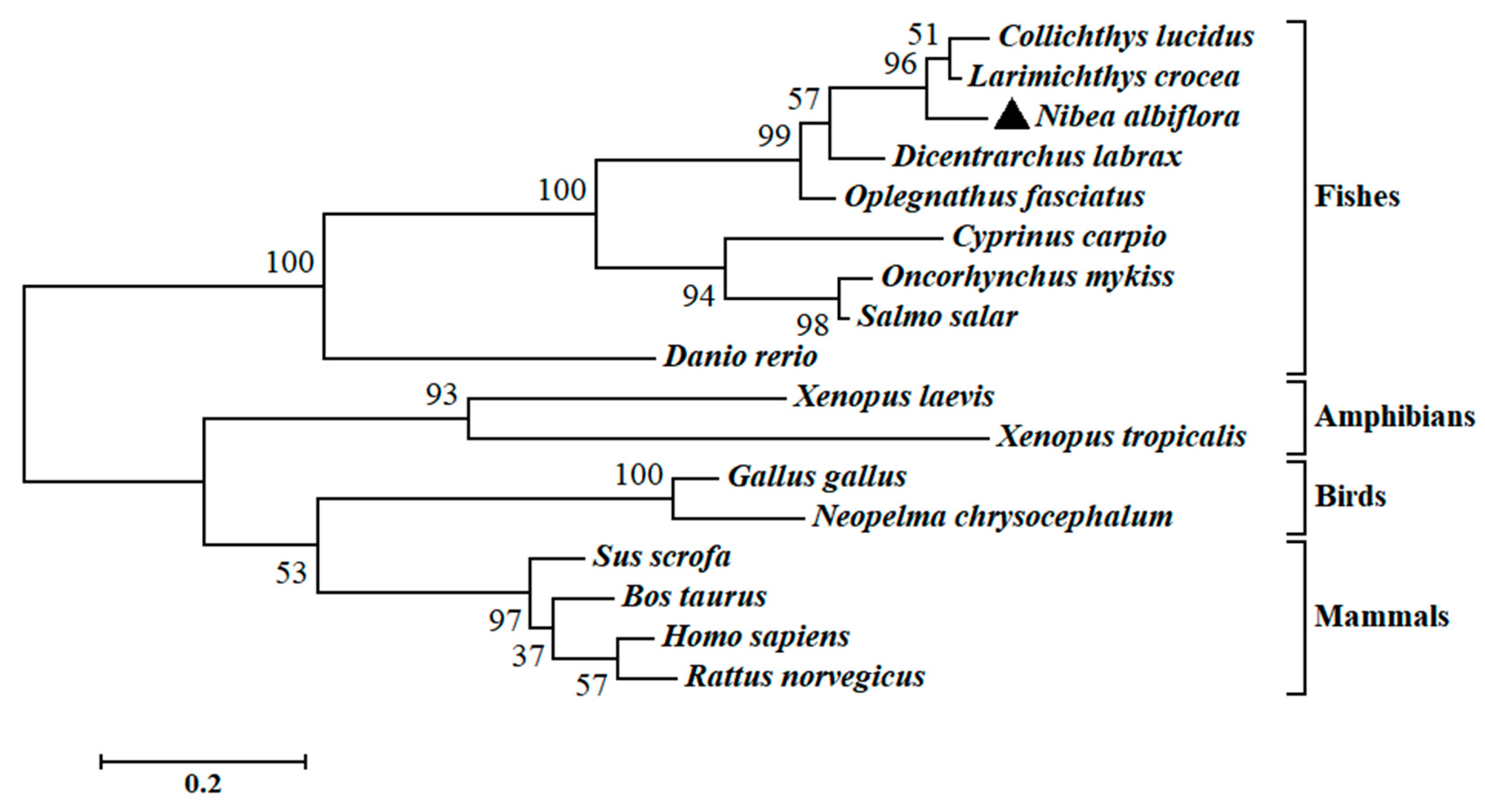
2.2. Multiple Sequence Alignments and Phylogenetic Analysis of NaGal-1
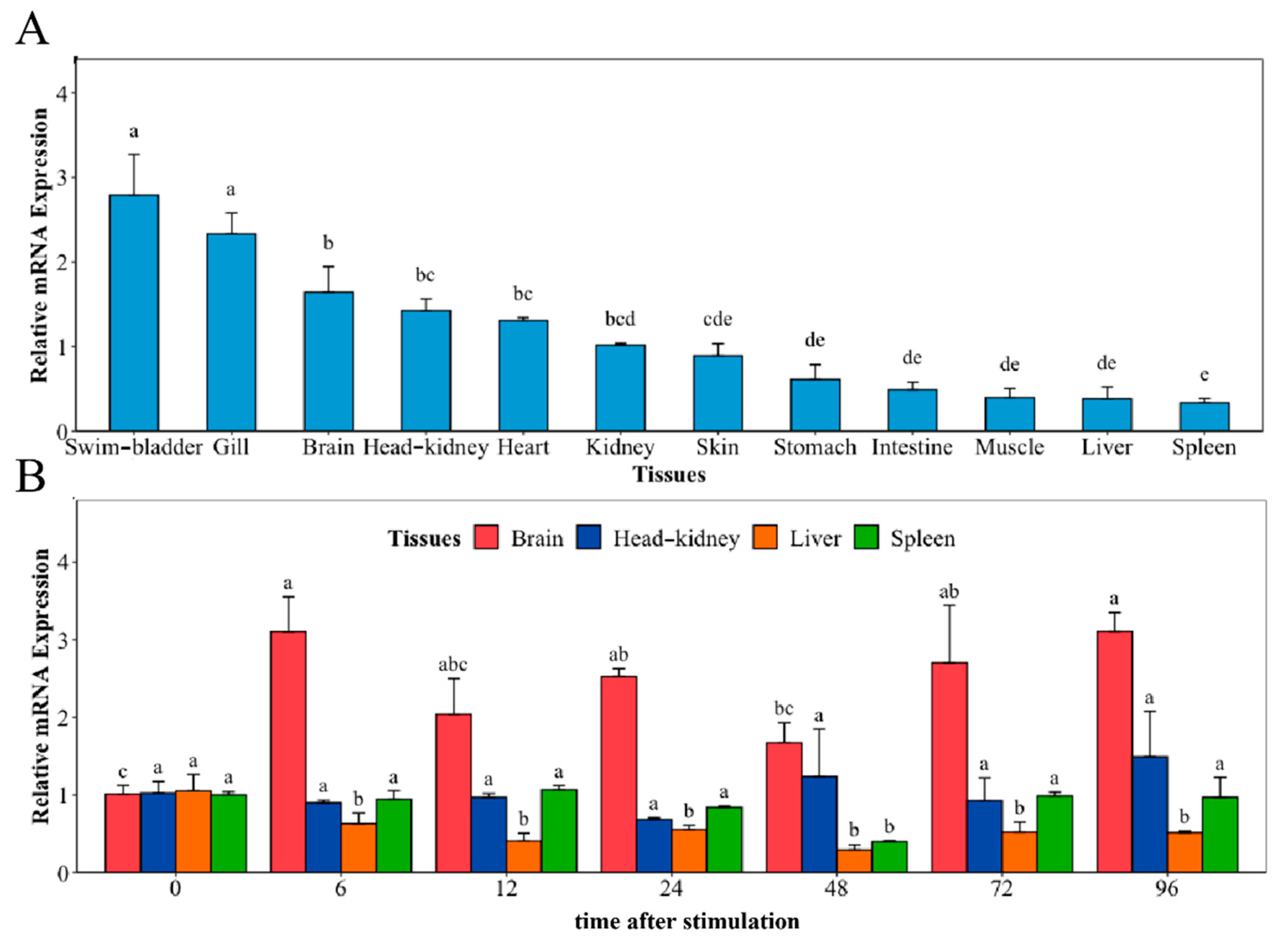
2.3. Tissue Distribution of NaGal-1 and Response to V. harveyi Infection
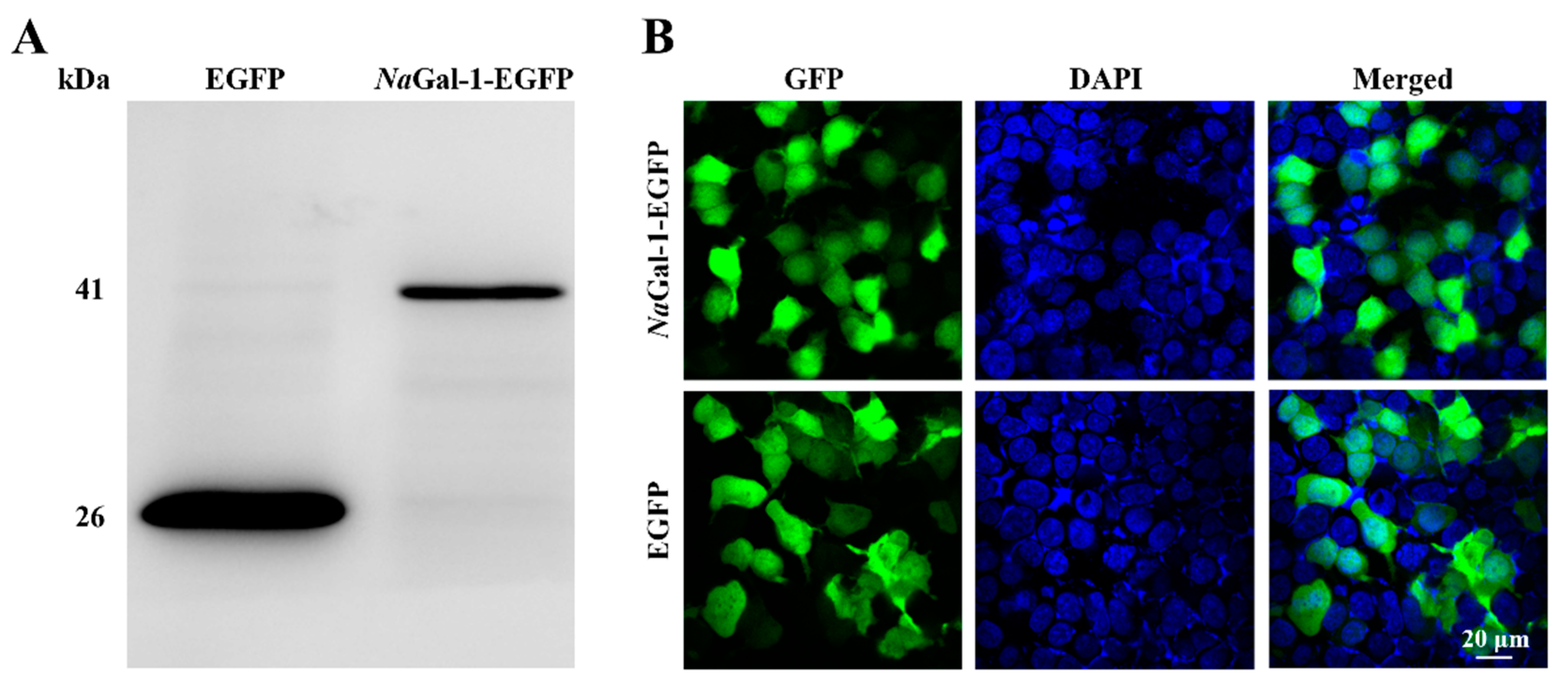
2.4. Expression of NaGal-1 in HEK 293T Cells
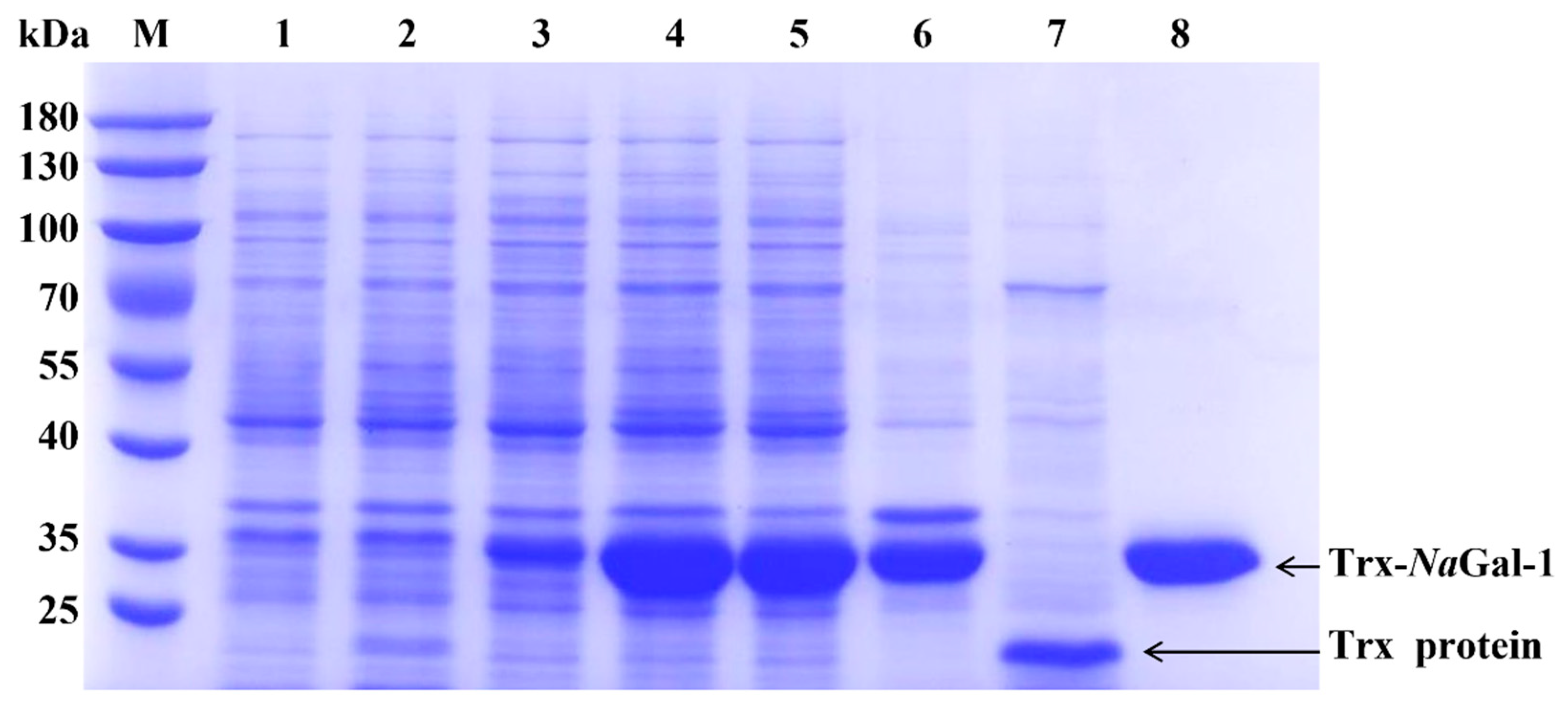
2.5. Prokaryotic Expression and Purification of NaGal-1
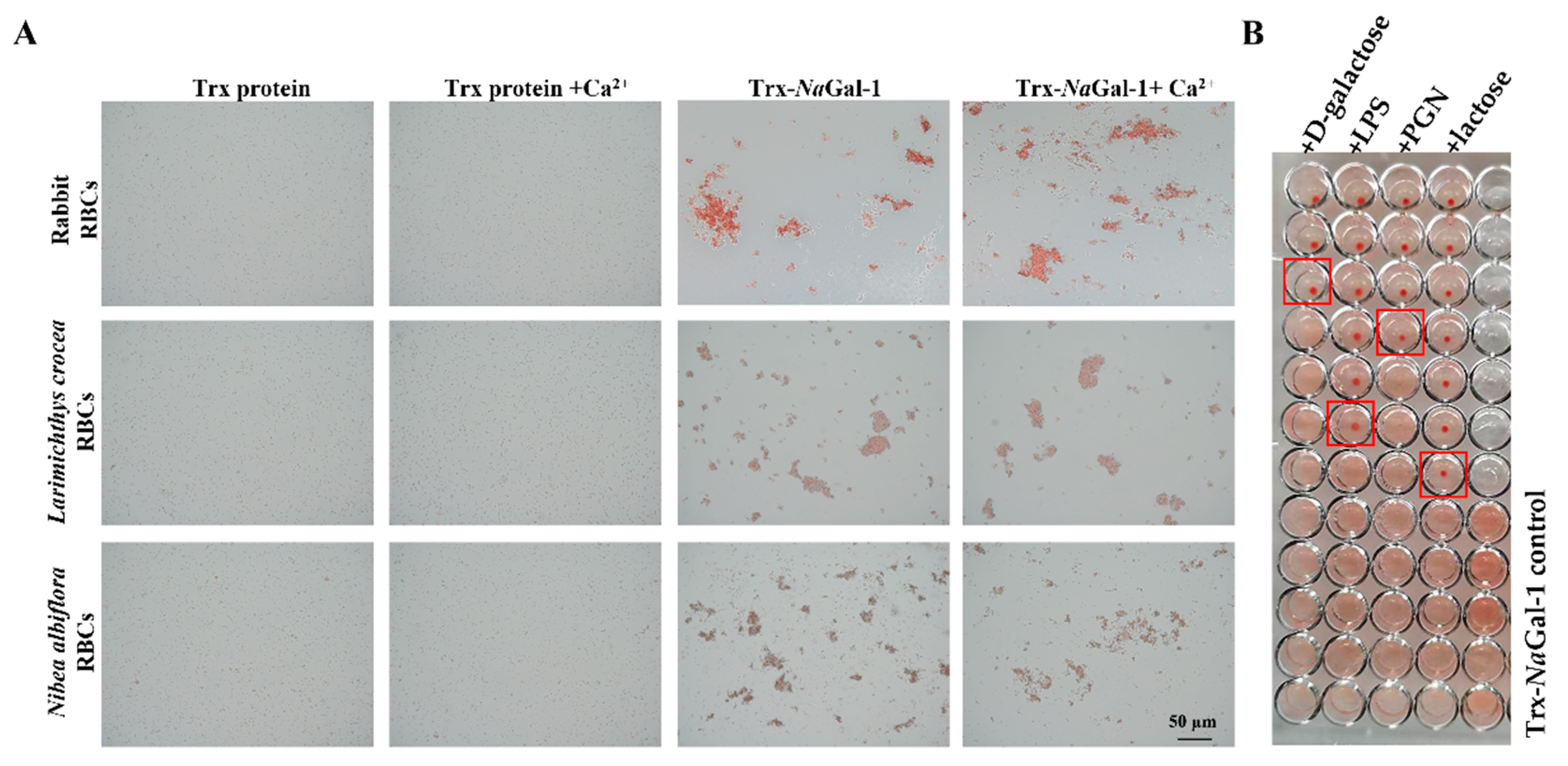
2.6. Hemagglutination and Sugar Inhibition Assay of NaGal-1
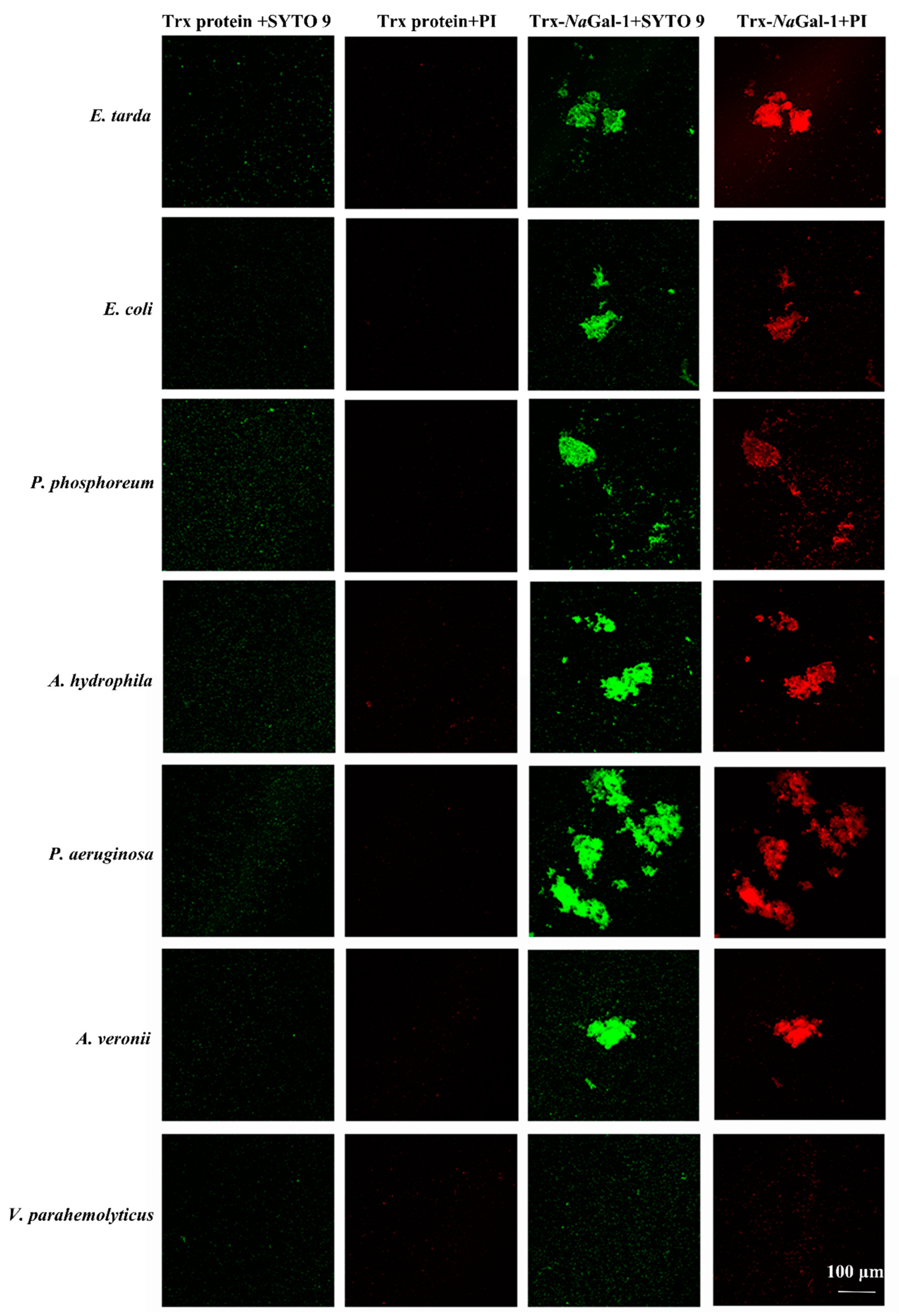
2.7. Bacterial Agglutination and Antibacterial Activity of NaGal-1
3. Discussion
4. Materials and Methods
4.1. Experimental Fish and Bacterial Infection Experiment
4.2. RNA Isolation and cDNA Synthesis
4.3. Cloning of NaGal-1 Gene
4.4. Bioinformatics Analysis
4.5. Tissue Distribution of NaGal-1 and Temporal Expression Pattern Post Infection
4.6. Expression of NaGal-1 in HEK 293T Cells
4.7. Prokaryotic Expression and Purification of NaGal-1 Protein
4.8. Hemagglutination and Sugar Inhibition Assays
4.9. Bacterial Agglutination and Live/Dead Tests
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fujita, T.; Matsushita, M.; Endo, Y. The Lectin-Complement Pathway—Its Role in Innate Immunity and Evolution. Immunol. Rev. 2004, 198, 185–202. [Google Scholar] [CrossRef] [PubMed]
- da Silva Lino, M.A.; Bezerra, R.F.; da Silva, C.D.C.; Carvalho, E.; Coelho, L. Fish Lectins: A Brief Review. In Advances in Zoology Research; Nova Science: Hauppauge, NY, USA, 2014; pp. 95–114. [Google Scholar]
- Cerliani, J.P.; Stowell, S.R.; Mascanfroni, I.D.; Arthur, C.M.; Cummings, R.D.; Rabinovich, G.A. Expanding the Universe of Cytokines and Pattern Recognition Receptors: Galectins and Glycans in Innate Immunity. J. Clin. Immunol. 2011, 31, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Davicino, R.C.; Eliçabe, R.J.; Di Genaro, M.S.; Rabinovich, G.A. Coupling Pathogen Recognition to Innate Immunity through Glycan-Dependent Mechanisms. Int. Immunopharmacol. 2011, 11, 1457–1463. [Google Scholar] [CrossRef] [PubMed]
- Barondes, S.H.; Cooper, D.N.; Gitt, M.A.; Leffler, H. Galectins. Structure and Function of a Large Family of Animal Lectins. J. Biol. Chem. 1994, 269, 20807–20810. [Google Scholar] [CrossRef] [PubMed]
- Vasta, G.R.; Nita-Lazar, M.; Giomarelli, B.; Ahmed, H.; Du, S.; Cammarata, M.; Parrinello, N.; Bianchet, M.A.; Amzel, L.M. Structural and Functional Diversity of the Lectin Repertoire in Teleost Fish: Relevance to Innate and Adaptive Immunity. Dev. Comp. Immunol. 2011, 35, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Teichberg, V.I.; Silman, I.; Beitsch, D.D.; Reshefft, G. A β-D-Galactoside Binding Protein from Electric Organ Tissue of Electrophorus Electricus. Proc. Nat. Acad. Sci. USA 1975, 5, 1383–1387. [Google Scholar] [CrossRef]
- Toscano, M.A.; Ilarregui, J.M.; Bianco, G.A.; Campagna, L.; Croci, D.O.; Salatino, M.; Rabinovich, G.A. Dissecting the Pathophysiologic Role of Endogenous Lectins: Glycan-Binding Proteins with Cytokine-like Activity? Cytokine Growth Factor Rev. 2007, 18, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Barrionuevo, P.; Beigier-Bompadre, M.; Ilarregui, J.M.; Toscano, M.A.; Bianco, G.A.; Isturiz, M.A.; Rabinovich, G.A. A Novel Function for Galectin-1 at the Crossroad of Innate and Adaptive Immunity: Galectin-1 Regulates Monocyte/Macrophage Physiology through a Nonapoptotic ERK-Dependent Pathway. J. Immunol. 2007, 178, 436–445. [Google Scholar] [CrossRef]
- Cedeno-Laurent, F.; Dimitroff, C.J. Galectins and Their Ligands: Negative Regulators of Anti-Tumor Immunity. Glycoconj. J. 2012, 29, 619–625. [Google Scholar] [CrossRef]
- Rajan, B.; Kiron, V.; Fernandes, J.M.O.; Brinchmann, M.F. Localization and Functional Properties of Two Galectin-1 Proteins in Atlantic Cod (Gadus Morhua) Mucosal Tissues. Dev. Comp. Immunol. 2013, 40, 83–93. [Google Scholar] [CrossRef]
- Thulasitha, W.S.; Umasuthan, N.; Wan, Q.; Nam, B.-H.; Kang, T.-W.; Lee, J. A Proto-Type Galectin-2 from Rock Bream (Oplegnathus Fasciatus): Molecular, Genomic, and Expression Analysis, and Recognition of Microbial Pathogens by Recombinant Protein. Dev. Comp. Immunol. 2017, 71, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Huang, Y.; Niu, J.; Wang, Z.; Tang, J.; Wang, B.; Lu, Y.; Cai, J.; Jian, J. Characterization of Galectin-2 from Nile Tilapia (Oreochromis Niloticus) Involved in the Immune Response to Bacterial Infection. Fish Shellfish Immunol. 2019, 87, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Thulasitha, W.S.; Umasuthan, N.; Whang, I.; Nam, B.-H.; Lee, J. Antimicrobial Response of Galectin-1 from Rock Bream Oplegnathus Fasciatus: Molecular, Transcriptional, and Biological Characterization. Fish Shellfish Immunol. 2016, 50, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Wei, X.; Yang, J.; Wang, S.; Zhang, Y. Galactoside-Binding Lectin in Solen Grandis as a Pattern Recognition Receptor Mediating Opsonization. Fish Shellfish Immunol. 2018, 82, 183–189. [Google Scholar] [CrossRef]
- Liu, S.; Hu, G.; Sun, C.; Zhang, S. Anti-Viral Activity of Galectin-1 from Flounder Paralichthys Olivaceus. Fish Shellfish Immunol. 2013, 34, 1463–1469. [Google Scholar] [CrossRef]
- Poisa-Beiro, L.; Dios, S.; Ahmed, H.; Vasta, G.R.; Martínez-López, A.; Estepa, A.; Alonso-Gutiérrez, J.; Figueras, A.; Novoa, B. Nodavirus Infection of Sea Bass (Dicentrarchus Labrax) Induces Up-Regulation of Galectin-1 Expression with Potential Anti-Inflammatory Activity. J. Immunol. 2009, 183, 6600–6611. [Google Scholar] [CrossRef]
- Liu, G.; Han, Z.; Jiang, D.; Li, W.; Zhang, W.; Ye, K.; Gu, L.; Dong, L.; Fang, M.; Wang, Z. Genome-Wide Association Study Identifies Loci for Traits Related to Swim Bladder in Yellow Drum (Nibea Albiflora). Aquaculture 2020, 526, 735327. [Google Scholar] [CrossRef]
- Tian, Q.; Li, W.; Li, J.; Xiao, Y.; Wu, B.; Wang, Z.; Han, F. Towards Understanding PRPS1 as a Molecular Player in Immune Response in Yellow Drum (Nibea Albiflora). Int. J. Mol. Sci. 2022, 23, 6475. [Google Scholar] [CrossRef]
- Dings, R.; Miller, M.; Griffin, R.; Mayo, K. Galectins as Molecular Targets for Therapeutic Intervention. Int. J. Mol. Sci. 2018, 19, 905. [Google Scholar] [CrossRef]
- Chen, M.; Liu, X.; Zhou, J.; Wang, X.; Liu, R.; Peng, H.; Li, B.; Cai, Z.; Jiang, C. Molecular Characterization and Expression Analysis of Galectins in Japanese Pufferfish (Takifugu Rubripes) in Response to Vibrio Harveyi Infection. Fish Shellfish Immunol. 2019, 86, 347–354. [Google Scholar] [CrossRef]
- Rivest, S. Regulation of Innate Immune Responses in the Brain. Nat. Rev. Immunol. 2009, 9, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Burguillos, M.A.; Svensson, M.; Schulte, T.; Boza-Serrano, A.; Garcia-Quintanilla, A.; Kavanagh, E.; Santiago, M.; Viceconte, N.; Oliva-Martin, M.J.; Osman, A.M.; et al. Microglia-Secreted Galectin-3 Acts as a Toll-like Receptor 4 Ligand and Contributes to Microglial Activation. Cell Rep. 2015, 10, 1626–1638. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, C.; Meng, J.; Li, N.; Xu, Z.; Liu, X.; Hou, S. Galectin-3 Regulates Microglial Activation and Promotes Inflammation through TLR4/MyD88/NF-KB in Experimental Autoimmune Uveitis. Clin. Immunol. 2022, 236, 108939. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Fu, P.; Huang, R.; Xiong, L.; Wang, Y.; He, L.; Liao, L.; Li, Y.; Zhu, Z.; Wang, Y. Molecular Characterization, Tissue Distribution and Functional Analysis of Galectin 1-like 2 in Grass Carp (Ctenopharyngodon Idella). Fish Shellfish Immunol. 2019, 94, 455–463. [Google Scholar] [CrossRef]
- Liu, F.-T.; Patterson, R.J.; Wang, J.L. Intracellular Functions of Galectins. Biochim. Biophys. Acta (BBA) Gen. Subj. 2002, 1572, 263–273. [Google Scholar] [CrossRef]
- Wu, B.; Song, Q.; Li, W.; Xie, Y.; Luo, S.; Tian, Q.; Zhao, R.; Liu, T.; Wang, Z.; Han, F. Characterization and Functional Study of a Chimera Galectin from Yellow Drum Nibea Albiflora. Int. J. Biol. Macromol. 2021, 187, 361–372. [Google Scholar] [CrossRef]
- Arasu, A.; Kumaresan, V.; Sathyamoorthi, A.; Chaurasia, M.K.; Bhatt, P.; Gnanam, A.J.; Palanisamy, R.; Marimuthu, K.; Pasupuleti, M.; Arockiaraj, J. Molecular Characterization of a Novel Proto-Type Antimicrobial Protein Galectin-1 from Striped Murrel. Microbiol. Res. 2014, 169, 824–834. [Google Scholar] [CrossRef]
- Chen, X.; Wei, J.; Xu, M.; Yang, M.; Li, P.; Wei, S.; Huang, Y.; Qin, Q. Molecular Cloning and Characterization of a Galectin-1 Homolog in Orange-Spotted Grouper, Epinephelus Coioides. Fish Shellfish Immunol. 2016, 54, 333–341. [Google Scholar] [CrossRef]
- Zhang, C.; Xue, Z.; Yu, Z.; Wang, H.; Liu, Y.; Li, H.; Wang, L.; Li, C.; Song, L. A Tandem-Repeat Galectin-1 from Apostichopus Japonicus with Broad PAMP Recognition Pattern and Antibacterial Activity. Fish Shellfish Immunol. 2020, 99, 167–175. [Google Scholar] [CrossRef]
- Mey, A.; Leffler, H.; Hmama, Z.; Normier, G.; Revillard, J.P. The Animal Lectin Galectin-3 Interacts with Bacterial Lipopolysaccharides via Two Independent Sites. J. Immunol. 1996, 156, 1572–1577. [Google Scholar] [CrossRef]
- Stowell, S.R.; Arthur, C.M.; McBride, R.; Berger, O.; Razi, N.; Heimburg-Molinaro, J.; Rodrigues, L.C.; Gourdine, J.-P.; Noll, A.J.; von Gunten, S.; et al. Microbial Glycan Microarrays Define Key Features of Host-Microbial Interactions. Nat. Chem. Biol. 2014, 10, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Stowell, S.R.; Arthur, C.M.; Dias-Baruffi, M.; Rodrigues, L.C.; Gourdine, J.-P.; Heimburg-Molinaro, J.; Ju, T.; Molinaro, R.J.; Rivera-Marrero, C.; Xia, B.; et al. Innate Immune Lectins Kill Bacteria Expressing Blood Group Antigen. Nat. Med. 2010, 16, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; St-Pierre, C.; Bhaumik, P.; Nieminen, J. Galectins in Innate Immunity: Dual Functions of Host Soluble β-Galactoside-Binding Lectins as Damage-Associated Molecular Patterns (DAMPs) and as Receptors for Pathogen-Associated Molecular Patterns (PAMPs). Immunol. Rev. 2009, 230, 172–187. [Google Scholar] [CrossRef] [PubMed]
- Vasta, G.R. Roles of Galectins in Infection. Nat. Rev. Microbiol. 2009, 7, 424–438. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Li, W.; Xie, Y.; Wu, B.; Sun, Y.; Tian, Q.; Wang, Z.; Han, F. A Molecular Insight into the Resistance of Yellow Drum to Vibrio Harveyi by Genome-Wide Association Analysis. Aquaculture 2021, 543, 736998. [Google Scholar] [CrossRef]
- Peng, J.; Li, W.; Wang, B.; Zhang, S.; Xiao, Y.; Han, F.; Wang, Z. UBE2G1 Is a Critical Component of Immune Response to the Infection of Pseudomonas Plecoglossicida in Large Yellow Croaker (Larimichthys Crocea). Int. J. Mol. Sci. 2022, 23, 8298. [Google Scholar] [CrossRef]

| GenBank Accession Numbers | Species | Identity (%) |
|---|---|---|
| XP_010736086.1 | Larimichthys crocea | 90.37 |
| TKS91807.1 | Collichthys lucidus | 85.93 |
| ADV35589.1 | Oplegnathus fasciatus | 80.74 |
| ACF77003.1 | Dicentrarchus labrax | 80.00 |
| NP_001134631.1 | Salmo salar | 61.48 |
| ACO07656.1 | Oncorhynchus mykiss | 60.74 |
| XP_018930790.1 | Cyprinus carpio | 60.00 |
| XP_005172121.1 | Danio rerio | 47.41 |
| AAK11514.1 | Xenopus laevis | 39.26 |
| NP_002296.1 | Homo sapiens | 37.78 |
| NP_786976.1 | Bos taurus | 37.04 |
| XP_027535424.1 | Neopelma chrysocephalum | 37.04 |
| NP_063969.1 | Rattus norvegicus | 37.04 |
| NP_990826.1 | Gallus gallus | 36.76 |
| NP_001001867.1 | Sus scrofa | 36.30 |
| XP_017949150.2 | Xenopus tropicalis | 31.25 |
| Primer Name | Sequence (5′-3′) | Purpose |
|---|---|---|
| Gal-1-F | GCTGATATCGGATCCGAATTCATGATGAAAGGTATGATCATAAAGAACA | ORF amplification |
| Gal-1-R | GTGGTGGTGGTGGTGCTCGAGTTACTTGATCTCAAAGCTTCTGACACG | |
| qGal-1-F | GAGGAATTTAAGATCGTCATTGAAT | qRT-PCR analysis |
| qGal-1-R | CTCAAAGCTTCTGACACGAACATCC | |
| β-actin-F | TTATGAAGGCTATGCCCTGCC | |
| β-actin-R | TGAAGGAGTAGCCACGCTCTGT | |
| sGal-1-F | CTACCGGACTCAGATCTCGAGATGATGAAAGGTATGATCATAAAGAACA | Subcellular localization |
| sGal-1-R | GTACCGTCGACTGCAGAATTCCCTTGATCTCAAAGCTTCTGACACG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, B.; Li, Q.; Li, W.; Luo, S.; Han, F.; Wang, Z. Molecular Cloning and Functional Characterization of Galectin-1 in Yellow Drum (Nibea albiflora). Int. J. Mol. Sci. 2023, 24, 3298. https://doi.org/10.3390/ijms24043298
Wu B, Li Q, Li W, Luo S, Han F, Wang Z. Molecular Cloning and Functional Characterization of Galectin-1 in Yellow Drum (Nibea albiflora). International Journal of Molecular Sciences. 2023; 24(4):3298. https://doi.org/10.3390/ijms24043298
Chicago/Turabian StyleWu, Baolan, Qiaoying Li, Wanbo Li, Shuai Luo, Fang Han, and Zhiyong Wang. 2023. "Molecular Cloning and Functional Characterization of Galectin-1 in Yellow Drum (Nibea albiflora)" International Journal of Molecular Sciences 24, no. 4: 3298. https://doi.org/10.3390/ijms24043298
APA StyleWu, B., Li, Q., Li, W., Luo, S., Han, F., & Wang, Z. (2023). Molecular Cloning and Functional Characterization of Galectin-1 in Yellow Drum (Nibea albiflora). International Journal of Molecular Sciences, 24(4), 3298. https://doi.org/10.3390/ijms24043298





