The Tumorigenic Role of Circular RNA-MicroRNA Axis in Cancer
Abstract
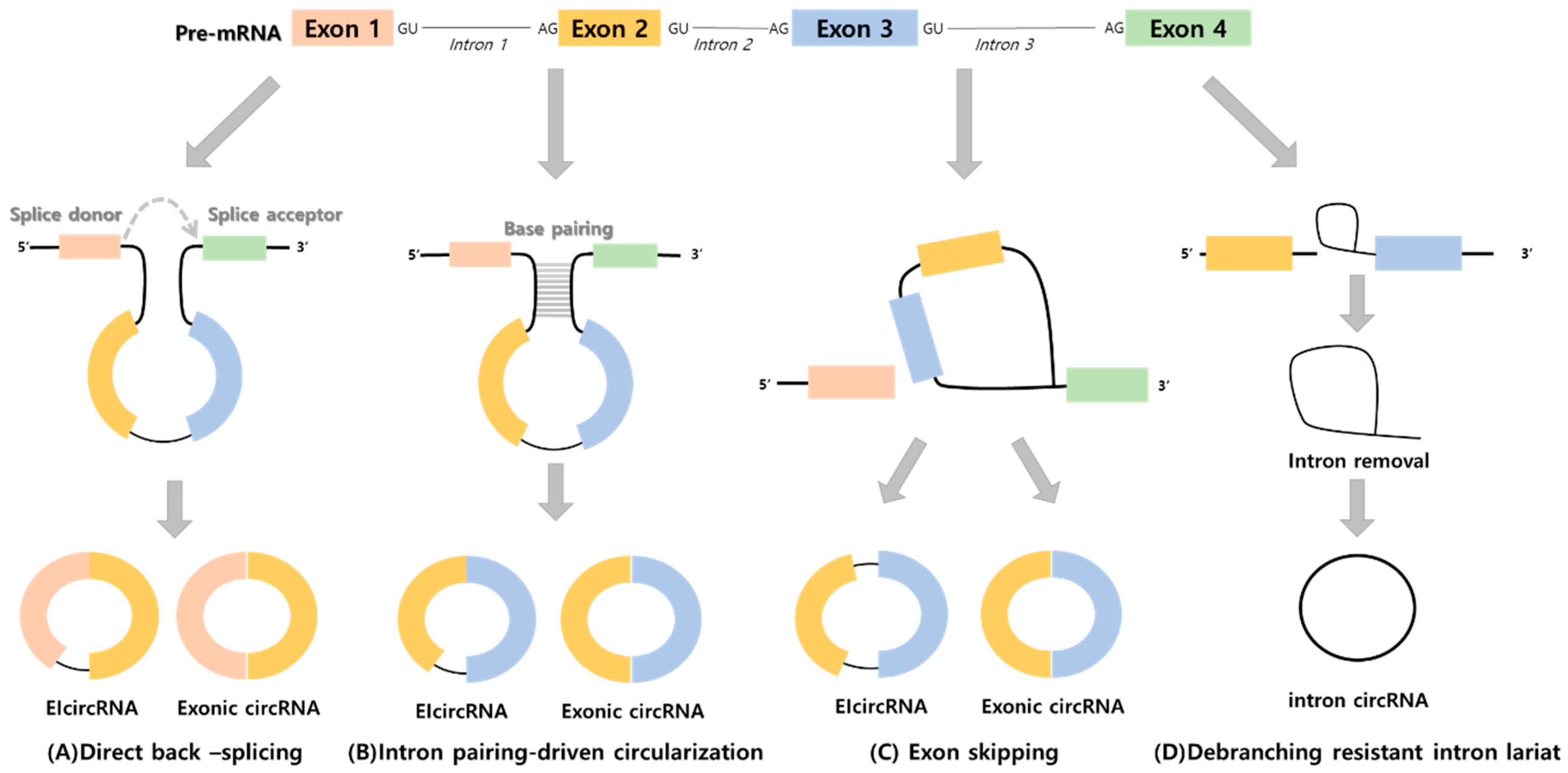
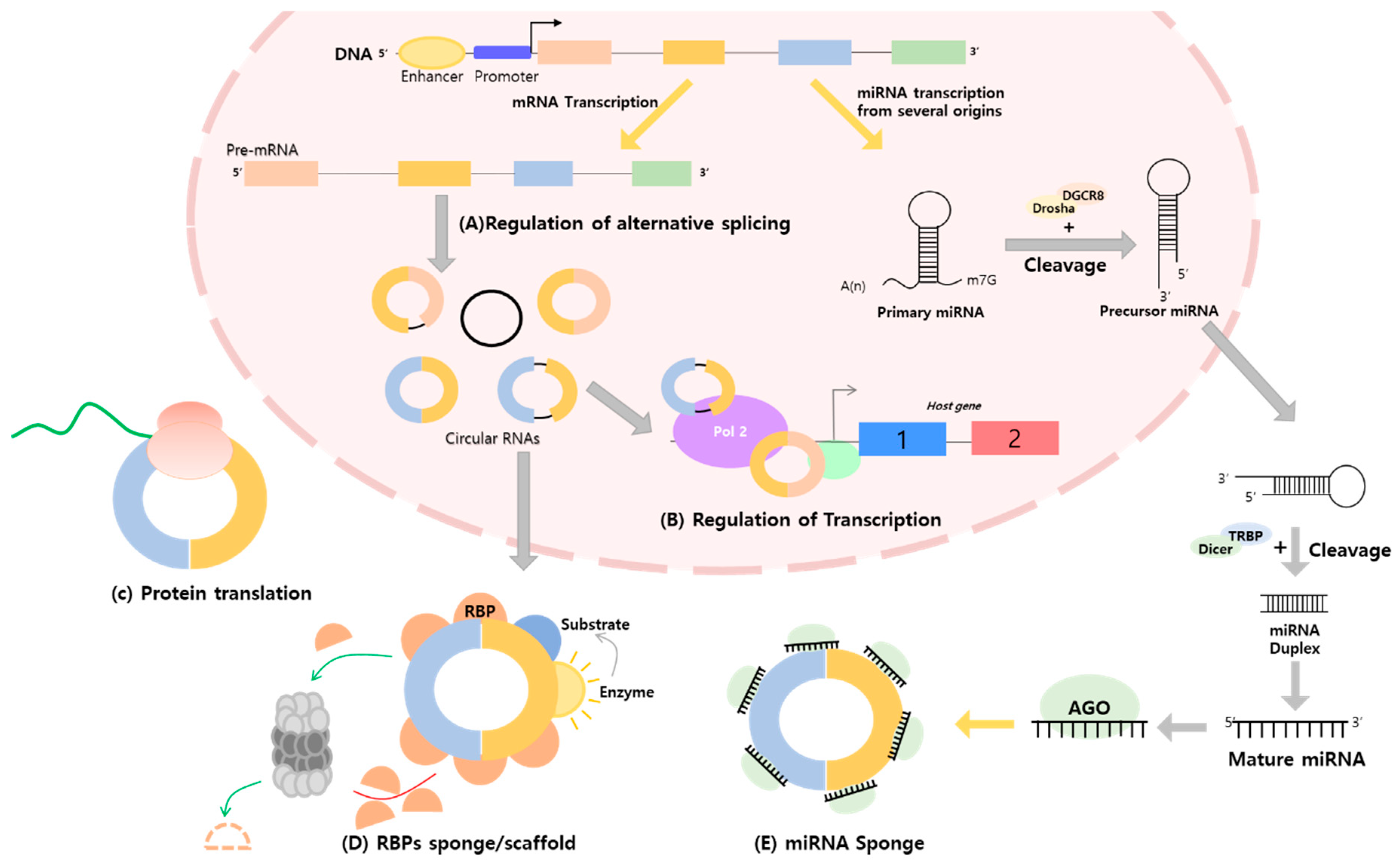
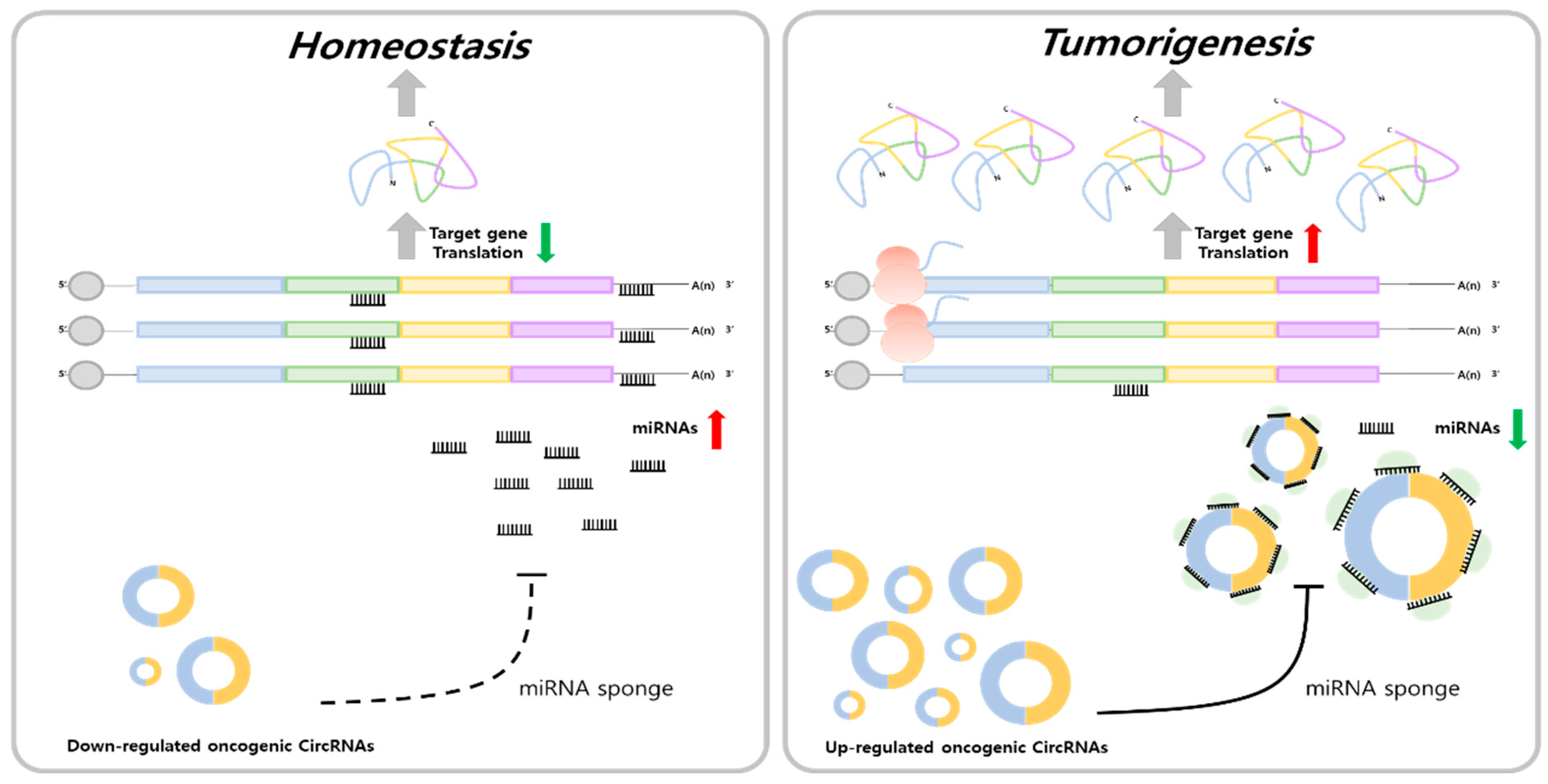
1. Introduction
2. Tumorigenic Regulation of Circular RNA–miRNA Axis in Five Major Mortality Cancers
2.1. Lung Cancer
2.2. Colon Cancer
2.3. Liver Cancer
2.4. Gastric Cancer
2.5. Breast Cancer
| Cancer Type | CircRNA Symbol /CircBase ID | Oncogenic /Tumor-Suppressive | MicroRNA | Target Gene | Cancer Related Biological/Regulatory Process | Ref. |
|---|---|---|---|---|---|---|
| Lung cancer | Circ_0000284 | oncogenic | miR-377-3p | PD-L1 | - | [47] |
| Circ_0000376 | oncogenic | miR-1182 | NOVA2 | glycolysis, viability, migration, and invasion | [93] | |
| Circ_0000677 | oncogenic | miR-106b-5p | CCND1 | proliferation | [94] | |
| Circ_0020123 | oncogenic | miR-144 | ZEB1, EZH2 | growth and metastasis | [95] | |
| Circ_0087862 | oncogenic | miR-1253 | RAB3D | tumor growth | [96] | |
| Circ_BANP | oncogenic | miR-503 | LARP1 | cancer growth, migration and invasion | [97] | |
| Circ_MAN2B2 | oncogenic | miR-1275 | FOXK1 | cell proliferation and invasion | [98] | |
| Circ_PIP5K1A | oncogenic | miR-600 | HIF-1α | proliferation and metastasis | [99] | |
| Circ_PVT1 | oncogenic | miR-125b | E2F2 | Proliferation and Invasion | [100] | |
| Circ_CDR1 | oncogenic | miR-641 | HOXA9 | cell stemness | [101] | |
| Circ_103809 | oncogenic | miR-4302 | MYC | cell proliferation and invasion | [63] | |
| ZNF121 | ||||||
| Circ_HIPK3 | oncogenic | miR-124 | SphK1, STAT3, CDK4 | cell survival, proliferation, cell death and apoptosis | [64] | |
| miR-149 | FOXM1 | cell proliferation and apoptosis | [65] | |||
| Circ_PVT1 | oncogenic | miR-497 | Bcl-2 | aggressive clinicopathological characteristics and poor prognosis | [66] | |
| Circ_ZNF208 | oncogenic | miR-7-5p | SNCA | radio-sensitivity of patients to X-rays | [67] | |
| Circ_0046264 | Tumor-suppressive | miR-1245 | BRCA2 | apoptosis, proliferation and invasion | [102] | |
| Circ_100395 | Tumor-suppressive | miR-1228 | TCF21 | proliferation, migration and invasion | [103] | |
| Circ_cESRP1 | Tumor-suppressive | miR-93-5p | CDKN1A | sensitivity to chemotherapy | [68] | |
| Circ_FOXO3 | Tumor-suppressive | miR-155 | FOXO3 | cell proliferation, migration and invasion | [104] | |
| Colon cancer | Circ_ERBIN | oncogenic | miR-125a-5p, miR-138-5p | 4EBP-1 | growth and metastasis | [105] |
| Circ_0000467 | oncogenic | miR-4766-5p | KLF12 | proliferation, metastasis, and angiogenesis | [106] | |
| Circ_0001313 | oncogenic | miR-510-5p | AKT2 | proliferation and apoptosis | [71] | |
| Circ_0001946 | oncogenic | miR-135a-5p | - | proliferation and metastasis | [107] | |
| Circ_0001982 | oncogenic | miR-144 | - | metastasis and poor prognosis | [69] | |
| Circ_0004277 | oncogenic | miR-512-5p | PTMA | cell apoptosis and cell proliferation | [108] | |
| Circ_0005963 | oncogenic | miR-122 | PKM2 | chemoresistance | [109] | |
| Circ_0007843 | oncogenic | mIR-518c-5p | MMP2 | migration and invasion | [110] | |
| Circ_000984 | oncogenic | miR-106b | CDK6 | cancer growth and metastasis | [75] | |
| Circ_0053277 | oncogenic | miR-2467-3p | MMP14 | cell proliferation, migration, and EMT | [111] | |
| Circ_0055625 | oncogenic | miR-106b-5p | ITGB8 | cell growth | [72] | |
| Circ_0060745 | oncogenic | miR-4736 | CSE1L | proliferation and metastasis | [112] | |
| Circ_CTNNA1 | oncogenic | miR-149-5p | FOXM1 | proliferation and invasion | [113] | |
| Circ_HIPK3 | oncogenic | miR-7 | c-Myb | cancer growth and metastasis | [114] | |
| Circ_HUWE1 | oncogenic | miR-486 | - | cell proliferation, migration and invasion | [70] | |
| Circ_NSD2 | oncogenic | miR-199b-5p | DDR1, JAG1 | migration and metastasis | [76] | |
| Circ_PRMT5 | oncogenic | miR-377 | E2F3 | proliferation | [115] | |
| Circ_PTK2 | oncogenic | miR-136-5p | YTHDF1 | proliferation, migration, invasion and chemoresistance | [116] | |
| Circ_UBAP2 | oncogenic | miR-199a | VEGFA | cell proliferation, migration, and invasion | [77] | |
| Circ_ACAP2 | Tumor-suppressive | miR-21-5p | Tiam1 | proliferation, migration, and invasion | [117] | |
| Liver cancer | Circ_0003141 | oncogenic | miR-1827 | UBAP2 | proliferation and invasion | [118] |
| Circ_0056836 | oncogenic | miR-766-3p | FOSL2 | cell migration, proliferation and invasion | [119] | |
| Circ_0061395 | oncogenic | miR-877-5p | PIK3R3 | proliferation, invasion, and migration | [120] | |
| Circ_FBLIM1 | oncogenic | miR-346 | FBLIM1 | cell proliferation, apoptosis and invasion | [121] | |
| Circ_MAST1 | oncogenic | miR-1299 | CTNND1 | cell proliferation and migration | [122] | |
| Circ_MET | oncogenic | miR-30-5p | Snail, DPP4, CXCL10 | epithelial to mesenchymal transition | [78] | |
| Circ_ZNF566 | oncogenic | miR-4738-3p | TDO2 | cell migration, invasion, and proliferation | [123] | |
| circ_0000673 | oncogenic | miR-767-3p | SET | cell proliferation and invasion | [124] | |
| Circ_0001955 | oncogenic | miR-516a-5p | TRAF6, MAPK11 | tumor growth | [79] | |
| Circ_0016788 | oncogenic | miR-486 | CDK4 | proliferation, invasion and apoptosis | [125] | |
| Circ_0067934 | oncogenic | miR-1324 | FZD5 | proliferation, migration and invasion | [126] | |
| Circ_103809 | oncogenic | miR-377-3p | FGFR1 | proliferation, cycle progression and migration | [127] | |
| Circ_104348 | oncogenic | miR-187-3p | RTKN2 | proliferation, migration, invasion and apoptosis | [128] | |
| Circ_Cdr1as | oncogenic | miR-7 | CCNE1, PIK3CD | proliferation and invasion | [129] | |
| Circ_GFRA1 | oncogenic | miR-498 | NAP1L3 | growth and invasion | [130] | |
| Circ_HIPK3 | oncogenic | miR-124 | AQP3 | cell proliferation and migration | [131] | |
| Circ_MAT2B | oncogenic | miR-338-3p | PKM2 | glycolysis | [132] | |
| Circ_5692 | Tumor-suppressive | miR-328-5p | DAB2IP | tumor growth | [80] | |
| Circ_C3P1 | Tumor-suppressive | miR-4641 | PCK1 | tumor growth and metastasis | [81] | |
| Circ_HIAT1 | Tumor-suppressive | miR-3171 | PTEN | cell growth | [82] | |
| Gastric cancer | ciRS-133 | oncogenic | miR-133 | PRDM16 | white adipose browning | [133] |
| Circ_0006282 | oncogenic | miR-155 | FBXO22 | cell growth, proliferation and metastasis | [134] | |
| Circ_0008035 | oncogenic | miR-375 | YBX1 | proliferation and invasion | [135] | |
| Circ_0002360 | oncogenic | miR-629-3p | PDLIM4 | proliferation, invasion and oxidative stress | [85] | |
| Circ_AKT3 | oncogenic | miR-198 | PIK3R1 | DNA damage repair and apoptosis | [136] | |
| Circ_CACTIN | oncogenic | miR-331-3p | TGFBR1 | tumor growth and EMT | [137] | |
| Circ_DLG1 | oncogenic | miR-141-3p | CXCL12 | proliferation, migration, invasion and immune evasion | [138] | |
| Circ_MTO1 | oncogenic | miR-199a-3p | PAWR | tumor growth, apoptosis, invasion and migration | [139] | |
| Circ_NHSL1 | oncogenic | miR-1306-3p | SIX1, vimentin | cell mobility, invasion and metastasis. | [86] | |
| Circ_NRIP1 | oncogenic | miR-149-5p | AKT1 | proliferation, migration and invasion | [140] | |
| Circ_PDSS1 | oncogenic | miR-186-5p | NEK2 | cell cycle and apoptosis | [141] | |
| Circ_PVRL3 | oncogenic | miR-203, miR-1272, miR-1283, miR-31, miR-638, miR-496, miR-485-3p, miR-766, and miR-876-3p | - | proliferation and migration | [84] | |
| Circ_0000039 | oncogenic | miR-1292-5p | DEK | proliferation, migration and invasion | [142] | |
| Circ_RanGAP1 | oncogenic | miR-877–3p | VEGFA | invasion and metastasis | [143] | |
| ciRS-7 | oncogenic | miR-7 | PTEN, PI3K | poor survival rate | [144] | |
| Circ_0026344 | Tumor-suppressive | miR-590-5p | PDCD4 | cell proliferation, migration and invasion | [145] | |
| Circ_CCDC9 | Tumor-suppressive | miR-6792-3p | CAV1 | tumor size, lymph node invasion, advanced clinical stage and survival rate | [87] | |
| Circ_LARP4 | Tumor-suppressive | miR-424-5p | LATS1 | cell proliferation and invasion | [146] | |
| Circ_MCTP2 | Tumor-suppressive | miR-99a-5p | MTMR3 | proliferation while promoting apoptosis of CDDP-resistant GC cells | [147] | |
| Circ_PSMC3 | Tumor-suppressive | miR-296-5p | - | the proliferation and metastasis | [148] | |
| Breast cancer | Circ_0001982 | oncogenic | miR-1287-5p | MUC19 | glycolysis, proliferation, migration, and invasion | [149] |
| Circ_0001429 | oncogenic | miR-205 | KDM4A | metastasis | [150] | |
| Circ_0005230 | oncogenic | miR-618 | CBX8 | prognostic predictor | [151] | |
| Circ_0008039 | oncogenic | miR-515-5p | CBX4 | proliferation, migration and invasion | [89] | |
| Circ_0008039 | oncogenic | miR-432-5p | E2F3 | proliferation, cell-cycle progression and migration | [90] | |
| Circ_0136666 | oncogenic | miR-1299 | CDK6 | proliferation, migration and invasion | [152] | |
| Circ_100219 | oncogenic | miR-485-3p | NTRK3 | proliferation and migration | [153] | |
| Circ_ANKS1B | oncogenic | miR-148a-3p, miR-152-3p | USF1 | metastasis | [154] | |
| Circ_BCBM1 | oncogenic | miR-125a | BRD4 | breast cancer brain metastasis | [92] | |
| Circ_CER | oncogenic | miR-136 | MMP13 | cell proliferation and migration | [155] | |
| Circ_FOXK2 | oncogenic | miR-370 | IGF2BP3 | migration and invasion | [156] | |
| Circ_MYO9B | oncogenic | miR-4316 | FOXP4 | cell proliferation and invasion | [157] | |
| Circ_PLK1 | oncogenic | miR-4500 | IGF1 | cell proliferation, migration and invasion | [158] | |
| Circ_RHOT1 | oncogenic | miR-106a-5p | STAT3 | malignant progression and ferroptosis | [159] | |
| Circ_RNF20 | oncogenic | miR-487a | HIF-1α | proliferation, Warburg effect | [91] | |
| Circ_RPPH1 | oncogenic | miR-556-5p | YAP1 | proliferation, migration, invasion, and angiogenesis | [160] | |
| Circ_HIPK3 | oncogenic | miR-193a | HMGB1, PI3K, AKT | cell proliferation and invasion | [161] | |
| Circ_0000442 | Tumor-suppressive | miR-148b-3p | PTEN | tumor growth | [162] | |
| Circ_CCDC85A | Tumor-suppressive | miR-550a-5p | MOB1A | proliferation, migration and invasion | [163] |
3. MDTEs Regulated by Oncogenic CircRNAs and Tumor-Suppressive circ RNAs in Various Cancers
3.1. MDTEs That Play a Common Role in Various Types of Cancer
3.2. MDTEs That Play Diverse Roles Depending on Regulation of Specific CircRNAs in Several Cancers
| Type of Circular RNA | Cancer Type | CircRNA Symbol /CircBase ID | MicroRNA | Subclass | Super Family | Target Gene | Cancer-Related Regulatory Process | Ref. |
|---|---|---|---|---|---|---|---|---|
| Oncogenic circular RNA | Bladder cancer | Circ_FARSA | miR-330-5p | SINE | MIR | - | cell proliferation, invasion, apoptosis and migration | [182] |
| Breast cancer | Circ_0007255 | miR-335-5p | SINE | MIR | SIX2 | inhibition of oxygen consumption, colony formation, cell migration and invasion | [183] | |
| Circ_0061825 (circ-TFF1) | miR-326 | DNA transposon | hAT-Tip100 | TFF1 | cell proliferation, migration, invasion and EMT | [170] | ||
| Circ_ABCB10 | miR-1271 | LINE | L2 | - | proliferation and apoptosis | [184] | ||
| Cervical cancer | Circ_0067934 | miR-545 | LINE | L2 | EIF3C | proliferation, colony formation, migration, invasion and EMT | [185] | |
| Circ_0141539 (Circ_8924) | miR-518d-5p | LINE | RTE-BovB | CBX8 | cell proliferation, migration and invasion | [186] | ||
| Cholangiocarcinoma | Circ_0000284 | miR-637 | LINE | L1 | - | migration, invasion and proliferation | [171] | |
| Chronic lymphocytic leukemia | Circ_CBFB | miR-607 | SINE | MIR | FZD3 | proliferation and apoptosis | [187] | |
| Colon cancer | Circ_PIP5K1A | miR-1273a | SINE | Alu | AP-1, IRF-4, CDX-2, Zic-1 | cell viability, cell invasion and migration | [188] | |
| Circ_FARSA | miR-330-5p | SINE | MIR | LASP1 | cell growth | [181] | ||
| Circ_ALG1 | miR-342-5p | SINE | tRNA-RTE | PGF | metastasis | [189] | ||
| Circ_102958 | miR-585 | LTR | ERVL-MaLR | CDC25B | growth, migration and invasion | [190] | ||
| Glioma | Circ_0001982 | miR-1205 | SINE | MIR | E2F1 | cell proliferation, migration, invasion and cell cycle progression | [191] | |
| Circ_0034642 | miR-1205 | SINE | MIR | BATF3 | cell proliferation and invasion | [192] | ||
| Liver cancer | Circ_0000517 | miR-326 | DNA transposon | hAT-Tip100 | IGF1R | cell viability, colony formation, migration, invasion and glycolysis | [169] | |
| Circ_G004213 | miR-513b-5p | DNA transposon | hAT-Tip100 | PRPF39 | cisplatin sensitivity and prognosis | [193] | ||
| Circ_ 001306 | miR-584-5p | DNA transposon | hAT-Blackjack | CDK16 | cell proliferation and growth | [194] | ||
| Circ_ 104075 | miR-582-3p | LINE | CR1 | HNF4a | YAP-dependent tumorigenesis | [195] | ||
| Lung cancer | Circ_ FOXM1 | miR-1304-5p | SINE | Alu | PPDPF, MACC1 | proliferation and invasion | [196] | |
| Circ_ ZKSCAN1 | miR-330-5p | SINE | MIR | FAM83A | tumor growth | [180] | ||
| Circ_ 0004015 | miR-1183 | LINE | L2 | PDPK1 | proliferation, invasion, TKI drug resistance | [197] | ||
| Circ_ 0014130 | miR-493-5p | LINE | L2 | - | - | [198] | ||
| Circ_ POLA2 | miR-326 | DNA transposon | hAT-Tip100 | GNB1 | cell stemness and progression | [168] | ||
| Circ_ 0003998 | miR-326 | DNA transposon | hAT-Tip100 | Notch1 | cell proliferation and invasion | [167] | ||
| Osteosarcoma | Circ_ HIPK3 | miR-637 | LINE | L1 | HDAC4 | proliferation and migration and invasion | [173] | |
| Papillary thyroid cancer | Circ_ ZFR | miR-1261 | DNA transposon | TcMar-Tigger | C8orf4 | cell proliferation and invasion | [199] | |
| Stomach cancer | Circ-LDLRAD3 | miR-224-5p | DNA transposon | DNA transposon | - | cell growth, migration invasion and apoptosis | [178] | |
| Circ_HIPK3 | miR-637 | LINE | L1 | AKT1 | cell growth and metastasis | [172] | ||
| Circ_ATXN7 | miR-4319 | SINE | MIR | ENTPD4 | proliferation, invasion and apoptosis | [200] | ||
| Circ_0008287 | miR-548c-3p | DNA transposon | TcMar-Mariner | CLIC1 | immune escape of cancer cell | [201] | ||
| Tumor- Suppressive circular RNA | Bladder cancer | Circ_ITCH | miR-224 | DNA transposon | DNA transposon | p21, PTEN | cell proliferation, migration, invasion and metastasis | [176] |
| Circ_HIPK3 | miR-558 | LTR | ERVL-MaLR | HPSE | migration, invasion, and angiogenesis | [202] | ||
| Breast cancer | Circ_AHNAK1 | miR-421 | LINE | L2 | RASA1 | proliferation and metastasis | [175] | |
| Colon cancer | Circ_SMARCA5 | miR-552 | LINE | L1 | - | growth, migration and invasion | [203] | |
| Liver cancer | Circ_SETD3 (circ_0000567) | miR-421 | LINE | L2 | MAPK14 | tumor cell growth | [174] | |
| Lung cancer | Circ_0078767 | miR-330-5p | SINE | MIR | RASSF1A | cancer cell viability, cell cycle progression and invasion | [179] | |
| Circ_0007059 | miR-378 | SINE | MIR | p53, CyclinD1, Bax, Cleaved-Caspase-3, E-cadherin, Vimentin, Twist, Zeb1 | proliferation and EMT | [204] | ||
| Osteosarcoma | Circ_0002052 | miR-1205 | SINE | MIR | APC2 | cell proliferation, migration, invasion and apoptosis | [205] | |
| Stomach cancer | Circ_FAT1(e2) | miR-548g | DNA transposon | TcMar-Mariner | YBX1 | cell proliferation, migration and invasion | [206] | |
| Circ_ZFR | miR-130a | LINE | RTE-BovB | PTEN | cell proliferation and apoptosis | [207] | ||
| Circ_0000096 | miR-224 | DNA transposon | DNA transposon | cyclin D1, CDK6, MMP-2, MMP-9 | cell growth and migration | [177] |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, Y.; Xu, S.; Liu, J.; Ponnusamy, M.; Zhao, Y.; Zhang, Y.; Wang, Q.; Li, P.; Wang, K. Non-coding RNA-linked epigenetic regulation in cardiac hypertrophy. Int. J. Biol. Sci. 2018, 14, 1133. [Google Scholar]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar]
- Lekka, E.; Hall, J. Noncoding RNA s in disease. FEBS Lett. 2018, 592, 2884–2900. [Google Scholar]
- Li, M.; Duan, L.; Li, Y.; Liu, B. Long noncoding RNA/circular noncoding RNA–miRNA–mRNA axes in cardiovascular diseases. Life Sci. 2019, 233, 116440. [Google Scholar]
- Su, Q.; Lv, X. Revealing new landscape of cardiovascular disease through circular RNA-miRNA-mRNA axis. Genomics 2020, 112, 1680–1685. [Google Scholar]
- Guo, L.; Jia, L.; Luo, L.; Xu, X.; Xiang, Y.; Ren, Y.; Ren, D.; Shen, L.; Liang, T. Critical roles of circular RNA in tumor metastasis via acting as a sponge of miRNA/isomiR. Int. J. Mol. Sci. 2022, 23, 7024. [Google Scholar]
- Cheng, D.; Wang, J.; Dong, Z.; Li, X. Cancer-related circular RNA: Diverse biological functions. Cancer Cell Int. 2021, 21, 1–16. [Google Scholar] [CrossRef]
- Marinescu, M.-C.; Lazar, A.-L.; Marta, M.M.; Cozma, A.; Catana, C.-S. Non-Coding RNAs: Prevention, Diagnosis, and Treatment in Myocardial Ischemia–Reperfusion Injury. Int. J. Mol. Sci. 2022, 23, 2728. [Google Scholar]
- Paul, S.; Ruiz-Manriquez, L.M.; Ledesma-Pacheco, S.J.; Benavides-Aguilar, J.A.; Torres-Copado, A.; Morales-Rodríguez, J.I.; De Donato, M.; Srivastava, A. Roles of microRNAs in chronic pediatric diseases and their use as potential biomarkers: A review. Arch. Biochem. Biophys. 2021, 699, 108763. [Google Scholar]
- Pandey, M.; Mukhopadhyay, A.; Sharawat, S.K.; Kumar, S. Role of microRNAs in regulating cell proliferation, metastasis and chemoresistance and their applications as cancer biomarkers in small cell lung cancer. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2021, 1876, 188552. [Google Scholar]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Campo-Paysaa, F.; Sémon, M.; Cameron, R.A.; Peterson, K.J.; Schubert, M. microRNA complements in deuterostomes: Origin and evolution of microRNAs. Evol. Dev. 2011, 13, 15–27. [Google Scholar] [CrossRef]
- Piriyapongsa, J.; Mariño-Ramírez, L.; Jordan, I.K. Origin and evolution of human microRNAs from transposable elements. Genetics 2007, 176, 1323–1337. [Google Scholar] [CrossRef]
- Rani, V.; Sengar, R.S. Biogenesis and mechanisms of microRNA-mediated gene regulation. Biotechnol. Bioeng. 2022, 119, 685–692. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. Global miRNA to miRNA interactions: Impacts for miR-21. Trends Cell Biol. 2021, 31, 3–5. [Google Scholar]
- Gong, Y.; Zhang, X. RNAi-based antiviral immunity of shrimp. Dev. Comp. Immunol. 2021, 115, 103907. [Google Scholar]
- Chen, C.; Zeng, Z.; Liu, Z.; Xia, R. Small RNAs, emerging regulators critical for the development of horticultural traits. Hortic. Res. 2018, 5, 63. [Google Scholar]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar]
- Fernández-Hernando, C.; Suárez, Y. MicroRNAs in endothelial cell homeostasis and vascular disease. Curr. Opin. Hematol. 2018, 25, 227. [Google Scholar]
- Zhang, Y.; Li, S.; Jin, P.; Shang, T.; Sun, R.; Lu, L.; Guo, K.; Liu, J.; Tong, Y.; Wang, J. Dual functions of microRNA-17 in maintaining cartilage homeostasis and protection against osteoarthritis. Nat. Commun. 2022, 13, 2447. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, D.-H.; Wu, N.; Xiao, J.-H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar]
- Panda, A.C. Circular RNAs act as miRNA sponges. Circ. RNAs 2018, 1087, 67–79. [Google Scholar]
- Cai, X.; Lin, L.; Zhang, Q.; Wu, W.; Su, A. Bioinformatics analysis of the circRNA–miRNA–mRNA network for non-small cell lung cancer. J. Int. Med. Res. 2020, 48, 0300060520929167. [Google Scholar]
- Ali, A.; Han, K.; Liang, P. Role of transposable elements in gene regulation in the human genome. Life 2021, 11, 118. [Google Scholar]
- Campo, S.; Sánchez-Sanuy, F.; Camargo-Ramírez, R.; Gómez-Ariza, J.; Baldrich, P.; Campos-Soriano, L.; Soto-Suárez, M.; San Segundo, B. A novel Transposable element-derived microRNA participates in plant immunity to rice blast disease. Plant Biotechnol. J. 2021, 19, 1798–1811. [Google Scholar]
- Lee, H.-E.; Huh, J.-W.; Kim, H.-S. Bioinformatics analysis of evolution and human disease related transposable element-derived microRNAs. Life 2020, 10, 95. [Google Scholar] [CrossRef]
- Anwar, S.L.; Wulaningsih, W.; Lehmann, U. Transposable elements in human cancer: Causes and consequences of deregulation. Int. J. Mol. Sci. 2017, 18, 974. [Google Scholar]
- Prats, A.-C.; David, F.; Diallo, L.H.; Roussel, E.; Tatin, F.; Garmy-Susini, B.; Lacazette, E. Circular RNA, the key for translation. Int. J. Mol. Sci. 2020, 21, 8591. [Google Scholar] [CrossRef]
- Meng, X.; Li, X.; Zhang, P.; Wang, J.; Zhou, Y.; Chen, M. Circular RNA: An emerging key player in RNA world. Brief. Bioinform. 2017, 18, 547–557. [Google Scholar]
- Li, W.; Liu, J.Q.; Chen, M.; Xu, J.; Zhu, D. Circular RNA in cancer development and immune regulation. J. Cell. Mol. Med. 2022, 26, 1785–1798. [Google Scholar]
- Kristensen, L.; Hansen, T.; Venø, M.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565. [Google Scholar]
- Li, M.; Ding, W.; Sun, T.; Tariq, M.A.; Xu, T.; Li, P.; Wang, J. Biogenesis of circular RNA s and their roles in cardiovascular development and pathology. FEBS J. 2018, 285, 220–232. [Google Scholar]
- Panda, A.C.; De, S.; Grammatikakis, I.; Munk, R.; Yang, X.; Piao, Y.; Dudekula, D.B.; Abdelmohsen, K.; Gorospe, M. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017, 45, e116. [Google Scholar] [CrossRef]
- Eger, N.; Schoppe, L.; Schuster, S.; Laufs, U.; Boeckel, J.-N. Circular RNA splicing. Circ. RNAs 2018, 1087, 41–52. [Google Scholar]
- Yao, T.; Chen, Q.; Fu, L.; Guo, J. Circular RNAs: Biogenesis, properties, roles, and their relationships with liver diseases. Hepatol. Res. 2017, 47, 497–504. [Google Scholar]
- Kelly, S.; Greenman, C.; Cook, P.R.; Papantonis, A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015, 427, 2414–2417. [Google Scholar]
- Li, J.; Yang, J.; Zhou, P.; Le, Y.; Zhou, C.; Wang, S.; Xu, D.; Lin, H.-K.; Gong, Z. Circular RNAs in cancer: Novel insights into origins, properties, functions and implications. Am. J. Cancer Res. 2015, 5, 472. [Google Scholar]
- Huang, M.-S.; Zhu, T.; Li, L.; Xie, P.; Li, X.; Zhou, H.-H.; Liu, Z.-Q. LncRNAs and CircRNAs from the same gene: Masterpieces of RNA splicing. Cancer Lett. 2018, 415, 49–57. [Google Scholar]
- Zhao, W.; Li, M.; Wang, S.; Li, Z.; Li, H.; Li, S. CircRNA SRRM4 affects glucose metabolism by regulating PKM alternative splicing via SRSF3 deubiquitination in epilepsy. Neuropathol. Appl. Neurobiol. 2022, e12850. [Google Scholar]
- Gao, Y.; Wang, J.; Zheng, Y.; Zhang, J.; Chen, S.; Zhao, F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat. Commun. 2016, 7, 12060. [Google Scholar] [CrossRef]
- Aufiero, S.; van den Hoogenhof, M.M.; Reckman, Y.J.; Beqqali, A.; van der Made, I.; Kluin, J.; Khan, M.A.; Pinto, Y.M.; Creemers, E.E. Cardiac circRNAs arise mainly from constitutive exons rather than alternatively spliced exons. RNA 2018, 24, 815–827. [Google Scholar] [CrossRef]
- Chen, L.-L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Chen, N.; Zhao, G.; Yan, X.; Lv, Z.; Yin, H.; Zhang, S.; Song, W.; Li, X.; Li, L.; Du, Z. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018, 19, 218. [Google Scholar]
- Wang, X.; Fang, L. Advances in circular RNAs and their roles in breast Cancer. J. Exp. Clin. Cancer Res. 2018, 37, 206. [Google Scholar]
- Liu, D.; Mewalal, R.; Hu, R.; Tuskan, G.A.; Yang, X. New technologies accelerate the exploration of non-coding RNAs in horticultural plants. Hortic. Res. 2017, 4, 17031. [Google Scholar]
- Abe, N.; Matsumoto, K.; Nishihara, M.; Nakano, Y.; Shibata, A.; Maruyama, H.; Shuto, S.; Matsuda, A.; Yoshida, M.; Ito, Y. Rolling circle translation of circular RNA in living human cells. Sci. Rep. 2015, 5, 16435. [Google Scholar]
- Wen, S.-y.; Qadir, J.; Yang, B.B. Circular RNA translation: Novel protein isoforms and clinical significance. Trends Mol. Med. 2022, 28, 405–420. [Google Scholar] [CrossRef]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E. Translation of circRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Zhu, L.-P.; He, Y.-J.; Hou, J.-C.; Chen, X.; Zhou, S.-Y.; Yang, S.-J.; Li, J.; Zhang, H.-D.; Hu, J.-H.; Zhong, S.-L. The role of circRNAs in cancers. Biosci. Rep. 2017, 37, BSR20170750. [Google Scholar]
- Zeng, Y.; Du, W.W.; Wu, Y.; Yang, Z.; Awan, F.M.; Li, X.; Yang, W.; Zhang, C.; Yang, Q.; Yee, A. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 2017, 7, 3842. [Google Scholar]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef]
- Chen, J.; Gu, J.; Tang, M.; Liao, Z.; Tang, R.; Zhou, L.; Su, M.; Jiang, J.; Hu, Y.; Chen, Y. Regulation of cancer progression by circRNA and functional proteins. J. Cell. Physiol. 2022, 237, 373–388. [Google Scholar]
- Liang, Z.-Z.; Guo, C.; Zou, M.-M.; Meng, P.; Zhang, T.-T. circRNA-miRNA-mRNA regulatory network in human lung cancer: An update. Cancer Cell Int. 2020, 20, 1–16. [Google Scholar] [CrossRef]
- Das, A.; Sinha, T.; Shyamal, S.; Panda, A.C. Emerging role of circular RNA–protein interactions. Non-Coding RNA 2021, 7, 48. [Google Scholar] [CrossRef]
- Bezzi, M.; Guarnerio, J.; Pandolfi, P.P. A circular twist on microRNA regulation. Cell Res. 2017, 27, 1401–1402. [Google Scholar]
- Zhang, M.; Xin, Y. Circular RNAs: A new frontier for cancer diagnosis and therapy. J. Hematol. Oncol. 2018, 11, 1–9. [Google Scholar] [CrossRef]
- He, Y.; Lin, J.; Ding, Y.; Liu, G.; Luo, Y.; Huang, M.; Xu, C.; Kim, T.K.; Etheridge, A.; Lin, M. A systematic study on dysregulated micro RNA s in cervical cancer development. Int. J. Cancer 2016, 138, 1312–1327. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Liu, W.; Ma, W.; Yuan, Y.; Zhang, Y.; Sun, S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem. Biophys. Res. Commun. 2018, 500, 846–851. [Google Scholar]
- Yu, H.; Chen, Y.; Jiang, P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem. Biophys. Res. Commun. 2018, 506, 455–462. [Google Scholar]
- Lu, H.; Han, X.; Ren, J.; Ren, K.; Li, Z.; Sun, Z. Circular RNA HIPK3 induces cell proliferation and inhibits apoptosis in non-small cell lung cancer through sponging miR-149. Cancer Biol. Ther. 2020, 21, 113–121. [Google Scholar] [CrossRef]
- Qin, S.; Zhao, Y.; Lim, G.; Lin, H.; Zhang, X.; Zhang, X. Circular RNA PVT1 acts as a competing endogenous RNA for miR-497 in promoting non-small cell lung cancer progression. Biomed. Pharmacother. 2019, 111, 244–250. [Google Scholar]
- Liu, B.; Li, H.; Liu, X.; Li, F.; Chen, W.; Kuang, Y.; Zhao, X.; Li, L.; Yu, B.; Jin, X. CircZNF208 enhances the sensitivity to X-rays instead of carbon-ions through the miR-7-5p/SNCA signal axis in non-small-cell lung cancer cells. Cell. Signal. 2021, 84, 110012. [Google Scholar] [CrossRef]
- Huang, W.; Yang, Y.; Wu, J.; Niu, Y.; Yao, Y.; Zhang, J.; Huang, X.; Liang, S.; Chen, R.; Chen, S. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-β signalling. Cell Death Differ. 2020, 27, 1709–1727. [Google Scholar]
- Deng, Q.; Wang, C.; Hao, R.; Yang, Q. Circ_0001982 accelerates the progression of colorectal cancer via sponging microRNA-144. Eur. Rev. Med. Pharm. Sci. 2020, 24, 1755–1762. [Google Scholar]
- Chen, H.-Y.; Li, X.-N.; Ye, C.-X.; Chen, Z.-L.; Wang, Z.-J. Circular RNA circHUWE1 is upregulated and promotes cell proliferation, migration and invasion in colorectal cancer by sponging miR-486. OncoTargets Ther. 2020, 13, 423. [Google Scholar] [CrossRef]
- Tu, F.-L.; Guo, X.-Q.; Wu, H.-X.; He, Z.-Y.; Wang, F.; Sun, A.-J.; Dai, X.-D. Circ-0001313/miRNA-510-5p/AKT2 axis promotes the development and progression of colon cancer. Am. J. Transl. Res. 2020, 12, 281. [Google Scholar]
- Zhang, J.; Liu, H.; Zhao, P.; Zhou, H.; Mao, T. Has_circ_0055625 from circRNA profile increases colon cancer cell growth by sponging miR-106b-5p. J. Cell. Biochem. 2019, 120, 3027–3037. [Google Scholar] [CrossRef]
- Xu, H.; Wang, C.; Song, H.; Xu, Y.; Ji, G. RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol. Cancer 2019, 18, 8. [Google Scholar]
- Ji, W.; Qiu, C.; Wang, M.; Mao, N.; Wu, S.; Dai, Y. Hsa_circ_0001649: A circular RNA and potential novel biomarker for colorectal cancer. Biochem. Biophys. Res. Commun. 2018, 497, 122–126. [Google Scholar] [CrossRef]
- Xu, X.-W.; Zheng, B.-A.; Hu, Z.-M.; Qian, Z.-Y.; Huang, C.-J.; Liu, X.-Q.; Wu, W.-D. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget 2017, 8, 91674. [Google Scholar]
- Chen, L.Y.; Zhi, Z.; Wang, L.; Zhao, Y.Y.; Deng, M.; Liu, Y.H.; Qin, Y.; Tian, M.M.; Liu, Y.; Shen, T. NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signalling. J. Pathol. 2019, 248, 103–115. [Google Scholar] [CrossRef]
- Dai, J.; Zhuang, Y.; Tang, M.; Qian, Q.; Chen, J. CircRNA UBAP2 facilitates the progression of colorectal cancer by regulating miR-199a/VEGFA pathway. Eur. Rev. Med. Pharm. Sci. 2020, 24, 7963–7971. [Google Scholar]
- Huang, X.-Y.; Zhang, P.-F.; Wei, C.-Y.; Peng, R.; Lu, J.-C.; Gao, C.; Cai, J.-B.; Yang, X.; Fan, J.; Ke, A.-W. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol. Cancer 2020, 19, 92. [Google Scholar] [CrossRef]
- Yao, Z.; Xu, R.; Yuan, L.; Xu, M.; Zhuang, H.; Li, Y.; Zhang, Y.; Lin, N. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019, 10, 945. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, Y.; Huang, Z.; Kong, Y.; Hu, X.; Xiao, W.; Quan, J.; Fan, X. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019, 10, 900. [Google Scholar]
- Zhong, L.; Wang, Y.; Cheng, Y.; Wang, W.; Lu, B.; Zhu, L.; Ma, Y. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem. Biophys. Res. Commun. 2018, 499, 1044–1049. [Google Scholar]
- Wang, Z.; Zhao, Y.; Wang, Y.; Jin, C. Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR-3171/PTEN axis. Biomed. Pharmacother. 2019, 116, 108932. [Google Scholar]
- Figueiredo, C.; Camargo, M.C.; Leite, M.; Fuentes-Pananá, E.M.; Rabkin, C.S.; Machado, J.C. Pathogenesis of gastric cancer: Genetics and molecular classification. Mol. Pathog. Signal Transduct. By Helicobacter Pylori 2017, 277–304. [Google Scholar]
- Sun, H.-D.; Xu, Z.-P.; Sun, Z.-Q.; Zhu, B.; Wang, Q.; Zhou, J.; Jin, H.; Zhao, A.; Tang, W.-W.; Cao, X.-F. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci. Rep. 2018, 8, 10111. [Google Scholar]
- Yu, Z.; Lan, J.; Li, W.; Jin, L.; Qi, F.; Yu, C.; Zhu, H. Circular RNA hsa_circ_0002360 promotes proliferation and invasion and inhibits oxidative stress in gastric cancer by sponging miR-629-3p and regulating the PDLIM4 expression. Oxidative Med. Cell. Longev. 2022, 2022, 2775433. [Google Scholar] [CrossRef]
- Zhu, Z.; Rong, Z.; Luo, Z.; Yu, Z.; Zhang, J.; Qiu, Z.; Huang, C. Circular RNA circNHSL1 promotes gastric cancer progression through the miR-1306-3p/SIX1/vimentin axis. Mol. Cancer 2019, 18, 126. [Google Scholar]
- Luo, Z.; Rong, Z.; Zhang, J.; Zhu, Z.; Yu, Z.; Li, T.; Fu, Z.; Qiu, Z.; Huang, C. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression. Mol. Cancer 2020, 19, 86. [Google Scholar]
- Seneviratne, S.; Lawrenson, R.; Scott, N.; Kim, B.; Shirley, R.; Campbell, I. Breast cancer biology and ethnic disparities in breast cancer mortality in New Zealand: A cohort study. PLoS ONE 2015, 10, e0123523. [Google Scholar] [CrossRef]
- Huang, F.; Dang, J.; Zhang, S.; Cheng, Z. Circular RNA hsa_circ_0008039 promotes proliferation, migration and invasion of breast cancer cells through upregulating CBX4 via sponging miR-515-5p. Eur. Rev. Med. Pharm. Sci. 2020, 24, 1887–1898. [Google Scholar]
- Liu, Y.; Lu, C.; Zhou, Y.; Zhang, Z.; Sun, L. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem. Biophys. Res. Commun. 2018, 502, 358–363. [Google Scholar] [CrossRef]
- Cao, L.; Wang, M.; Dong, Y.; Xu, B.; Chen, J.; Ding, Y.; Qiu, S.; Li, L.; Karamfilova Zaharieva, E.; Zhou, X. Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1α/HK2. Cell Death Dis. 2020, 11, 145. [Google Scholar] [CrossRef]
- Fu, B.; Liu, W.; Zhu, C.; Li, P.; Wang, L.; Pan, L.; Li, K.; Cai, P.; Meng, M.; Wang, Y. Circular RNA circBCBM1 promotes breast cancer brain metastasis by modulating miR-125a/BRD4 axis. Int. J. Biol. Sci. 2021, 17, 3104. [Google Scholar]
- Li, C.; Liu, H.; Niu, Q.; Gao, J. Circ_0000376, a novel circRNA, promotes the progression of non-small cell lung cancer through regulating the miR-1182/NOVA2 network. Cancer Manag. Res. 2020, 12, 7635. [Google Scholar]
- Hu, X.; Wang, P.; Qu, C.; Zhang, H.; Li, L. Circular RNA Circ_0000677 promotes cell proliferation by regulating microRNA-106b-5p/CCND1 in non-small cell lung cancer. Bioengineered 2021, 12, 6229–6239. [Google Scholar] [CrossRef]
- Qu, D.; Yan, B.; Xin, R.; Ma, T. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am. J. Cancer Res. 2018, 8, 1387. [Google Scholar]
- Li, L.; Wan, K.; Xiong, L.; Liang, S.; Tou, F.; Guo, S. CircRNA hsa_circ_0087862 acts as an oncogene in non-small cell lung cancer by targeting miR-1253/RAB3D axis. OncoTargets Ther. 2020, 13, 2873. [Google Scholar]
- Han, J.; Zhao, G.; Ma, X.; Dong, Q.; Zhang, H.; Wang, Y.; Cui, J. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem. Biophys. Res. Commun. 2018, 503, 2429–2435. [Google Scholar] [CrossRef]
- Ma, X.; Yang, X.; Bao, W.; Li, S.; Liang, S.; Sun, Y.; Zhao, Y.; Wang, J.; Zhao, C. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem. Biophys. Res. Commun. 2018, 498, 1009–1015. [Google Scholar] [CrossRef]
- Chi, Y.; Luo, Q.; Song, Y.; Yang, F.; Wang, Y.; Jin, M.; Zhang, D. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1α regulation. J. Cell. Biochem. 2019, 120, 19019–19030. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Z.; Jiang, H.; Li, Q.; Wang, R.; Pan, H.; Niu, Y.; Liu, F.; Gu, H.; Fan, X. Circular RNA circPVT1 promotes proliferation and invasion through sponging miR-125b and activating E2F2 signaling in non-small cell lung cancer. Cell. Physiol. Biochem. 2018, 51, 2324–2340. [Google Scholar] [CrossRef]
- Zhao, Y.; Zheng, R.; Chen, J.; Ning, D. CircRNA CDR1as/miR-641/HOXA9 pathway regulated stemness contributes to cisplatin resistance in non-small cell lung cancer (NSCLC). Cancer Cell Int. 2020, 20, 289. [Google Scholar]
- Yang, L.; Wang, J.; Fan, Y.; Yu, K.; Jiao, B.; Su, X. Hsa_circ_0046264 up-regulated BRCA2 to suppress lung cancer through targeting hsa-miR-1245. Respir. Res. 2018, 19, 115. [Google Scholar]
- Chen, D.; Ma, W.; Ke, Z.; Xie, F. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle 2018, 17, 2080–2090. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, H.; Zhang, L. Identification of the tumor-suppressive function of circular RNA FOXO3 in non-small cell lung cancer through sponging miR-155. Mol. Med. Rep. 2018, 17, 7692–7700. [Google Scholar]
- Chen, L.-Y.; Wang, L.; Ren, Y.-X.; Pang, Z.; Liu, Y.; Sun, X.-D.; Tu, J.; Zhi, Z.; Qin, Y.; Sun, L.-N. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol. Cancer 2020, 19, 164. [Google Scholar] [CrossRef]
- Chen, H.; Wu, C.; Luo, L.; Wang, Y.; Peng, F. circ_0000467 promotes the proliferation, metastasis, and angiogenesis in colorectal cancer cells through regulating KLF12 expression by sponging miR-4766-5p. Open Med. 2021, 16, 1415–1427. [Google Scholar] [CrossRef]
- Deng, Z.; Li, X.; Wang, H.; Geng, Y.; Cai, Y.; Tang, Y.; Wang, Y.; Yu, X.; Li, L.; Li, R. Dysregulation of CircRNA_0001946 contributes to the proliferation and metastasis of colorectal cancer cells by targeting MicroRNA-135a-5p. Front. Genet. 2020, 11, 357. [Google Scholar]
- Yang, L.; Sun, H.; Liu, X.; Chen, J.; Tian, Z.; Xu, J.; Xiang, B.; Qin, B. Circular RNA hsa_circ_0004277 contributes to malignant phenotype of colorectal cancer by sponging miR-512-5p to upregulate the expression of PTMA. J. Cell. Physiol. 2020. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Yang, H.; Bai, M.; Ning, T.; Deng, T.; Liu, R.; Fan, Q.; Zhu, K.; Li, J. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020, 14, 539–555. [Google Scholar] [CrossRef]
- He, J.H.; Han, Z.P.; Luo, J.G.; Jiang, J.W.; Zhou, J.B.; Chen, W.M.; Lv, Y.B.; He, M.L.; Zheng, L.; Li, Y.G. Hsa_Circ_0007843 acts as a mIR-518c-5p sponge to regulate the migration and invasion of colon cancer SW480 cells. Front. Genet. 2020, 11, 9. [Google Scholar] [CrossRef]
- Xiao, H.; Liu, M. Circular RNA hsa_circ_0053277 promotes the development of colorectal cancer by upregulating matrix metallopeptidase 14 via miR-2467-3p sequestration. J. Cell. Physiol. 2020, 235, 2881–2890. [Google Scholar]
- Wang, X.; Ren, Y.; Ma, S.; Wang, S. Circular RNA 0060745, a novel circRNA, promotes colorectal cancer cell proliferation and metastasis through miR-4736 sponging. OncoTargets Ther. 2020, 13, 1941. [Google Scholar]
- Chen, P.; Yao, Y.; Yang, N.; Gong, L.; Kong, Y.; Wu, A. Circular RNA circCTNNA1 promotes colorectal cancer progression by sponging miR-149-5p and regulating FOXM1 expression. Cell Death Dis. 2020, 11, 557. [Google Scholar] [CrossRef]
- Zeng, K.; Chen, X.; Xu, M.; Liu, X.; Hu, X.; Xu, T.; Sun, H.; Pan, Y.; He, B.; Wang, S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018, 9, 417. [Google Scholar] [CrossRef]
- Yang, B.; Du, K.; Yang, C.; Xiang, L.; Xu, Y.; Cao, C.; Zhang, J.; Liu, W. CircPRMT5 circular RNA promotes proliferation of colorectal cancer through sponging miR-377 to induce E2F3 expression. J. Cell. Mol. Med. 2020, 24, 3431–3437. [Google Scholar]
- Jiang, Z.; Hou, Z.; Liu, W.; Yu, Z.; Liang, Z.; Chen, S. Circular RNA protein tyrosine kinase 2 (circPTK2) promotes colorectal cancer proliferation, migration, invasion and chemoresistance. Bioengineered 2022, 13, 810–823. [Google Scholar]
- He, J.-H.; Li, Y.-G.; Han, Z.-P.; Zhou, J.-B.; Chen, W.-M.; Lv, Y.-B.; He, M.-L.; Zuo, J.-D.; Zheng, L. The CircRNA-ACAP2/Hsa-miR-21-5p/Tiam1 regulatory feedback circuit affects the proliferation, migration, and invasion of colon cancer SW480 cells. Cell. Physiol. Biochem. 2018, 49, 1539–1550. [Google Scholar]
- Wang, Y.; Gao, R.; Li, J.; Tang, S.; Li, S.; Tong, Q.; Mao, Y. Circular RNA hsa_circ_0003141 promotes tumorigenesis of hepatocellular carcinoma via a miR-1827/UBAP2 axis. Aging 2020, 12, 9793. [Google Scholar]
- Li, Z.; Liu, Y.; Yan, J.; Zeng, Q.; Hu, Y.; Wang, H.; Li, H.; Li, J.; Yu, Z. Circular RNA hsa_circ_0056836 functions an oncogenic gene in hepatocellular carcinoma through modulating miR-766-3p/FOSL2 axis. Aging 2020, 12, 2485. [Google Scholar] [CrossRef]
- Yu, Y.; Bian, L.; Liu, R.; Wang, Y.; Xiao, X. Circular RNA hsa_circ_0061395 accelerates hepatocellular carcinoma progression via regulation of the miR-877-5p/PIK3R3 axis. Cancer Cell Int. 2021, 21, 10. [Google Scholar]
- Bai, N.; Peng, E.; Qiu, X.; Lyu, N.; Zhang, Z.; Tao, Y.; Li, X.; Wang, Z. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J. Exp. Clin. Cancer Res. 2018, 37, 172. [Google Scholar] [CrossRef]
- Yu, X.; Sheng, P.; Sun, J.; Zhao, X.; Zhang, J.; Li, Y.; Zhang, Y.; Zhang, W.; Wang, J.; Liu, K. The circular RNA circMAST1 promotes hepatocellular carcinoma cell proliferation and migration by sponging miR-1299 and regulating CTNND1 expression. Cell Death Dis. 2020, 11, 340. [Google Scholar] [CrossRef]
- Li, S.; Weng, J.; Song, F.; Li, L.; Xiao, C.; Yang, W.; Xu, J. Circular RNA circZNF566 promotes hepatocellular carcinoma progression by sponging miR-4738-3p and regulating TDO2 expression. Cell Death Dis. 2020, 11, 452. [Google Scholar] [CrossRef]
- Jiang, W.; Wen, D.; Gong, L.; Wang, Y.; Liu, Z.; Yin, F. Circular RNA hsa_circ_0000673 promotes hepatocellular carcinoma malignance by decreasing miR-767-3p targeting SET. Biochem. Biophys. Res. Commun. 2018, 500, 211–216. [Google Scholar] [CrossRef]
- Guan, Z.; Tan, J.; Gao, W.; Li, X.; Yang, Y.; Li, X.; Li, Y.; Wang, Q. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J. Cell. Physiol. 2019, 234, 500–508. [Google Scholar] [CrossRef]
- Zhu, Q.; Lu, G.; Luo, Z.; Gui, F.; Wu, J.; Zhang, D.; Ni, Y. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochem. Biophys. Res. Commun. 2018, 497, 626–632. [Google Scholar] [CrossRef]
- Zhan, W.; Liao, X.; Chen, Z.; Li, L.; Tian, T.; Yu, L.; Wang, W.; Hu, Q. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J. Cell. Physiol. 2020, 235, 1733–1745. [Google Scholar] [CrossRef]
- Huang, G.; Liang, M.; Liu, H.; Huang, J.; Li, P.; Wang, C.; Zhang, Y.; Lin, Y.; Jiang, X. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis. 2020, 11, 1065. [Google Scholar] [CrossRef]
- Yu, L.; Gong, X.; Sun, L.; Zhou, Q.; Lu, B.; Zhu, L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS ONE 2016, 11, e0158347. [Google Scholar] [CrossRef]
- Lv, S.; Li, Y.; Ning, H.; Zhang, M.; Jia, Q.; Wang, X. CircRNA GFRA1 promotes hepatocellular carcinoma progression by modulating the miR-498/NAP1L3 axis. Sci. Rep. 2021, 11, 386. [Google Scholar]
- Chen, G.; Shi, Y.; Liu, M.; Sun, J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 175. [Google Scholar] [CrossRef]
- Li, Q.; Pan, X.; Zhu, D.; Deng, Z.; Jiang, R.; Wang, X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology 2019, 70, 1298–1316. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, L.; Bai, M.; Liu, Y.; Zhan, Y.; Deng, T.; Yang, H.; Sun, W.; Wang, X.; Zhu, K. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int. J. Cancer 2019, 144, 2501–2515. [Google Scholar]
- He, Y.; Wang, Y.; Liu, L.; Liu, S.; Liang, L.; Chen, Y.; Zhu, Z. Circular RNA circ_0006282 contributes to the progression of gastric cancer by sponging miR-155 to upregulate the expression of FBXO22. OncoTargets Ther. 2020, 13, 1001. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, X.; Guan, B.; Sun, P.; Hong, C.T.; Peng, J.; Tang, S.; Yang, J. A novel circular RNA hsa_circ_0008035 contributes to gastric cancer tumorigenesis through targeting the miR-375/YBX1 axis. Am. J. Transl. Res. 2019, 11, 2455. [Google Scholar]
- Huang, X.; Li, Z.; Zhang, Q.; Wang, W.; Li, B.; Wang, L.; Xu, Z.; Zeng, A.; Zhang, X.; Zhang, X. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol. Cancer 2019, 18, 71. [Google Scholar]
- Zhang, L.; Song, X.; Chen, X.; Wang, Q.; Zheng, X.; Wu, C.; Jiang, J. Circular RNA CircCACTIN promotes gastric cancer progression by sponging MiR-331-3p and regulating TGFBR1 expression. Int. J. Biol. Sci. 2019, 15, 1091–1103. [Google Scholar]
- Chen, D.-L.; Sheng, H.; Zhang, D.-S.; Jin, Y.; Zhao, B.-T.; Chen, N.; Song, K.; Xu, R.-H. The circular RNA circDLG1 promotes gastric cancer progression and anti-PD-1 resistance through the regulation of CXCL12 by sponging miR-141-3p. Mol. Cancer 2021, 20, 166. [Google Scholar] [CrossRef]
- Song, R.; Li, Y.; Hao, W.; Yang, L.; Chen, B.; Zhao, Y.; Sun, B.; Xu, F. Circular RNA MTO1 inhibits gastric cancer progression by elevating PAWR via sponging miR-199a-3p. Cell Cycle 2020, 19, 3127–3139. [Google Scholar]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef]
- Ouyang, Y.; Li, Y.; Huang, Y.; Li, X.; Zhu, Y.; Long, Y.; Wang, Y.; Guo, X.; Gong, K. CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR-186-5p and modulating NEK2. J. Cell. Physiol. 2019, 234, 10458–10469. [Google Scholar]
- Fan, D.; Wang, C.; Wang, D.; Zhang, N.; Yi, T. Circular RNA circ_0000039 enhances gastric cancer progression through miR-1292-5p/DEK axis. Cancer Biomark. 2021, 30, 167–177. [Google Scholar]
- Lu, J.; Wang, Y.-h.; Yoon, C.; Huang, X.-y.; Xu, Y.; Xie, J.-w.; Wang, J.-b.; Lin, J.-x.; Chen, Q.-y.; Cao, L.-l. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877–3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020, 471, 38–48. [Google Scholar]
- Pan, H.; Li, T.; Jiang, Y.; Pan, C.; Ding, Y.; Huang, Z.; Yu, H.; Kong, D. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J. Cell. Biochem. 2018, 119, 440–446. [Google Scholar] [CrossRef]
- Lv, L.; Du, J.; Wang, D.; Yan, Z. Circular RNA hsa_circ_0026344 suppresses gastric cancer cell proliferation, migration and invasion via the miR-590-5p/PDCD4 axis. J. Pharm. Pharmacol. 2022, 74, 1193–1204. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, H.; Hou, L.; Wang, G.; Zhang, R.; Huang, Y.; Chen, X.; Zhu, J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol. Cancer 2017, 16, 151. [Google Scholar]
- Sun, G.; Li, Z.; He, Z.; Wang, W.; Wang, S.; Zhang, X.; Cao, J.; Xu, P.; Wang, H.; Huang, X. Circular RNA MCTP2 inhibits cisplatin resistance in gastric cancer by miR-99a-5p-mediated induction of MTMR3 expression. J. Exp. Clin. Cancer Res. 2020, 39, 246. [Google Scholar] [CrossRef]
- Rong, D.; Lu, C.; Zhang, B.; Fu, K.; Zhao, S.; Tang, W.; Cao, H. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol. Cancer 2019, 18, 25. [Google Scholar]
- Qiu, Z.; Wang, L.; Liu, H. Hsa_circ_0001982 promotes the progression of breast cancer through miR-1287-5p/MUC19 axis under hypoxia. World J. Surg. Oncol. 2021, 19, 161. [Google Scholar]
- Xu, Y.; Qian, C.; Liu, C.; Fu, Y.; Zhu, K.; Niu, Z.; Liu, J. Investigation of the Mechanism of hsa_circ_000 1429 Adsorbed miR-205 to Regulate KDM4A and Promote Breast Cancer Metastasis. Contrast Media Mol. Imaging 2022, 2022, 4657952. [Google Scholar] [CrossRef]
- Xu, Y.; Yao, Y.; Leng, K.; Ji, D.; Qu, L.; Liu, Y.; Cui, Y. Increased expression of circular RNA circ_0005230 indicates dismal prognosis in breast cancer and regulates cell proliferation and invasion via miR-618/CBX8 signal pathway. Cell. Physiol. Biochem. 2018, 51, 1710–1722. [Google Scholar]
- Liu, L.H.; Tian, Q.Q.; Liu, J.; Zhou, Y.; Yong, H. Upregulation of hsa_circ_0136666 contributes to breast cancer progression by sponging miR-1299 and targeting CDK6. J. Cell. Biochem. 2019, 120, 12684–12693. [Google Scholar] [CrossRef]
- Yao, Y.; Fang, Z. Circular RNA-100219 promotes breast cancer progression by binding to microRNA-485-3p. J BUON 2019, 24, 501–508. [Google Scholar]
- Zeng, K.; He, B.; Yang, B.B.; Xu, T.; Chen, X.; Xu, M.; Liu, X.; Sun, H.; Pan, Y.; Wang, S. The pro-metastasis effect of circANKS1B in breast cancer. Mol. Cancer 2018, 17, 160. [Google Scholar]
- Qu, Y.; Dou, P.; Hu, M.; Xu, J.; Xia, W.; Sun, H. circRNA-CER mediates malignant progression of breast cancer through targeting the miR-136/MMP13 axis. Mol. Med. Rep. 2019, 19, 3314–3320. [Google Scholar]
- Zhang, W.; Liu, H.; Jiang, J.; Yang, Y.; Wang, W.; Jia, Z. CircRNA circFOXK2 facilitates oncogenesis in breast cancer via IGF2BP3/miR-370 axis. Aging 2021, 13, 18978. [Google Scholar]
- Wang, N.; Gu, Y.; Li, L.; Wang, F.; Lv, P.; Xiong, Y.; Qiu, X. Circular RNA circMYO9B facilitates breast cancer cell proliferation and invasiveness via upregulating FOXP4 expression by sponging miR-4316. Arch. Biochem. Biophys. 2018, 653, 63–70. [Google Scholar]
- Lin, G.; Wang, S.; Zhang, X.; Wang, D. Circular RNA circPLK1 promotes breast cancer cell proliferation, migration and invasion by regulating miR-4500/IGF1 axis. Cancer Cell Int. 2020, 20, 593. [Google Scholar]
- Zhang, H.; Ge, Z.; Wang, Z.; Gao, Y.; Wang, Y.; Qu, X. Circular RNA RHOT1 promotes progression and inhibits ferroptosis via mir-106a-5p/STAT3 axis in breast cancer. Aging 2021, 13, 8115. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, X.; Lan, J.; Wan, Y.; Zhu, X. Circular RNA circRPPH1 promotes triple-negative breast cancer progression via the miR-556-5p/YAP1 axis. Am. J. Transl. Res. 2020, 12, 6220. [Google Scholar]
- Chen, Z.G.; Zhao, H.J.; Lin, L.; Liu, J.B.; Bai, J.Z.; Wang, G.S. Circular RNA CirCHIPK3 promotes cell proliferation and invasion of breast cancer by sponging miR-193a/HMGB1/PI3K/AKT axis. Thorac. Cancer 2020, 11, 2660–2671. [Google Scholar]
- Zhang, X.-Y.; Mao, L. Circular RNA Circ_0000442 acts as a sponge of MiR-148b-3p to suppress breast cancer via PTEN/PI3K/Akt signaling pathway. Gene 2021, 766, 145113. [Google Scholar] [CrossRef]
- Meng, L.; Chang, S.; Sang, Y.; Ding, P.; Wang, L.; Nan, X.; Xu, R.; Liu, F.; Gu, L.; Zheng, Y. Circular RNA circCCDC85A inhibits breast cancer progression via acting as a miR-550a-5p sponge to enhance MOB1A expression. Breast Cancer Res. 2022, 24, 1. [Google Scholar] [CrossRef]
- Liu, W.-L.; Wang, H.-x.; Shi, C.-x.; Shi, F.-y.; Zhao, L.-y.; Zhao, W.; Wang, G.-h. MicroRNA-1269 promotes cell proliferation via the AKT signaling pathway by targeting RASSF9 in human gastric cancer. Cancer Cell Int. 2019, 19, 308. [Google Scholar]
- Wu, F.; Liu, F.; Dong, L.; Yang, H.; He, X.; Li, L.; Zhao, L.; Jin, S.; Li, G. miR-1273g silences MAGEA3/6 to inhibit human colorectal cancer cell growth via activation of AMPK signaling. Cancer Lett. 2018, 435, 1–9. [Google Scholar] [CrossRef]
- Xu, X.; Cao, L.; Zhang, Y.; Lian, H.; Sun, Z.; Cui, Y. MicroRNA-1246 inhibits cell invasion and epithelial mesenchymal transition process by targeting CXCR4 in lung cancer cells. Cancer Biomark. 2018, 21, 251–260. [Google Scholar] [CrossRef]
- Yu, W.; Jiang, H.; Zhang, H.; Li, J. Hsa_circ_0003998 promotes cell proliferation and invasion by targeting miR-326 in non-small cell lung cancer. OncoTargets Ther. 2018, 11, 5569. [Google Scholar]
- Fan, Z.; Bai, Y.; Zhang, Q.; Qian, P. CircRNA circ_POLA2 promotes lung cancer cell stemness via regulating the miR-326/GNB1 axis. Environ. Toxicol. 2020, 35, 1146–1156. [Google Scholar] [CrossRef]
- He, S.; Yang, J.; Jiang, S.; Li, Y.; Han, X. Circular RNA circ_0000517 regulates hepatocellular carcinoma development via miR-326/IGF1R axis. Cancer Cell Int. 2020, 20, 404. [Google Scholar] [CrossRef]
- Pan, G.; Mao, A.; Liu, J.; Lu, J.; Ding, J.; Liu, W. Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 2020, 53, e12720. [Google Scholar]
- Wang, S.; Hu, Y.; Lv, X.; Li, B.; Gu, D.; Li, Y.; Sun, Y.; Su, Y. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin. Sci. 2019, 133, 1935–1953. [Google Scholar] [CrossRef]
- Yang, D.; Hu, Z.; Zhang, Y.; Zhang, X.; Xu, J.; Fu, H.; Zhu, Z.; Feng, D.; Cai, Q. CircHIPK3 promotes the tumorigenesis and development of gastric cancer through miR-637/AKT1 pathway. Front. Oncol. 2021, 11, 637761. [Google Scholar]
- Wen, Y.; Li, B.; He, M.; Teng, S.; Sun, Y.; Wang, G. circHIPK3 promotes proliferation and migration and invasion via regulation of miR-637/HDAC4 signaling in osteosarcoma cells. Oncol. Rep. 2021, 45, 169–179. [Google Scholar] [CrossRef]
- Xu, L.; Feng, X.; Hao, X.; Wang, P.; Zhang, Y.; Zheng, X.; Li, L.; Ren, S.; Zhang, M.; Xu, M. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J. Exp. Clin. Cancer Res. 2019, 38, 98. [Google Scholar]
- Xiao, W.; Zheng, S.; Zou, Y.; Yang, A.; Xie, X.; Tang, H.; Xie, X. CircAHNAK1 inhibits proliferation and metastasis of triple-negative breast cancer by modulating miR-421 and RASA1. Aging 2019, 11, 12043. [Google Scholar]
- Yang, C.; Yuan, W.; Yang, X.; Li, P.; Wang, J.; Han, J.; Tao, J.; Li, P.; Yang, H.; Lv, Q. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol. Cancer 2018, 17, 19. [Google Scholar] [CrossRef]
- Li, P.; Chen, H.; Chen, S.; Mo, X.; Li, T.; Xiao, B.; Yu, R.; Guo, J. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br. J. Cancer 2017, 116, 626–633. [Google Scholar]
- Wang, Y.; Yin, H.; Chen, X. Circ-LDLRAD3 enhances cell growth, migration, and invasion and inhibits apoptosis by regulating MiR-224-5p/NRP2 axis in gastric cancer. Dig. Dis. Sci. 2021, 66, 3862–3871. [Google Scholar] [CrossRef]
- Chen, T.; Yang, Z.; Liu, C.; Wang, L.; Yang, J.; Chen, L.; Li, W. Circ_0078767 suppresses non-small-cell lung cancer by protecting RASSF1A expression via sponging miR-330-3p. Cell Prolif. 2019, 52, e12548. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, R.; Zhang, D.; Lu, T.; Yu, W.; Wo, Y.; Liu, A.; Sui, T.; Cui, J.; Qin, Y. Circ-ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR-330-5p to promote non-small cell lung cancer progression. Transl. Lung Cancer Res. 2019, 8, 862. [Google Scholar]
- Lu, C.; Fu, L.; Qian, X.; Dou, L.; Cang, S. Knockdown of circular RNA circ-FARSA restricts colorectal cancer cell growth through regulation of miR-330-5p/LASP1 axis. Arch. Biochem. Biophys. 2020, 689, 108434. [Google Scholar] [CrossRef]
- Fang, C.; Huang, X.; Dai, J.; He, W.; Xu, L.; Sun, F. The circular RNA circFARSA sponges microRNA-330-5p in tumor cells with bladder cancer phenotype. BMC Cancer 2022, 22, 1–14. [Google Scholar] [CrossRef]
- Jia, Q.; Ye, L.; Xu, S.; Xiao, H.; Xu, S.; Shi, Z.; Li, J.; Chen, Z. Circular RNA 0007255 regulates the progression of breast cancer through miR-335-5p/SIX2 axis. Thorac. Cancer 2020, 11, 619–630. [Google Scholar] [CrossRef]
- Liang, H.-F.; Zhang, X.-Z.; Liu, B.-G.; Jia, G.-T.; Li, W.-L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am. J. Cancer Res. 2017, 7, 1566. [Google Scholar]
- Hu, C.; Wang, Y.; Li, A.; Zhang, J.; Xue, F.; Zhu, L. Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR-545/EIF3C axis. J. Cell. Physiol. 2019, 234, 9225–9232. [Google Scholar]
- Liu, J.; Wang, D.; Long, Z.; Liu, J.; Li, W. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p/519-5p family and modulating the expression of CBX8. Cell. Physiol. Biochem. 2018, 48, 173–184. [Google Scholar] [CrossRef]
- Xia, L.; Wu, L.; Bao, J.; Li, Q.; Chen, X.; Xia, H.; Xia, R. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/β-catenin pathway. Biochem. Biophys. Res. Commun. 2018, 503, 385–390. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, C.; Ma, J.-X.; Ren, H.; Sun, Y.; Xu, J.-Z. Circular RNA PIP5K1A promotes colon cancer development through inhibiting miR-1273a. World J. Gastroenterol. 2019, 25, 5300. [Google Scholar]
- Lin, C.; Ma, M.; Zhang, Y.; Li, L.; Long, F.; Xie, C.; Xiao, H.; Liu, T.; Tian, B.; Yang, K. The N6-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol. Cancer 2022, 21, 373. [Google Scholar]
- Li, R.; Wu, B.; Xia, J.; Ye, L.; Yang, X. Circular RNA hsa_circRNA_102958 promotes tumorigenesis of colorectal cancer via miR-585/CDC25B axis. Cancer Manag. Res. 2019, 11, 6887. [Google Scholar] [CrossRef]
- Ma, Z.; Ma, J.; Lang, B.; Xu, F.; Zhang, B.; Wang, X. Circ_0001982 Up-regulates the Expression of E2F1 by Adsorbing miR-1205 to Facilitate the Progression of Glioma. Mol. Biotechnol. 2022, 1–11. [Google Scholar] [CrossRef]
- Yi, C.; Li, H.; Li, D.; Qin, X.; Wang, J.; Liu, Y.; Liu, Z.; Zhang, J. Upregulation of circular RNA circ_0034642 indicates unfavorable prognosis in glioma and facilitates cell proliferation and invasion via the miR-1205/BATF3 axis. J. Cell. Biochem. 2019, 120, 13737–13744. [Google Scholar] [CrossRef]
- Qin, L.; Zhan, Z.; Wei, C.; Li, X.; Zhang, T.; Li, J. Hsa-circRNA-G004213 promotes cisplatin sensitivity by regulating miR-513b-5p/PRPF39 in liver cancer Corrigendum in/10.3892/mmr. 2021.12359. Mol. Med. Rep. 2021, 23, 1–12. [Google Scholar]
- Liu, Q.; Wang, C.; Jiang, Z.; Li, S.; Li, F.; Tan, H.B.; Yue, S.Y. circRNA 001306 enhances hepatocellular carcinoma growth by up-regulating CDK16 expression via sponging miR-584-5p. J. Cell. Mol. Med. 2020, 24, 14306–14315. [Google Scholar]
- Zhang, X.; Xu, Y.; Qian, Z.; Zheng, W.; Wu, Q.; Chen, Y.; Zhu, G.; Liu, Y.; Bian, Z.; Xu, W. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 1091. [Google Scholar] [CrossRef]
- Liu, G.; Shi, H.; Deng, L.; Zheng, H.; Kong, W.; Wen, X.; Bi, H. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2019, 513, 207–212. [Google Scholar]
- Zhou, Y.; Zheng, X.; Xu, B.; Chen, L.; Wang, Q.; Deng, H.; Jiang, J. Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem. Biophys. Res. Commun. 2019, 508, 527–535. [Google Scholar]
- Zhang, S.; Zeng, X.; Ding, T.; Guo, L.; Li, Y.; Ou, S.; Yuan, H. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci. Rep. 2018, 8, 2878. [Google Scholar]
- Wei, H.; Pan, L.; Tao, D.; Li, R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem. Biophys. Res. Commun. 2018, 503, 56–61. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, H.; Chen, Z.; Li, G.; Liu, B. Circular RNA ATXN7 promotes the development of gastric cancer through sponging miR-4319 and regulating ENTPD4. Cancer Cell Int. 2020, 20, 25. [Google Scholar]
- Li, B.; Liang, L.; Chen, Y.; Liu, J.; Wang, Z.; Mao, Y.; Zhao, K.; Chen, J. Circ_0008287 promotes immune escape of gastric cancer cells through impairing microRNA-548c-3p-dependent inhibition of CLIC1. Int. Immunopharmacol. 2022, 111, 108918. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, F.; Xiao, X.; Xie, F.; Tao, D.; Huang, C.; Liu, D.; Wang, M.; Wang, L.; Zeng, F. Circ HIPK 3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017, 18, 1646–1659. [Google Scholar]
- Yang, S.; Gao, S.; Liu, T.; Liu, J.; Zheng, X.; Li, Z. Circular RNA SMARCA5 functions as an anti-tumor candidate in colon cancer by sponging microRNA-552. Cell Cycle 2021, 20, 689–701. [Google Scholar]
- Gao, S.; Yu, Y.; Liu, L.; Meng, J.; Li, G. Circular RNA hsa_circ_0007059 restrains proliferation and epithelial-mesenchymal transition in lung cancer cells via inhibiting microRNA-378. Life Sci. 2019, 233, 116692. [Google Scholar] [CrossRef]
- Wu, Z.; Shi, W.; Jiang, C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis. Biochem. Biophys. Res. Commun. 2018, 502, 465–471. [Google Scholar]
- Fang, J.; Hong, H.; Xue, X.; Zhu, X.; Jiang, L.; Qin, M.; Liang, H.; Gao, L. A novel circular RNA, circFAT1 (e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019, 442, 222–232. [Google Scholar]
- Liu, T.; Liu, S.; Xu, Y.; Shu, R.; Wang, F.; Chen, C.; Zeng, Y.; Luo, H. Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR-130a/miR-107 and modulating PTEN. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2018, 50, 1396–1417. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, W.R.; Park, E.G.; Lee, D.H.; Lee, Y.J.; Bae, W.H.; Kim, H.-S. The Tumorigenic Role of Circular RNA-MicroRNA Axis in Cancer. Int. J. Mol. Sci. 2023, 24, 3050. https://doi.org/10.3390/ijms24033050
Kim WR, Park EG, Lee DH, Lee YJ, Bae WH, Kim H-S. The Tumorigenic Role of Circular RNA-MicroRNA Axis in Cancer. International Journal of Molecular Sciences. 2023; 24(3):3050. https://doi.org/10.3390/ijms24033050
Chicago/Turabian StyleKim, Woo Ryung, Eun Gyung Park, Du Hyeong Lee, Yun Ju Lee, Woo Hyeon Bae, and Heui-Soo Kim. 2023. "The Tumorigenic Role of Circular RNA-MicroRNA Axis in Cancer" International Journal of Molecular Sciences 24, no. 3: 3050. https://doi.org/10.3390/ijms24033050
APA StyleKim, W. R., Park, E. G., Lee, D. H., Lee, Y. J., Bae, W. H., & Kim, H.-S. (2023). The Tumorigenic Role of Circular RNA-MicroRNA Axis in Cancer. International Journal of Molecular Sciences, 24(3), 3050. https://doi.org/10.3390/ijms24033050







