1. Introduction
Enzymes are biocatalysts with high substrate specificity, sensitivity and exceptional catalytic activity under mild physiological conditions [
1]. They are used in many industrial and pharmaceutical areas for both diagnostics and compensation therapy of inherited and metabolic diseases [
1,
2,
3,
4]. Currently, exogenous enzymes are intravenously infused into the bloodstream of patients with enzyme deficiency. Mandatory frequent administration of enzymes leads to high costs and negatively impacts patient compliance [
1,
2,
3]. Moreover, proteolytic hydrolysis, oxidation and partial denaturation in the presence of ligands and on surfaces may lower the performance of enzyme-based analytical devices or limit industrial efficiency, thus hampering the further implementation of natural enzymes [
1,
2,
3,
4,
5]. Poor membrane permeability and biological barrier-crossing capacities of free enzymes constrain the treatment of diseases [
1,
2,
3,
4].
Great expectations of nanomedicine are caused by enhanced pharmacokinetic stability of NP, reduced toxicity, improved bioavailability and biodistribution. Successful enzyme delivery systems must be able to overcome several physiological barriers and release bioactive enzymes at the target organs or tissues, often inside the living cells. Currently, there are many types of nanocarriers of bioactive enzymes and enzyme-responsive drug delivery systems capable of releasing drugs in a controlled manner [
2,
3,
4]. Liposomes, polymeric and inorganic NP can deliver encapsulated drugs on demand [
2,
3]. To increase stability, protein or medium engineering and chemical modification were used [
4]. Immobilization of enzymes onto carriers, or entrapment within a matrix, framework or NP can also delay their degradation [
1,
4], but partial denaturation of proteins after attachment to solid support [
5], aggregation or agglomeration cannot be excluded. Multiple enzyme molecules can be contained within nanomaterials or matrices of different origins. ENP consisting of glucose oxidase, HRP, uricase, cholesterol oxidase and hemoglobin were immobilized onto a matrix (electrode or membrane) in order to construct diagnostic biosensors [
6]. A cell membrane-coated porous metal–organic zeolitic imidazolate framework has been suggested to encapsulate recombinant enzyme–uricase for intracellular delivery [
7]. However, only 38% of the initial activity of the inputted uricase was retained in the metal–organic framework [
7]. Moreover, an induced immune response may destroy foreign nanomaterials. The same concerns can be relevant to metallic or organic NP loaded with proteins or drugs [
8,
9,
10].
ENP are the stable water-insoluble aggregates of enzymes or NP with surface enzymes of 10–500 nm in sizes capable of (1) maintaining enzymatic activity; (2) ensuring long-term stability against temperature, dehydration, organic solvents, and aggressive pH; and (3) enabling a tuning or reversible switching of enzyme activity. Enzymes can be stabilized by thin shells consisting of polymers prepared either by
in situ polymerization or by assembling preformed polymers [
4].
ENP provides many benefits, including increased enzyme stability, decreased degradation during circulation and targeting to the desired locations. Moreover, nanocarriers have the advantage of accumulating preferentially in solid tumors through the enhanced penetration and retention (EPR) effect. Other tumor-associated features include altered gene expression of enzymes and leaky vasculature [
2]. Moreover, enzyme-containing nanostructures may be used for enzyme prodrug therapy for cancer. In particular, the enzyme HRP (which oxidizes the plant growth phytohormone—prodrug indole-3-acetic acid (IAA) to release toxic oxidative species) can be conjugated with Au NP for intracellular delivery in breast cancer cells. The nanodevice is biocompatible and effectively internalized by breast cancer cell lines. Co-treatment with HRP-Au NP and IAA reduced the viability of breast cancer cells below 5%, whereas the free formulated components (HRP, IAA) had no effect [
11]. Overall, the ability of nanotechnology to improve the pharmacologic profile of a drug promises to increase efficacy while decreasing unspecific side effects.
Besides inorganic and organic NPs, virus-like particles (VLP) can be decorated with different drugs and enzymes. The attachment of the α-glucosidase Ima1p of
Saccharomyces cerevisiae to the surface of parvovirus B19 VLP resulted in increased V
max toward p-nitrophenyl-α-D-glucopyranoside, a notorious 10 °C shift in its optimal temperature, from 35 °C for the non-attached enzyme to 45 °C for the enzyme attached to the NP. The decorated VLP may be further developed as part of therapy for the treatment of lysosomal storage diseases derived from defects in the human acid α-glucosidase [
12].
A deoxyribonuclease (DNase) is a generic term for enzymes that catalyze the hydrolysis of phosphodiester bonds in the DNA. There are two main types of DNases found in metazoans: deoxyribonuclease I (
EC 3.1.21.1) and deoxyribonuclease II. In biotechnology, DNase is used to degrade DNA from highly viscous bacterial lysates for the isolation of recombinant proteins. In medicine, DNase combined with intrapleural tissue plasminogen activator can be used to increase pleural drainage, decrease hospital length of stay, and decrease the need for surgery in parapneumonic effusions and empyema. DNase may also be used together with mucolytic agents like N-acetyl cysteine (NAC) to lower the viscosity of pulmonary mucosal secretion. They break the disulfide bonds linking the mucin monomers and depolarize the mucin network [
13]. DNases derived from pathogens are necessary for wound infection monitoring.
Ribonuclease (RNase) is a type of nuclease that catalyzes the hydrolytic cleavage of RNA into shorter fragments. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. All known organisms contain many RNases of both classes, showing that RNA degradation is an evolutionary ancient and important process. RNases are required for the maturation of all types of RNA and their clearance. In addition, RNA degradation systems are the first defense lines against RNA-containing viruses and RNA interference. RNase A (EC 3.1.27.5) is specific to single-stranded RNAs. It cleaves the 3′-end of unpaired C and U residues, ultimately forming a 3′-phosphorylated product via a 2′,3′-cyclic monophosphate intermediate. RNase A from bovine pancreas is widely used in biomedical research and genetic engineering due to the developed methods of isolation and amazing stability. RNase 7, belonging to the RNase A superfamily is secreted by human skin and serves as a potent antipathogen defense. RNases take part in the unspecific protection of the respiratory tract due to antiviral and bactericidal activity.
Serine protease trypsin (EC 3.4.21.4) is abundant in pancreases and can be easily purified. Trypsin cuts peptide chains mainly on the carboxyl side of the amino acids lysine or arginine. Due to the developed procedure of isolation and known biochemical properties, trypsin is widely used in biotechnological processes (for resuspension and harvesting adhesive cells; for destruction of casein in breast milk to produce hypoallergenic food; in the extraction and control aroma formation in cheese and milk products; to tenderize meat) and medicine (in proteomics to digest proteins before mass spectrometry; to dissolve blood clots in its recombinant form and to treat inflammation in its pancreatic form). In veterinary, trypsin is an ingredient in wound spray to dissolve dead tissue and pus in wounds. Trypsin reduces the barrier properties of sputum and can be used for the combined therapy of respiratory viral infections [
13,
14].
Chymotrypsin (EC 3.4.21.1) is also a serine endopeptidase secreted by the pancreas. Chymotrypsin precursor—chymotrypsinogen is activated by trypsin. Chymotrypsin preferentially cleaves peptide bonds where the N-terminal amino acid is a large hydrophobic amino acid (tyrosine, tryptophan and phenylalanine) but can also hydrolyze other amide bonds in peptides at slower rates, particularly those containing leucine and methionine.
Antioxidant enzyme catalase (EC 1.11.1.6) is Mn III containing hemoprotein with four iron-containing heme groups which catalyzes the decomposition of hydrogen peroxide to water and oxygen in almost all aerobic organisms for protection from oxidative damage by reactive oxygen species (ROS). Catalase is also involved in aging, inflammation, apoptosis, ethanol metabolism and cancer. H2O2 interferes with the production of melanin, the pigment that gives hair its color. Therefore, low levels of catalase may play a role in hair graying. Catalase is used in the food industry for removing hydrogen peroxide from milk before cheese production and in food wrappers to prevent food oxidizing; in the textile industry to make peroxide-free fabrics; and in contact lens hygiene. The catalase test is important for identifying bacterial species.
Horseradish peroxidase (HRP) (EC 1.11.1.7) is a redox glycoenzyme with a ferroprotoporphyrin group at the active site, which is naturally found in horseradish roots. It catalyzes the oxidation of various protons with peroxide or alkylperoxides. Due to its low cost and broad substrate specificity, HRP is widely used in immunodiagnostics for the detection of proteins and nucleic acids. HRP, in combination with the naturally occurring plant growth phytohormone IAA, has demonstrated antitumor activity in vitro and in vivo [
11]. It is suitable for removing diluted emulsified oil and dissolved oil from water [
15].
Lipases from both animal and bacterial sources catalyze the hydrolysis of fats and are involved in metabolism of dietary triglycerides in most organisms, cell signaling and inflammation. Besides that, lipases are widely used in laundry detergents. Acidic adipocyte triacylglycerol lipase (EC 3.1.1.3) is found in animals, plants, fungi and bacteria. It hydrolyses triglycerides into diglycerides and subsequently into monoglycerides and free fatty acids. The pancreatic enzyme requires a protein cofactor, namely colipase, to counteract the inhibitory effects of bile salts. Blood tests for lipase may be used to diagnose acute pancreatitis and other disorders of the pancreas.
To construct stable protein NP without any cross-linking, a novel method of nanoprecipitation of proteins from their solutions in fluoroalcohol was developed and applied for fabrication of NP from albumin, fibrinogen, immunoglobulin and lysozyme [
16]. The current research was aimed at ENP construction, analysis of their conformation stability, enzyme activity and possible toxicity for tissue cultures.
4. Discussion
Protein NP can be constructed by different methods of two categories: emulsification and precipitation. Nanoprecipitation is based on the gradual reduction of the solvent in which the main constituent of the NP is dissolved. Depending on the approach used to reduce the solvent quality, nanoprecipitation can be classified into different subtypes, such as non-solvent precipitation, desolvation, coacervation, salting out and albumin-bound technology [
22]. Our method of protein nanoprecipitation is based on non-solvent precipitation [
16]. Non-solvent precipitation includes three steps: generation of supersaturation, nucleation and growth of NP. Proteins were dissolved in fluoroalcohol as a solvent, while 40% ethanol was used as a poor solvent for proteins. By vigorous mixing of alcohols with water, protein supersaturation permits the formation of protein nuclei with subsequent condensation of free protein molecules around the nuclei, thus creating protein NP [
16,
22]. This method does not require the use of toxic cross-linkers, such as glutaraldehyde. The intrinsic property of proteins to form structural motifs, such as α-helices and β-sheets leads to aggregation in NP [
17]. Protein nanoprecipitation was previously developed for albumin. The protein NP were shown to retain their abilities to bind with ligands and specific antibodies [
16]. Enzymatic activity was earlier shown for lysozyme NP capable of destroying bacterial cell walls [
16]. It corresponds to the metastability of alcohol-denatured lysozyme [
23]. One possible reason is known to be the stability of α-helixes of the lysozyme active center in fluoroalcohol solution. The active site of lysozyme consists of a deep crevice, which divides the protein into two domains linked by an alpha helix [
23]. The structure of the lysozyme is consistent under a variety of conditions. Lysozyme appeared to be thermally stable and active at pH 6–9. To our knowledge, the structures and functions of the ENP consisting of nucleases, proteases, lipase and antioxidant enzymes have not been previously studied.
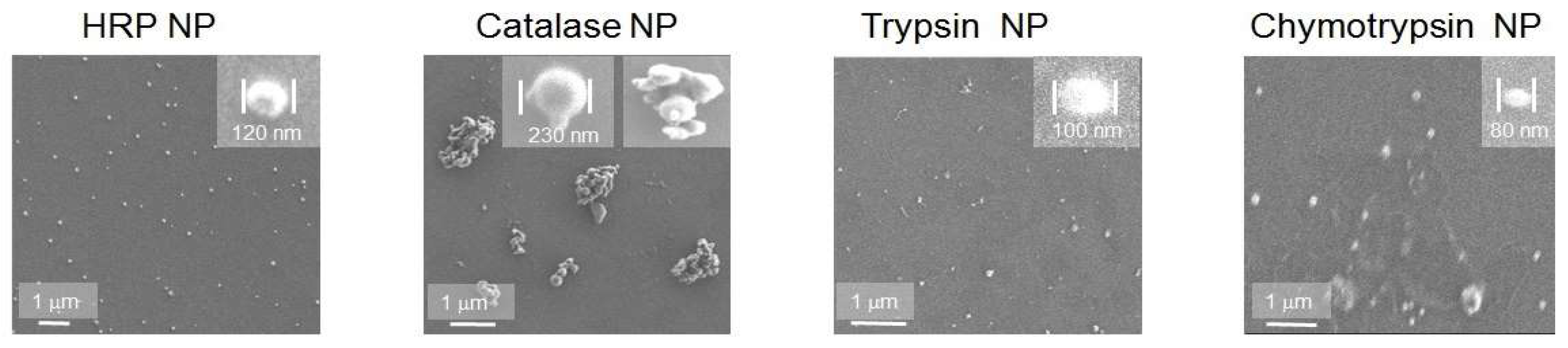
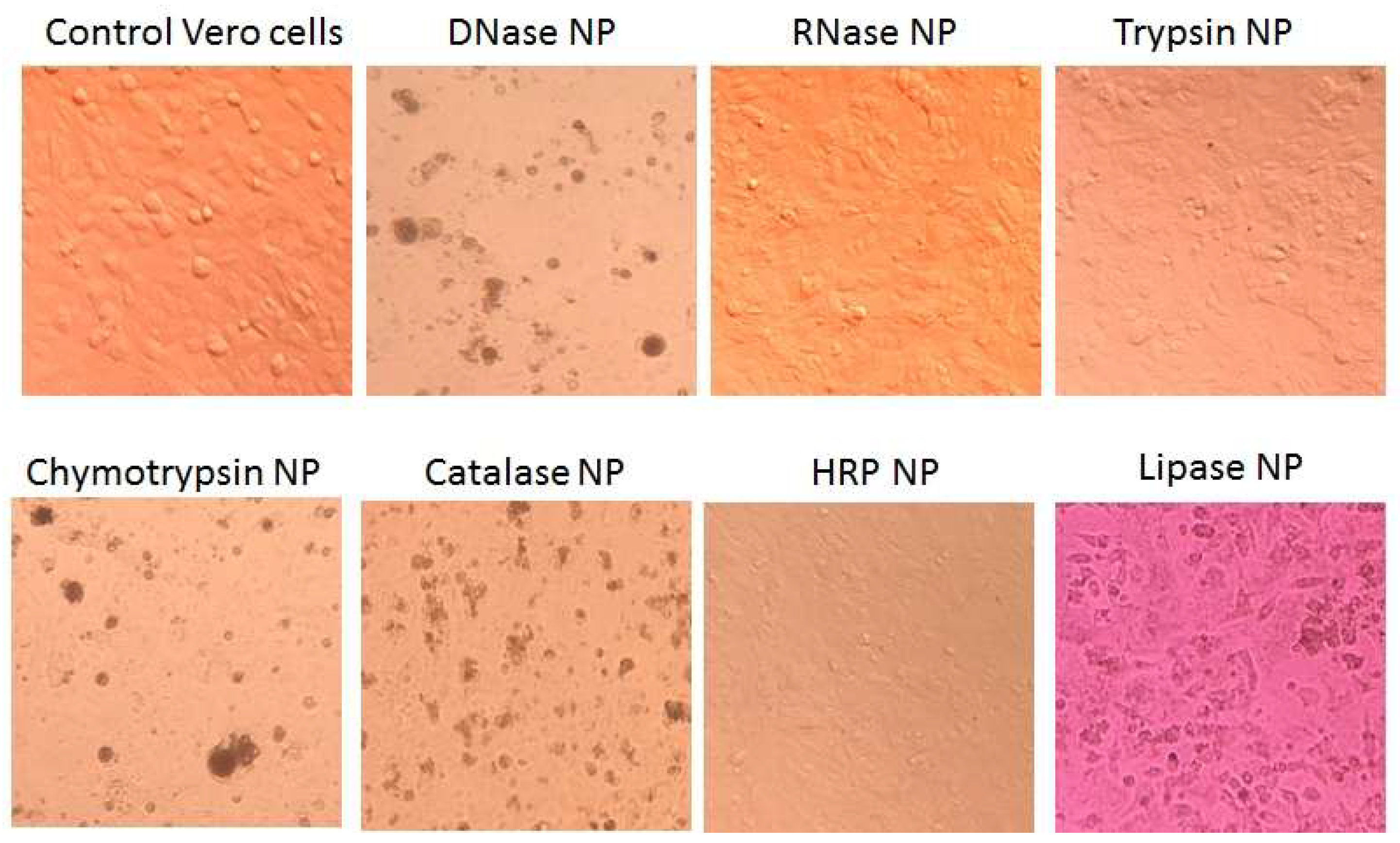
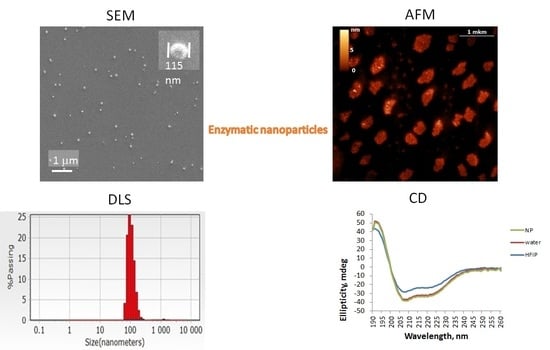
SEM, AFM and DLS demonstrated ENP of irregular shapes with broad overlapping sizes ranging from 50 to 300 nm, similar to the sizes of protein NP from BSA, fibrinogen and lysozyme, without strict correlation between original enzyme molecular weight or protein concentrations and ENP diameters (
Figure 1,
Figure 2,
Figure 3 and
Figures S1 and S2) [
16]. The smallest ENP were fabricated from lysozyme (MW 14.7 kDa, NP sizes 10–40 nm) [
16] and chymotrypsin (MW 499.5 kDa, NP sizes 50–100 nm) (
Figure 1). Usually, NP sizes range from a few nanometers to several hundreds nm, which enables endocytosis-mediated cellular uptake and an enhanced permeability and retention (EPR) effect in tumor tissues [
3].
CD spectra of the enzymes in HFIP solutions (
Supplementary Materials Figure S3) confirmed the stability of α-helixes in fluoroalcohols [
18,
19]. It is noteworthy that CD spectra (
Supplementary Materials Figure S3) and their deconvolution analysis (
Table 1) differed for the enzymes in HFIP used for the nanoprecipitation of the proteins and in the resulting ENP. Trypsin, chymotrypsin and catalase NP had similar secondary structures in their water solutions and in the corresponding ENP (
Table 1,
Supplementary Materials Figure S3), but for DNase NP, HRP NP (
Table 1) and especially for lipase NP, a significant decrease in content of α-helixes was found despite known stability of α-helixes in HFIP solution [
18,
19]. RNase A, with a small molecular weight of 13.7 kDa, is composed of three α-helixes and seven β-strands arranged in two “lobes”. Conformations of RNase A in water solutions and NP were shown to differ significantly (
Table 1,
Supplementary Materials Figure S3). Nevertheless, due to the well-known remarkable stability of RNase active center RNase, NP produced by nanoprecipitation did not lose their catalytic properties (
Figure 5B,
Supplementary Materials Table S1).
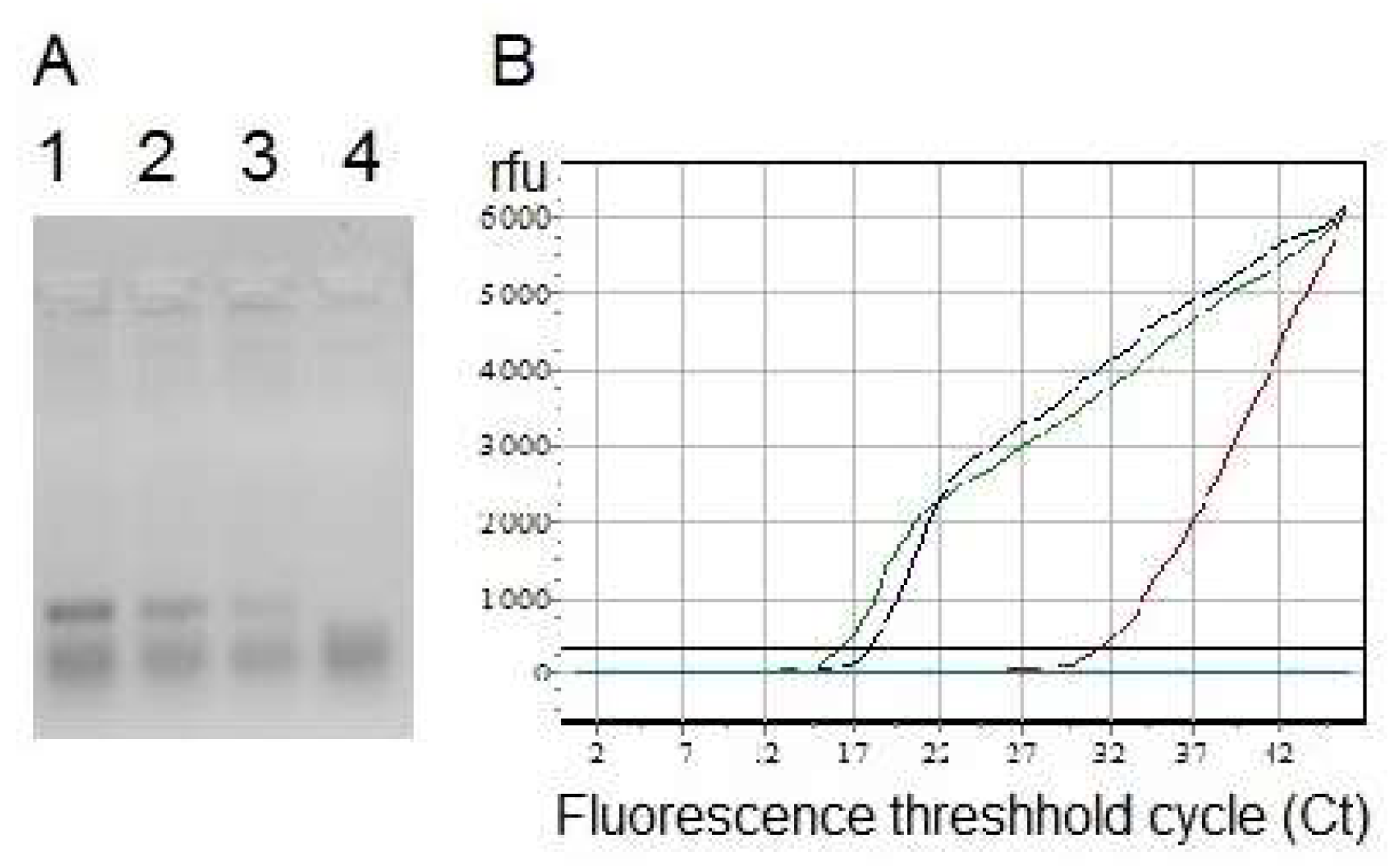
Nuclease activities of DNase and RNase NP were shown by means of gel electrophoresis of DNA (
Figure 5A) and RNA before and after treatment with the enzymes in water and in NP, followed by ethidium bromide staining and by the most sensitive detection method—reverse transcription with real-time PCR (
Figure 5B,
Supplementary Materials Table S1). The last approach confirmed the exhaustive hydrolysis of cDNA and RNA in the presence of nuclease NP.
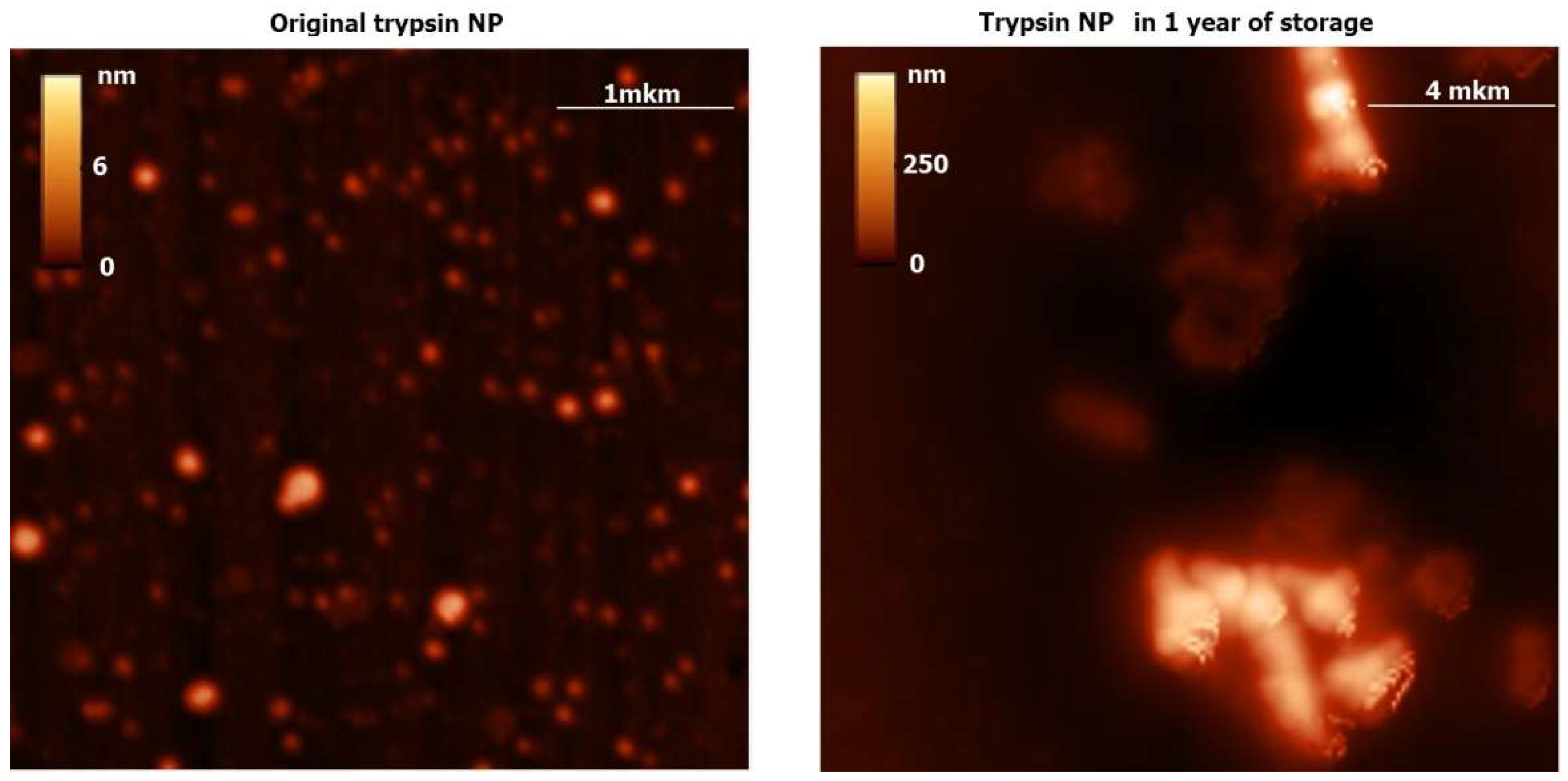
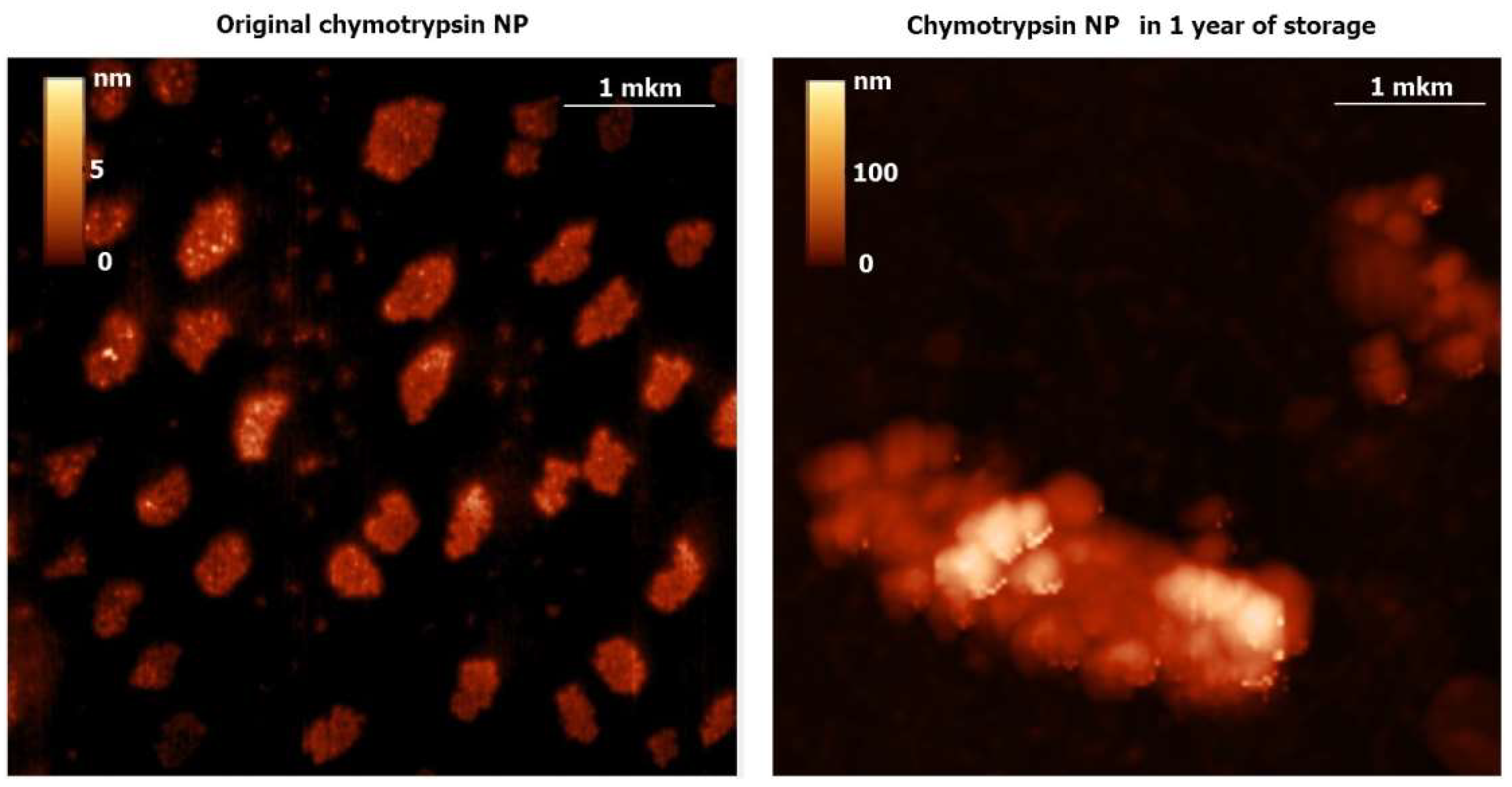
Proteolytic hydrolysis after addition of the trypsin NP (
Supplementary Materials Figure S4) and chymotrypsin NP despite long-term stability of both protease ENP (
Figure 2,
Figure 3 and
Figure S1) could be explained by storage of the ENP at cold temperatures (+4 °C) in water, thus preventing protease autocleavage.
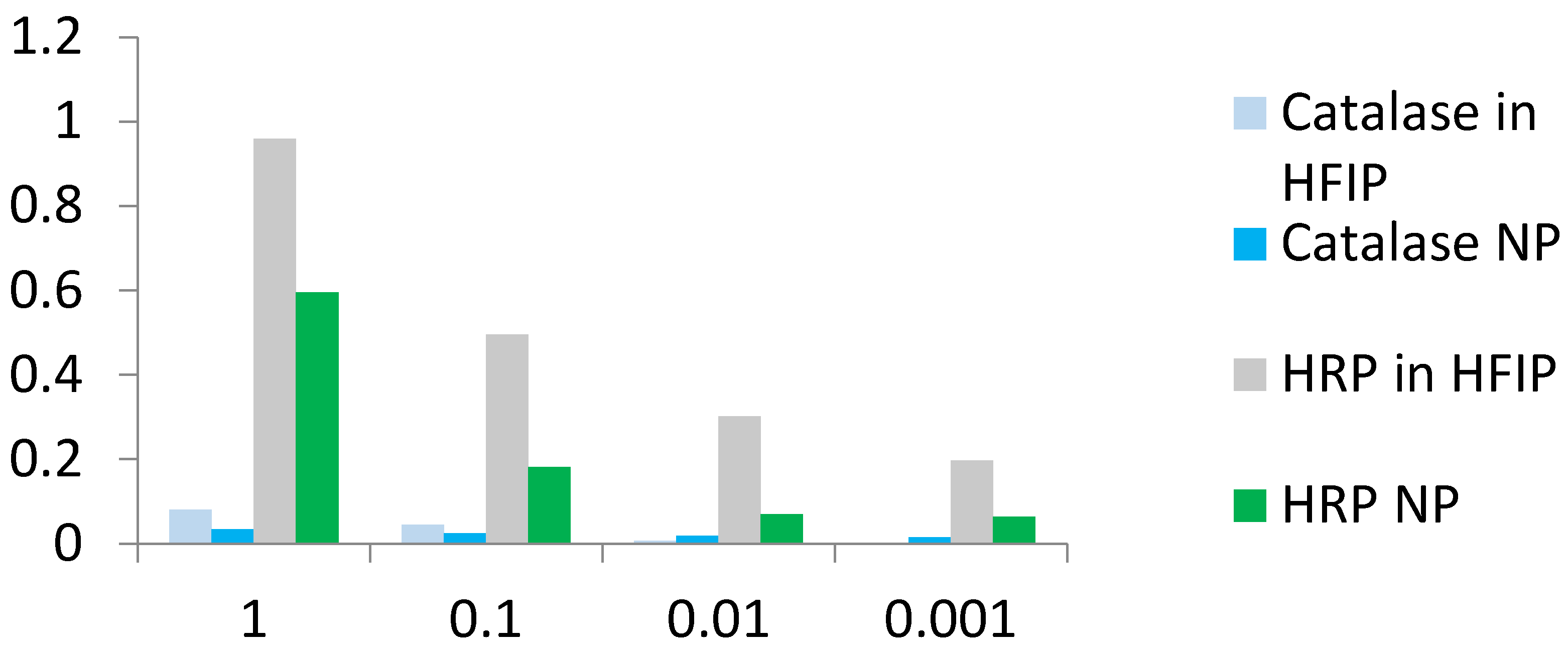
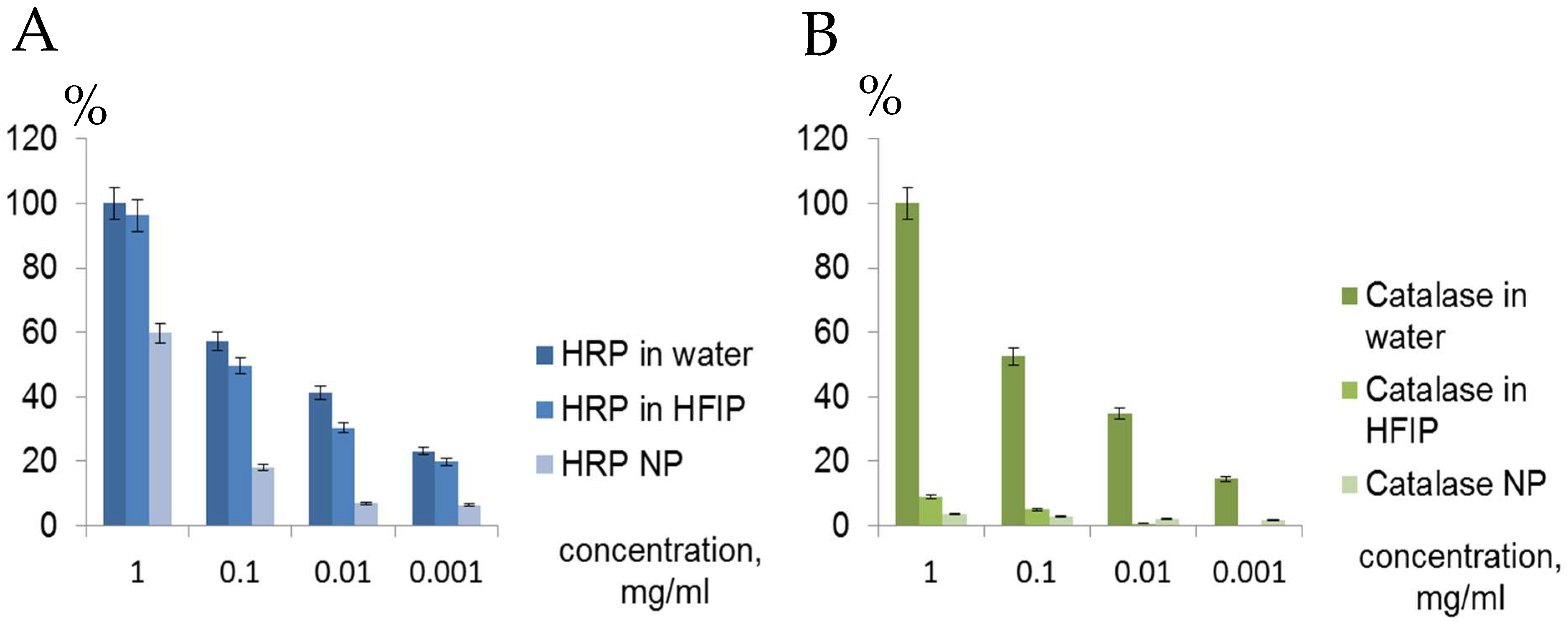
CD spectra of catalase in water, HFIP and NP coincided well (
Supplementary Materials Figure S3), but catalase activity in HFIP solution and especially in the ENP with hydrogen peroxide and TMB was weak compared to HRP solution in HFIP and HRP NP (
Figure 6 and
Figure 7). Catalase is a tetramer of four polypeptide chains, each over 500 amino acids long with four iron-containing heme groups which permit the destruction of hydrogen peroxide. The core of each subunit is generated by an eight-stranded antiparallel β-barrel (β1-8), with nearest neighbor connectivity capped by β-barrel loops on one side and α9 loops on the other. A helical domain at one face of the β-barrel is composed of four C-terminal helices (α16, α17, α18, and α19) and four helices derived from residues between β4 and β5 (α4, α5, α6, and α7). The optimum pH for human catalase is approximately 7, and the rate of reaction does not change appreciably between pH 6.8 and 7.5. Catalase has one of the highest turnover numbers of all enzymes; one catalase molecule can convert millions of hydrogen peroxide molecules to water and oxygen each second. Catalase solution in HFIP remained colored, suggesting the presence of hemoprotein, but pellets of the catalase NP were white. The stable secondary structure of the catalase in fluoroalcohol HFIP and in the catalase NP (
Supplementary Materials Figure S3,
Table 1) did not provide significant catalytic activity (
Figure 6 and
Figure 7). Possible reasons for the limited activity and low stability of the catalase NP are the aggregation in microparticles of various shapes and sizes (
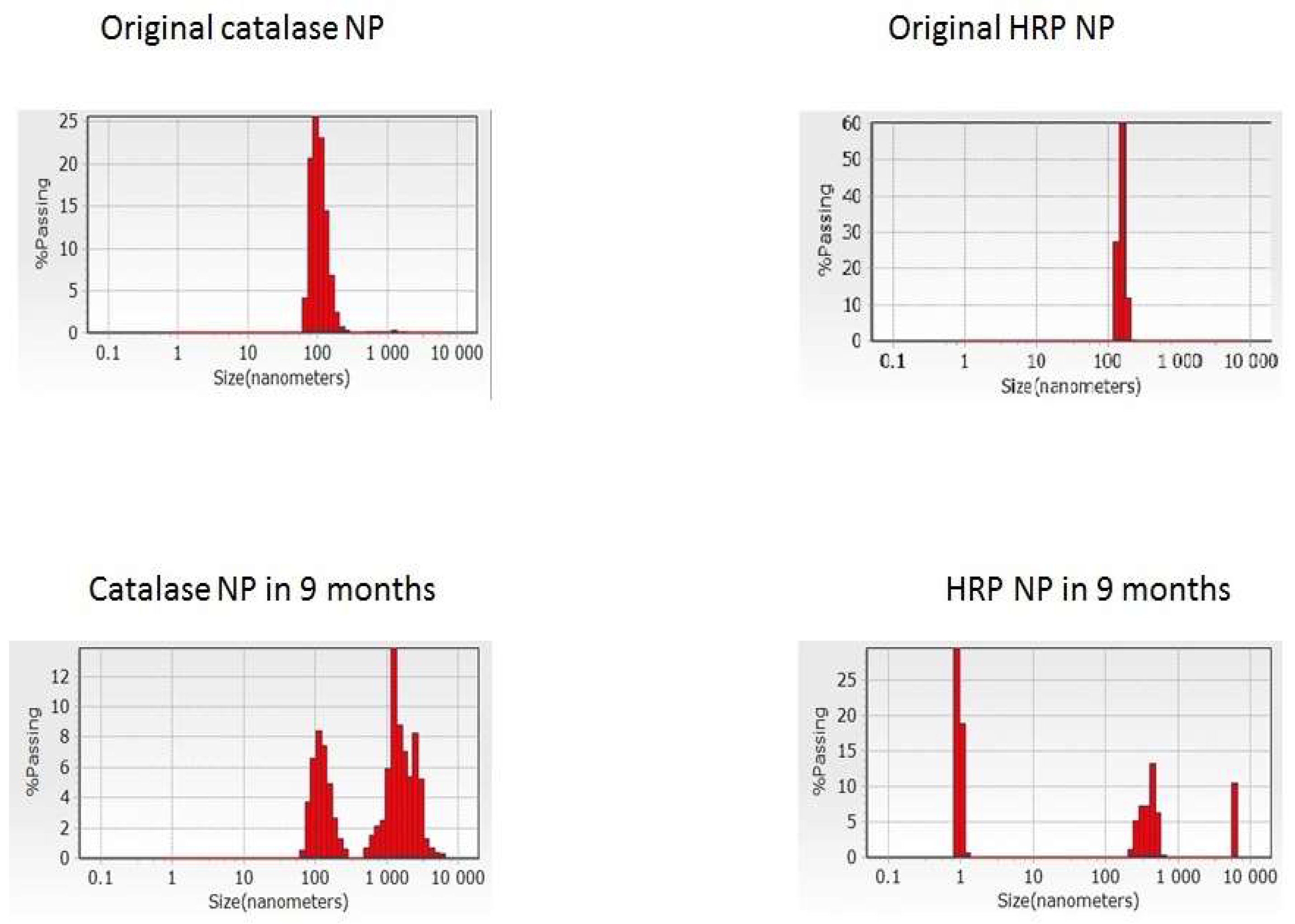
Figure 1), the loss of the iron-containing heme groups during the catalase NP precipitation and probable unavailability of the substrate-binding and catalytic centers.
HRP is a large α-helical brown protein with a molecular weight of 40 kDa that binds heme as a redox cofactor. It consists of a colorless protein (apo-enzyme) combined with an iron-porphyrin. HRP is stable up to 69 °C and from pH 4.5 to 12. By adding the sugars and zinc ions, the peroxidase retains an activity until ~75 °C. Due to the high stability of the HRP, the corresponding ENP remained active after storage at +4 °C for 1 year despite the conformational rearrangements (
Supplementary Materials Figure S3,
Table 1). From a biomedical point of view, HRP presents numerous advantageous features, namely, biocompatibility, high stability, high catalytic activity at neutral pH, and the possibility of conjugation to antibodies and NP.
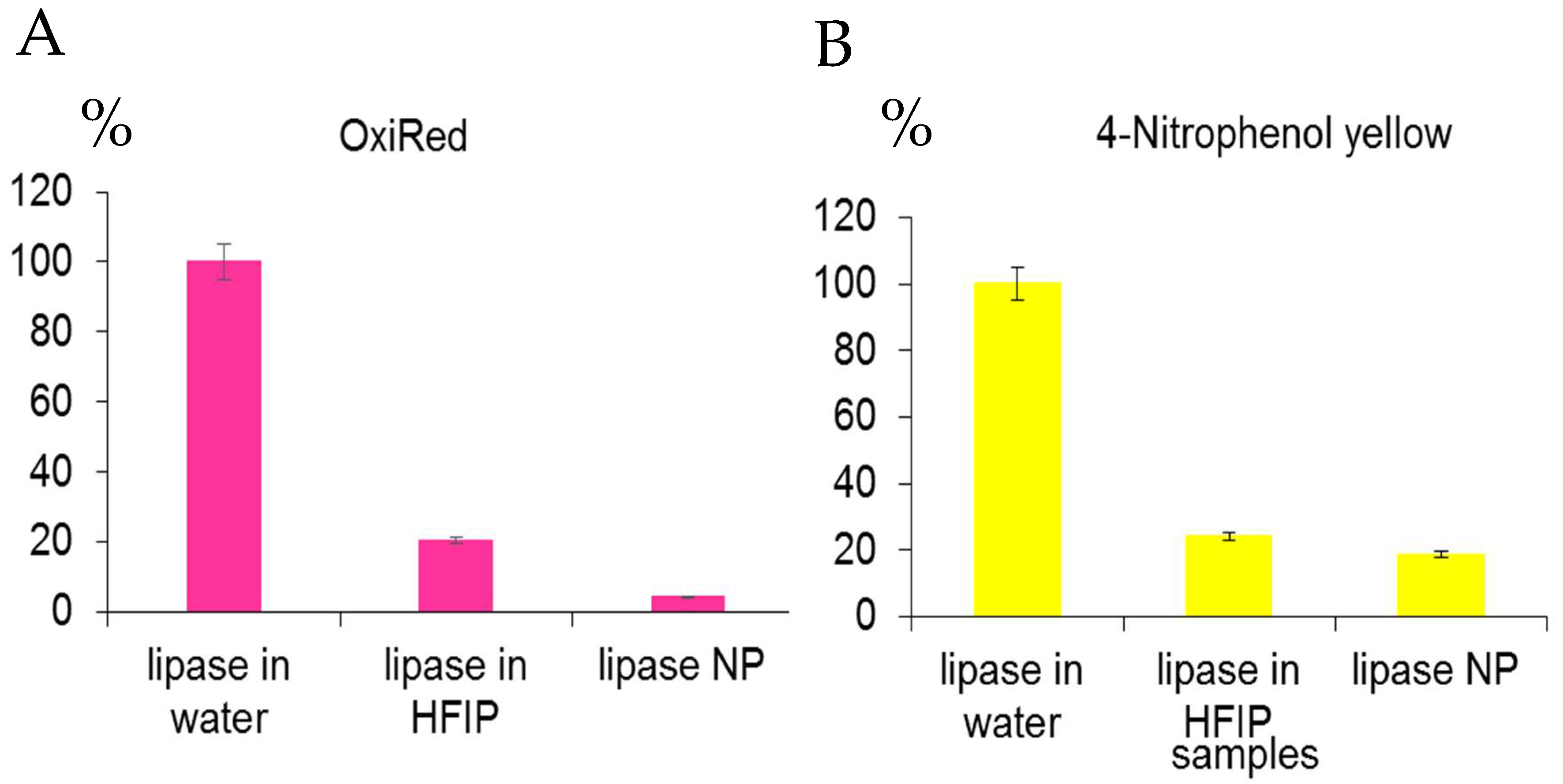
Lipase is a highly soluble enzyme in water and acts on the surface of oil droplets. Access to the active site is controlled by the opening of a lid, which, when closed, hides the hydrophobic surface surrounding the active site. The lid opens when the enzyme contacts an oil–water interface (interfacial activation). The lipase activity with two different specific substrates remained low even in the fresh solution in HFIP (
Figure 8A,B) and in the resulting lipase NP was in a range 4.3–18.6% from the original fresh lipase solution in water. Consequently, the method of protein nanoprecipitation is not convenient for the nanotechnological construction of the lipase NP.
Bioactive enzymes interact with their specific substrates and other ingredients involved in cellular metabolism under mild conditions (temperature near 36.7 °C, aqueous media, pH 7.5–8.0) [
3]. In the absence of specific receptors, clathrin-caveolar independent endocytosis is involved. NP internalization is based on the endocytic pathway through which the particles remain trapped in endosomes and lysosomes. Unspecific endocytosis-mediated cellular uptake of protein NP includes early and late endosomes and lysosomes, where proteolytic hydrolysis of foreign proteins forms oligopeptides necessary for antigen presentation and immune response induction [
24]. All the protection mechanisms against nanomaterials of different origin should be taken into consideration in order to improve endosomal escape, to trigger either intracellular or extracellular controlled drug release [
3] and to optimize targeted delivery, relative stability and biodistribution of novel biomaterials such as the ENP. Currently, tissue distribution, cellular internalization, intracellular trafficking, and drug release for a wide variety of already developed bionanomaterials, with some of them under clinical trials, can be rationally controlled [
2]. Concerns about their biosafety should be a key issue [
3].
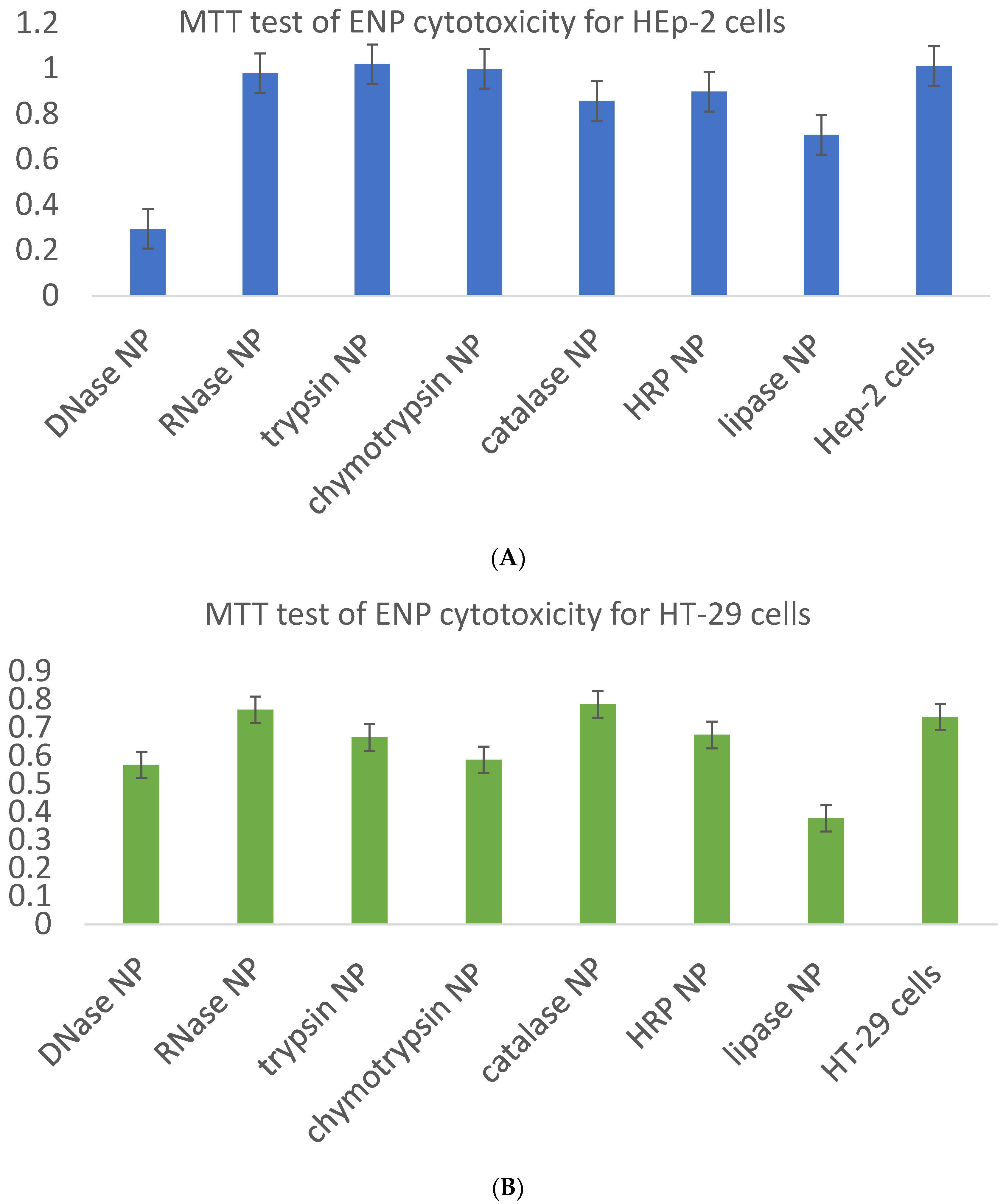
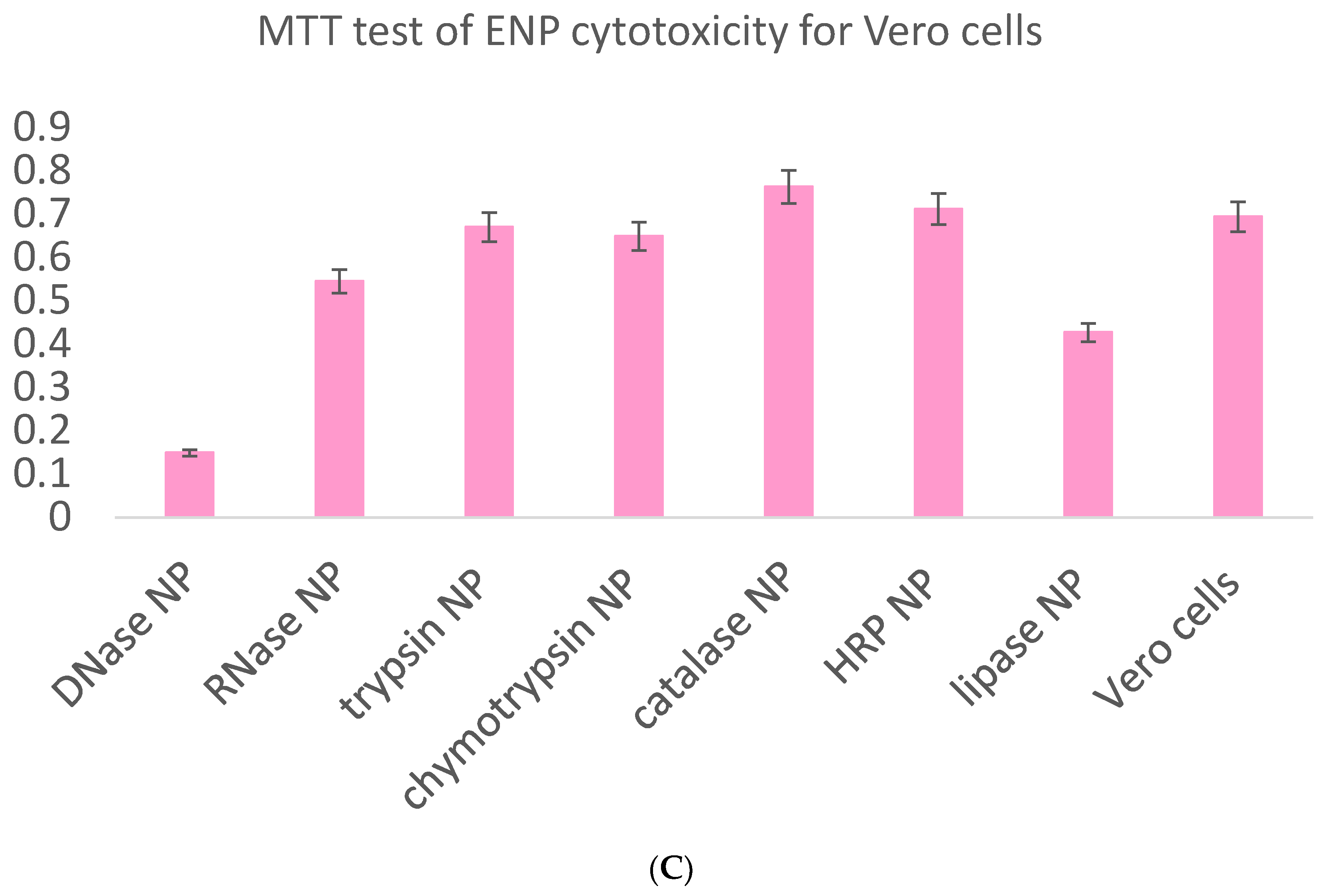
Besides the required evaluation of ENP biocompatibility with intracellular enzymes within physiologically acceptable concentration limits on the basis of the MTT test (
Figure 10,
Table 2) and sulforhodamine B test in order to estimate long-term cell killing [
8], biodegradability of the nanocarriers has to be taken into consideration [
2]. Since enzymatic activities vary among different patients and diseases, and many enzymes may share similar cleavage sites for compensation of possible deficiency, then the correlation between the certain enzyme and pathology should be carefully explored before clinical implementations.