Construction of Fusion Protein with Carbohydrate-Binding Module and Leaf-Branch Compost Cutinase to Enhance the Degradation Efficiency of Polyethylene Terephthalate
Abstract
:1. Introduction
2. Results and Discussion
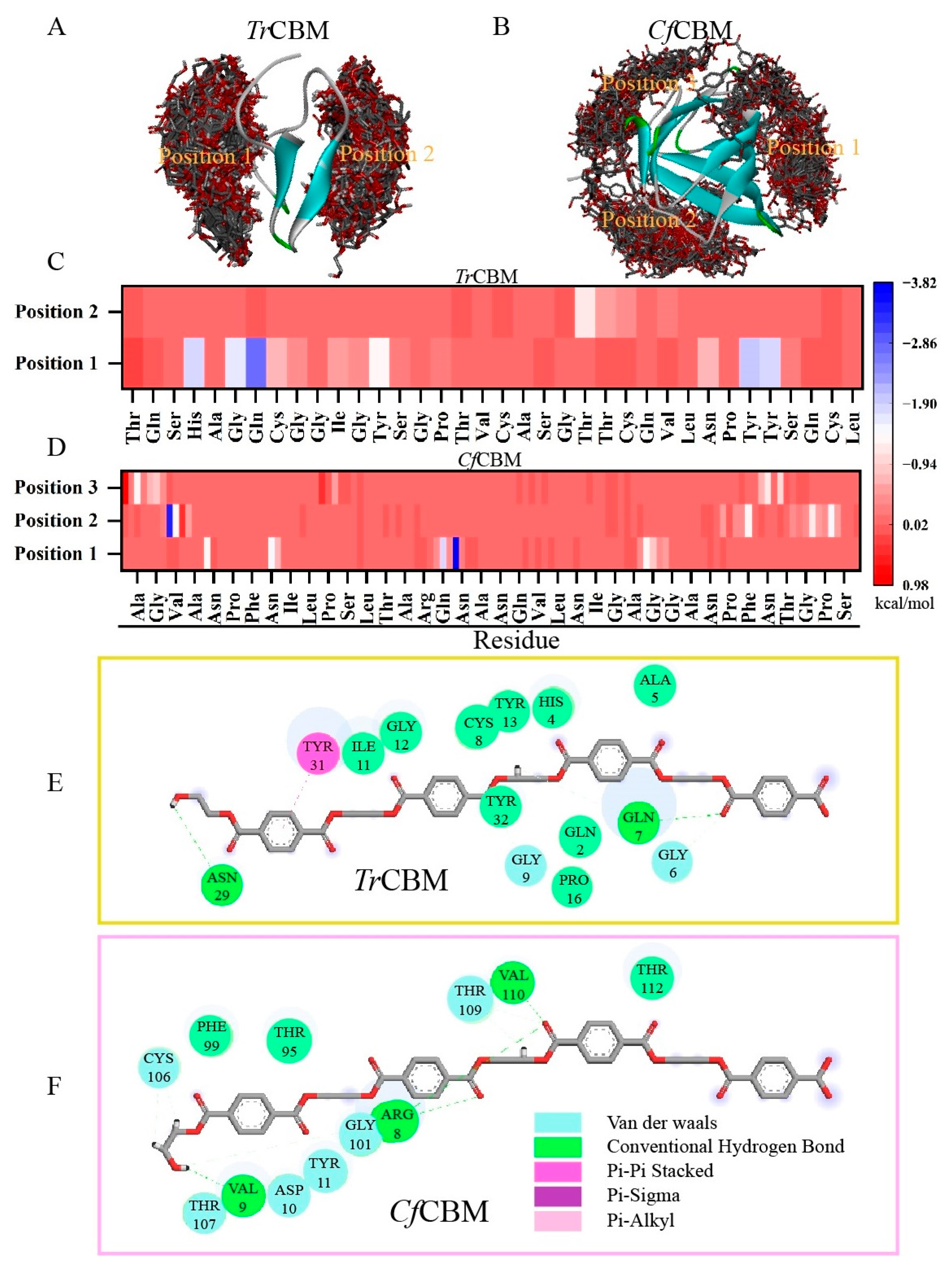
2.1. Rational Screening of CBM Domains and Their Binding Mechanism Evaluation
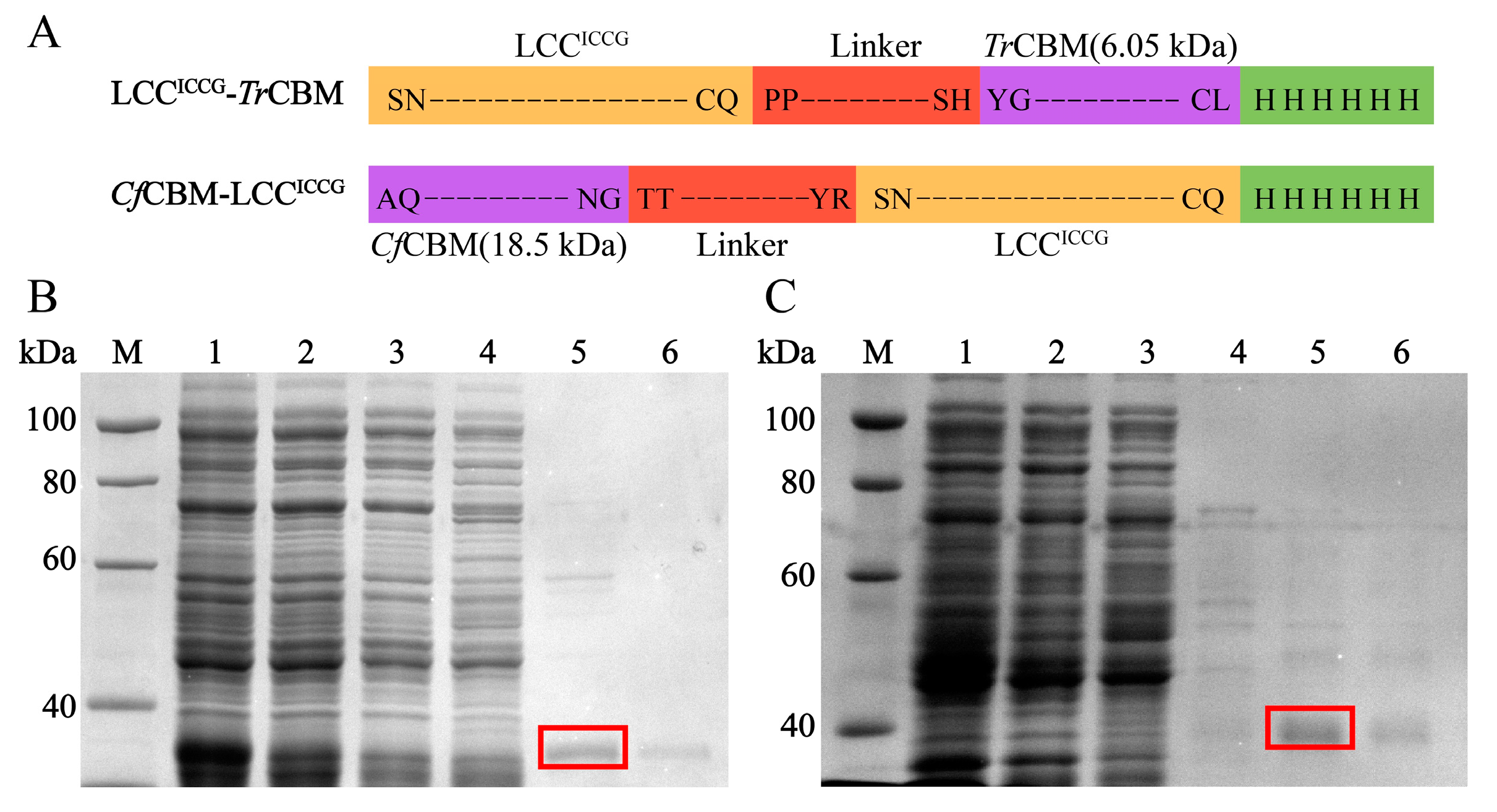
2.2. Expression and Purification of LCC, LCCICCG, LCCICCG-TrCBM and CfCBM-LCCICCG
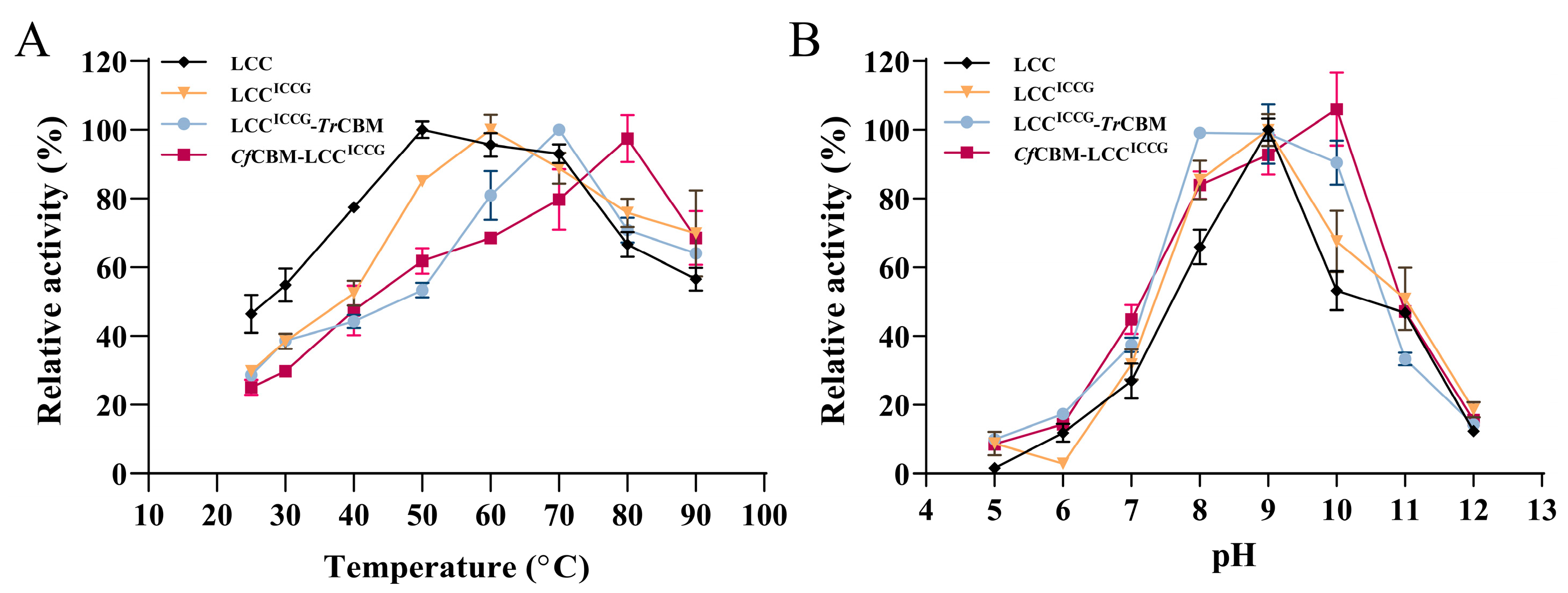
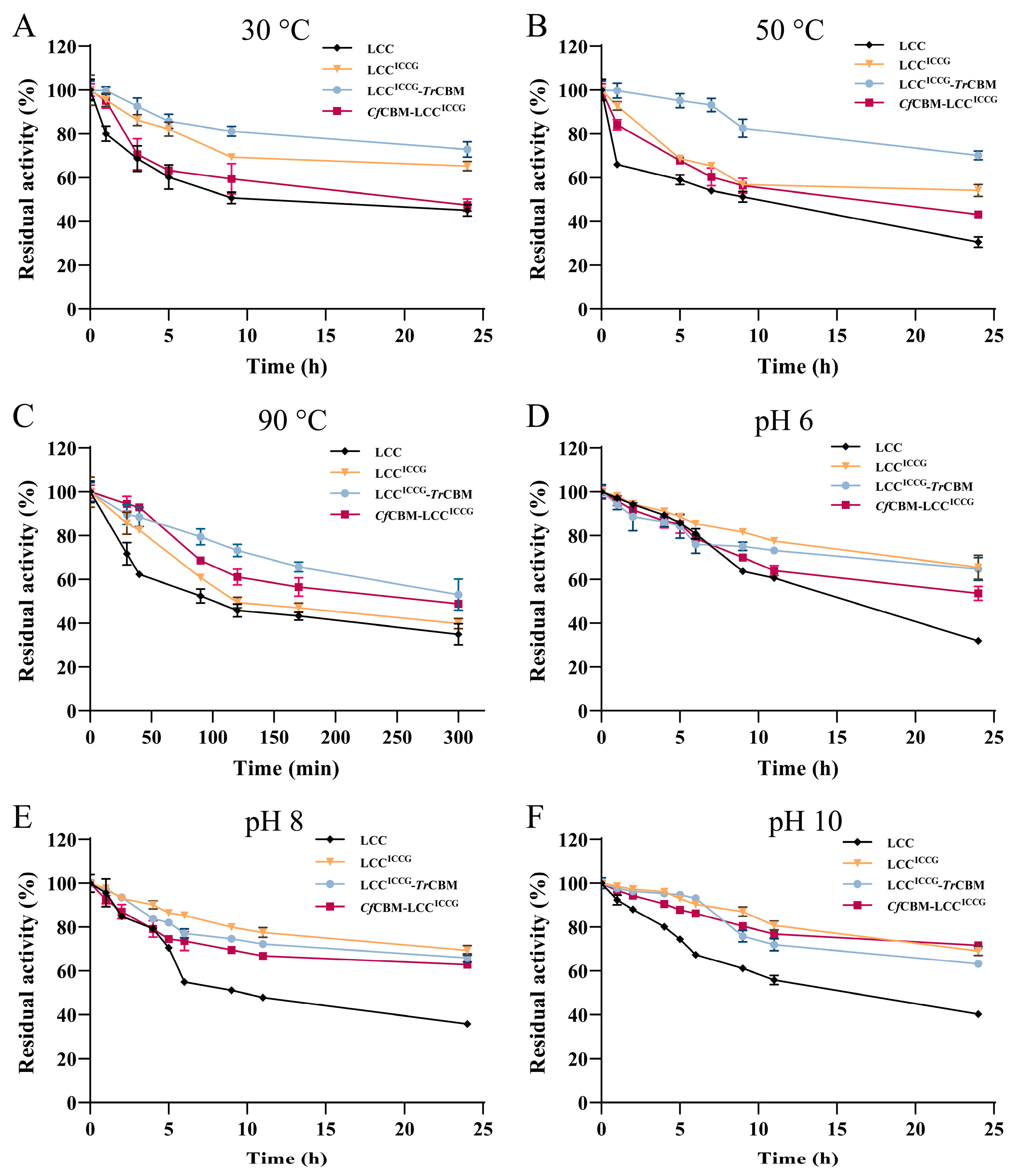
2.3. Characterization of Fusion Proteins
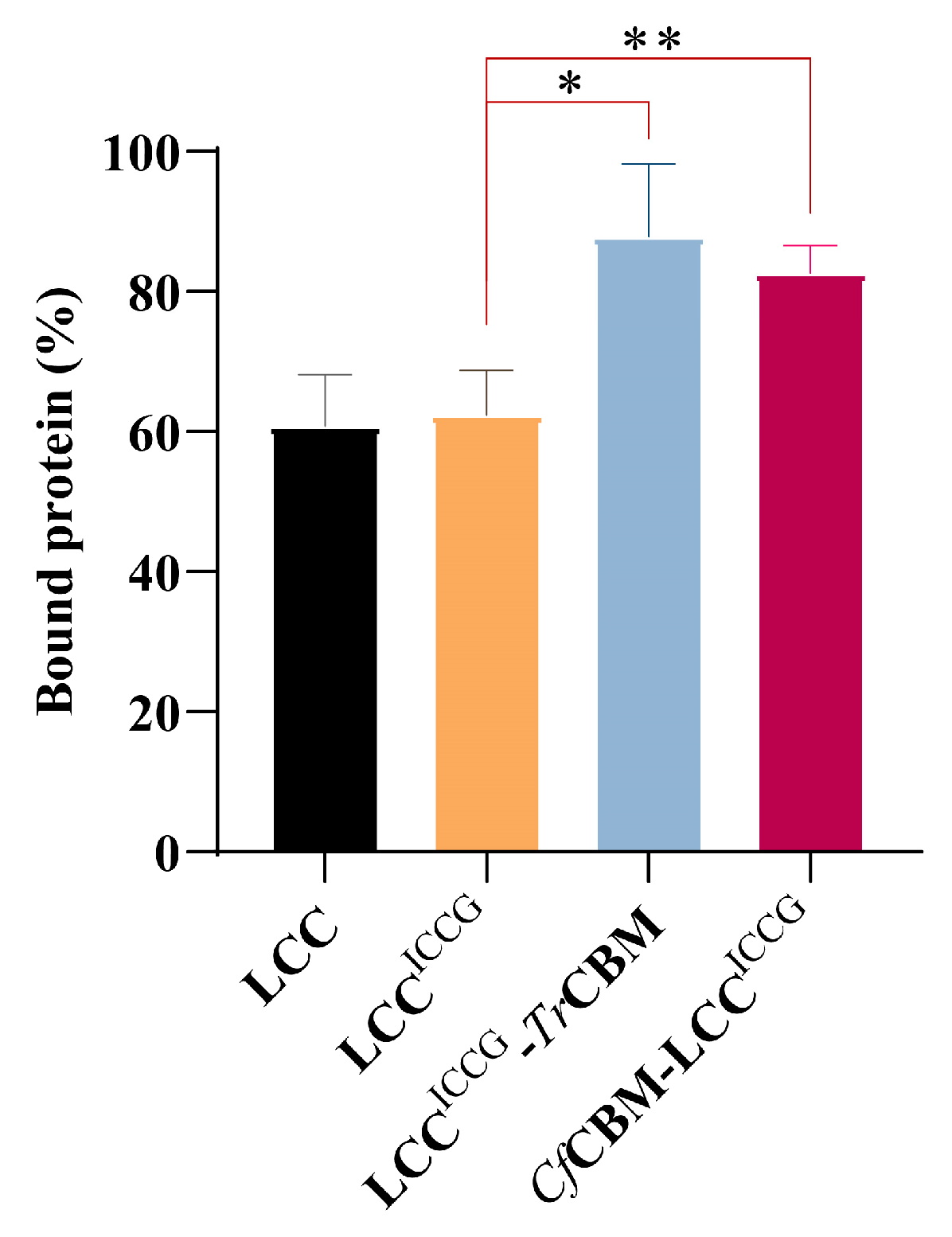
2.4. Adsorption Capacity Analysis of Fusion Proteins on PET Films
2.5. Degradation Performance Analysis of Fusion Proteins on PET Films
3. Materials and Methods
3.1. Materials
3.2. Rational Screening of CBM Domains and Their Binding Mechanism Evaluation
3.3. Expression and Purification of LCC, LCCICCG, LCCICCG-TrCBM and CfCBM-LCCICCG
3.4. Characterization of Fusion Proteins
3.5. Adsorption Capacity Analysis of Fusion Proteins on PET Films
3.6. Degradation Performance Analysis of Fusion Proteins on PET Films
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Webb, H.K.; Arnott, J.; Crawford, R.J.; Ivanova, E.P. Plastic degradation and its environmental implications with special reference to poly(ethylene terephthalate). Polymers 2013, 5, 1–18. [Google Scholar] [CrossRef]
- Samak, N.A.; Jia, Y.; Sharshar, M.M.; Mu, T.; Yang, M.; Peh, S.; Xing, J. Recent advances in biocatalysts engineering for polyethylene terephthalate plastic waste green recycling. Environ. Int. 2020, 145, 106144. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Sun, C. A marine bacterial community capable of degrading poly(ethylene terephthalate) and polyethylene. J. Hazard Mater. 2021, 8, 125928. [Google Scholar] [CrossRef]
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef] [PubMed]
- Baath, J.A.; Novy, V.; Carneiro, L.V.; Guebitz, G.M.; Olsson, L.; Westh, P.; Ribitsch, D. Structure-function analysis of two closely related cutinases from Thermobifida cellulosilytica. Biotechnol. Bioeng. 2021, 119, 470–481. [Google Scholar] [CrossRef] [PubMed]
- White, E.M.; Clark, S.; Manire, C.A.; Crawford, B.; Wang, S.; Loklin, J.; Ritchiw, B.W. Ingested micronizing plastic particle compositions and size distributions within stranded post-hatchling sea turtles. Environ. Sci. Technol. 2018, 52, 10307–10316. [Google Scholar] [CrossRef]
- Nicholson, S.R.; Rorrer, N.A.; Carpenter, A.C.; Beckham, G.T. Manufacturing energy and greenhouse gas emissions associated with plastics consumption. Joule 2021, 5, 673–686. [Google Scholar] [CrossRef]
- Demetrious, A.; Crossin, E. Life cycle assessment of paper and plastic packaging waste in landfill, incineration, and gasification-pyrolysis. J. Mater. Cycles Waste Manag. 2019, 21, 850–860. [Google Scholar] [CrossRef]
- Zink, T.; Geyer, R. Material recycling and the myth of landfill diversion. J. Ind. Ecol. 2018, 23, 541–548. [Google Scholar] [CrossRef]
- Ragaret, K.; Delva, L.; Geem, K.V. Mechanical and chemical recycling of solid plastic waste. Waste Manag. 2017, 69, 24–58. [Google Scholar] [CrossRef]
- Ali, S.S.; Elsamahy, T.; Koutra, E.; Kornaros, M.; EI-Sheekh, M.; Abdelkarim, E.A.; Zhu, D.; Sun, J. Degradation of conventional plastic wastes in the environment: A review on current status of knowledge and future perspectives of disposal. Sci. Total Environ. 2021, 771, 144719. [Google Scholar] [CrossRef]
- Qin, Z.; Mou, J.; Chao, C.Y.H.; Chopra, S.S.; Daoud, W.; Leu, S.; Ning, Z.; Tso, C.Y.; Chan, C.K.; Tang, S.; et al. Biotechnology of plastic waste degradation, recycling, and valorization: Current advances and future perspectives. ChemSusChem 2021, 14, 4103–4114. [Google Scholar] [CrossRef]
- Ellis, L.D.; Rorrer, N.A.; Sullivan, K.P.; Otto, M.; McGreehan, J.E.; Roman-Leshkov, Y.; Wierckx, N.; Beckham, G.T. Chemical and biological catalysis for plastics recycling and upcycling. Nat. Catal. 2021, 4, 539–556. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; von Haugwitz, G.; Pfaff, L.; Mican, J.; Badenhorst, C.P.S.; Liu, W.; Weber, G.; Austin, H.P.; Bednar, D.; Damborsky, J.; et al. Mechanism-based design of efficient pet hydrolases. ACS Catal. 2022, 12, 3382–3396. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Tiso, T.; Bertling, J.; O’Connor, K.; Blank, L.M.; Bornscheuer, U.T. Possibilities and limitations of biotechnological plastic degradation and recycling. Nat. Catal. 2020, 3, 867–871. [Google Scholar] [CrossRef]
- Sulaiman, S.; Yamato, S.; Kanaya, E.; Kim, J.J.; Koga, Y.; Takano, K.; Kanaya, S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microb. 2012, 78, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Shirke, A.N.; White, C.; Englaender, J.A.; Zwarycz, A.; Butterfoss, G.L.; Linhardt, R.J.; Gross, R.A. Stabilizing leaf and branch compost cutinase (LCC) with glycosylation: Mechanism and effect on pet hydrolysis. Biochemistry 2018, 57, 1190–1200. [Google Scholar] [CrossRef]
- Anuar, N.F.S.K.; Huyop, F.; Ur-Rehman, G.; Abdullah, F.; Normi, Y.M.; Sabullah, M.K.; Wahab, R.A. An overview into polyethylene terephthalate (PET) hydrolases and efforts in tailoring enzymes for improved plastic degradation. Int. J. Mol. Sci. 2022, 23, 12644. [Google Scholar] [CrossRef]
- Neumann, A.P.; Suen, G. The phylogenomic diversity of herbivore-associated fibrobacter spp. is correlated to lignocellulose-degrading potential. mSphere 2018, 3, e00593-18. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, C.; Cesaro, P.; Spertino, S.; Icardi, S.; Cavaletto, M. Enhanced features of Dictyoglomus turgidum cellulase A engineered with carbohydrate binding module 11 from Clostridium thermocellum. Sci. Rep. 2018, 8, 4402. [Google Scholar] [CrossRef] [PubMed]
- Madland, E.; Forsberg, Z.; Wang, Y.; Lindorff-Larsen, K.; Niebisch, A.; Modregger, J.; Eijsink, V.G.H.; Aachmann, F.L.; Courtade, G. Structural and functional variation of chitin-binding domains of a lytic polysaccharide monooxygenase from Cellvibrio japonicus. J. Biol. Chem. 2021, 297, 101084. [Google Scholar] [CrossRef]
- Chen, C.Q.; Dai, L.; Ma, L.; Guo, R.T. Enzymatic degradation of plant biomass and synthetic polymers. Nat. Rev. Chem. 2020, 4, 114–126. [Google Scholar] [CrossRef]
- Janeček, Š.; Mareček, F.; MacGregor, E.A.; Svensson, B. Starch-binding domains as CBM families-history, occurrence, structure, function and evolution. Biotechnol. Adv. 2019, 37, 107451. [Google Scholar] [CrossRef]
- Dai, L.H.; Qu, Y.; Huang, J.W.; Hu, Y.; Hu, H.; Li, S.; Chen, C.Q.; Guo, R.T. Enhancing PET hydrolytic enzyme activity by fusion of the cellulose-binding domain of cellobiohydrolase I from Trichoderma reesei. J. Biotechnol. 2021, 334, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Xue, R.; Chen, Y.; Rong, H.; Wei, R.; Cui, Z.; Zhou, J.; Dong, W.L.; Jiang, M. Fusion of chitin-binding domain from Chitinolyticbacter meiyuanensis SYBC-H1 to the leaf-branch compost cutinase for enhanced PET hydrolysis. Front. Bioeng. Biotechnol. 2021, 9, 762854. [Google Scholar] [CrossRef]
- Graham, R.; Erickson, E.; Brizendine, R.K.; Salvachúa, D.; Michener, W.E.; Li, Y.; Tan, Z.; Beckham, G.T.; McGeehan, J.E.; Pickford, A.R. The role of binding modules in enzymatic poly(ethylene terephthalate) hydrolysis at high-solids loadings. Chem. Catal. 2022, 2, 2644–2657. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Wu, J. Enhancement of PET biodegradation by anchor peptide-cutinase fusion protein. Enzyme Microb. Technol. 2022, 156, 110004. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2021, 50, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Lai, G.; Wang, F.; Nie, D.; Lei, S.; Wu, Z.; Cao, J. Identifying active substances and the pharmacological mechanism of Houttuynia cordata Thunb. in treating radiation-induced lung injury based on network pharmacology and molecular docking verification. Evid. Based Complement Alternat. Med. 2022, 2022, 3776340. [Google Scholar] [CrossRef]
- Weber, J.; Petrović, D.; Strodel, B.; Smits, S.H.J.; Kolkenbrock, S.; Leggewie, C.; Jaeger, K.E. Interaction of carbohydrate-binding modules with poly(ethylene terephthalate). Appl. Microbiol. Biotechnol. 2019, 103, 4801–4812. [Google Scholar] [CrossRef]
- Deng, J.J.; Zhang, M.S.; Li, Z.W.; Lu, D.L.; Mao, H.H.; Zhu, M.J.; Li, J.Z.; Luo, X.C. One-step processing of shrimp shell waste with a chitinase fused to a carbohydrate-binding module. Green Chem. 2020, 22, 6862–6873. [Google Scholar] [CrossRef]
- Baptista, R.P.; Chen, L.Y.; Paixão, A.; Cabral, J.M.S.; Melo, E.P. A novel pathway to enzyme deactivation: The cutinase model. Biotechnol. Bioeng. 2003, 82, 851–857. [Google Scholar] [CrossRef]
- Voutilainen, S.P.; Nurmi-Rantala, S.; Penttilä, M.; Koivula, A. Engineering chimeric thermostable GH7 cellobiohydrolases in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2014, 98, 2991–3001. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Ma, Y.; Zhe, Y.; Tang, X.; Wu, Q.; Huang, Z.; Han, N. Improving the thermostability of a fungal GH11 xylanase via fusion of a submodule (C2) from hyperthermophilic CBM9_1-2. Int. J. Mol. Sci. 2022, 23, 463. [Google Scholar] [CrossRef]
- Cheng, R.; Chen, J.; Yu, X.; Wang, Y.; Wang, S.; Zhang, J. Recombinant production and characterization of full-length and truncated β-1,3-glucanase PglA from Paenibacillus sp. S09. BMC Biotechnol. 2013, 13, 105. [Google Scholar] [CrossRef]
- Wang, J.; Lisanza, S.; Juergens, D.; Tischer, D.; Watson, J.L.; Castro, K.M.; Ragotte, R.; Saragovi, A.; Milles, L.F.; Baek, M.; et al. Scaffolding protein functional sites using deep learning. Science 2022, 377, 387–394. [Google Scholar] [CrossRef]
- Wei, R.; Breite, D.; Song, C.; Gräsing, D.; PLoSs, T.; Hille, P.; Schwerdtfeger, R.; Matysik, J.; Schulze, A.; Zimmermann, W. Biocatalytic degradation efficiency of postconsumer polyethylene terephthalate packaging determined by their polymer microstructures. Adv. Sci. 2019, 6, 1900491. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.; Darden, T.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Götz, A.W.; Homeyer, N.; et al. Amber 16; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Özpınar, G.A.; Peukert, W.; Clark, T. An improved generalized AMBER force field (GAFF) for urea. J. Mol. Model. 2010, 16, 1427–1440. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. FF14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089. [Google Scholar] [CrossRef]
- Matioli, G.; Zanin, G.M.; De Moraes, F.F. Characterization of cyclodextrin glycosyltransferase from Bacillus firmus strain No. 37. Appl. Biochem. Biotechnol. 2001, 91–93, 643–654. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, Y.; Liu, X.; Dong, S.; Tian, Y.; Qiao, Y.; Mitra, R.; Han, J.; Li, C.; Han, X.; et al. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy. ACS Catal. 2021, 11, 1340–1350. [Google Scholar] [CrossRef]
- Alshammari, B.A.; Al-Mubaddel, F.S.; Karim, M.R.; Hossain, M.; Mutairi, A.S.A.; Wilkinson, A.N. Addition of graphite filler to enhance electrical, morphological, thermal, and mechanical properties in poly (ethylene terephthalate): Experimental characterization and material modeling. Polymers 2019, 11, 1411. [Google Scholar] [CrossRef]
- Hwang, D.H.; Lee, M.E.; Cho, B.H.; Oh, J.W.; You, S.K.; Ko, Y.J.; Hyeon, J.E.; Han, S.O. Enhanced biodegradation of waste poly(ethylene terephthalate) using a reinforced plastic degrading enzyme complex. Sci. Total Environ. 2022, 842, 156890. [Google Scholar] [CrossRef]
- Son, H.F.; Cho, I.J.; Joo, S.; Seo, H.; Sagong, H.Y.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Rational protein engineering of thermo-stable petase from Ideonella sakaiensis for highly efficient PET degradation. ACS Catal. 2019, 9, 3519–3526. [Google Scholar] [CrossRef]
- Lu, H.; Diaz, D.J.; Czarnecki, N.J.; Zhu, C.; Kim, W.; Shroff, R.; Acosta, D.J.; Alexander, B.R.; Cole, H.O.; Zhang, Y.; et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 2022, 604, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Zhong-Johnson, E.Z.L.; Voigt, C.A.; Sinskey, A.J. An absorbance method for analysis of enzymatic degradation kinetics of poly(ethylene terephthalate) films. Sci. Rep. 2021, 11, 928. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.L.; Smithson, R.; Kilbride, S.; Foster, J.; Hardy, F.J.; Ramachandran, S.; Tedstone, A.A.; Haigh, S.J.; Garforth, A.A.; Day, P.J.R.; et al. Directed evolution of an efficient and thermostable PET depolymerase. Nat. Catal. 2022, 5, 673–681. [Google Scholar] [CrossRef]
- Acero, E.H.; Ribitsch, D.; Steinkellner, G.; Gruber, K.; Greimel, K.; Eiteljoerg, I.; Trotscha, E.; Wei, R.; Zimmermann, W.; Zinn, M.; et al. Enzymatic surface hydrolysis of pet: Effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules 2011, 44, 4632–4640. [Google Scholar] [CrossRef]
- Furukawa, M.; Kawakami, N.; Tomizawa, A.; Miyamoto, K. Efficient degradation of poly(ethylene terephthalate) with Thermobifida fusca cutinase exhibiting improved catalytic activity generated using mutagenesis and additive-based approaches. Sci. Rep. 2019, 9, 16038. [Google Scholar] [CrossRef]
- Danso, D.; Schmeisser, C.; Chow, J.; Zimmermann, W.; Wei, R.; Leggewie, C.; Li, X.; Hazen, T.; Streit, W.R. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl. Environ. Microbiol. 2018, 84, e02773-17. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Kobayashi, N.; Koga, N.; Iino, R. Positive charge introduction on the surface of thermostabilized PET hydrolase facilitates PET binding and degradation. ACS Catal. 2021, 11, 8550–8564. [Google Scholar] [CrossRef]
- Ronkvist, Å.M.; Xie, W.; Lu, W.; Gross, R.A. Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules 2009, 42, 5128–5138. [Google Scholar] [CrossRef]
- Carniel, A.; Valoni, É.; Nicomedes, J.; Gomes, A.C.; Castro, A.M. Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid. Process Biochem. 2017, 59, 84–90. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Zhang, S.; Zhai, Z.; Zhang, S.; Ma, J.; Liang, X.; Li, Q. Construction of Fusion Protein with Carbohydrate-Binding Module and Leaf-Branch Compost Cutinase to Enhance the Degradation Efficiency of Polyethylene Terephthalate. Int. J. Mol. Sci. 2023, 24, 2780. https://doi.org/10.3390/ijms24032780
Chen Y, Zhang S, Zhai Z, Zhang S, Ma J, Liang X, Li Q. Construction of Fusion Protein with Carbohydrate-Binding Module and Leaf-Branch Compost Cutinase to Enhance the Degradation Efficiency of Polyethylene Terephthalate. International Journal of Molecular Sciences. 2023; 24(3):2780. https://doi.org/10.3390/ijms24032780
Chicago/Turabian StyleChen, Yingxuan, Shudi Zhang, Zhenyu Zhai, Shuo Zhang, Jun Ma, Xiao Liang, and Quanshun Li. 2023. "Construction of Fusion Protein with Carbohydrate-Binding Module and Leaf-Branch Compost Cutinase to Enhance the Degradation Efficiency of Polyethylene Terephthalate" International Journal of Molecular Sciences 24, no. 3: 2780. https://doi.org/10.3390/ijms24032780
APA StyleChen, Y., Zhang, S., Zhai, Z., Zhang, S., Ma, J., Liang, X., & Li, Q. (2023). Construction of Fusion Protein with Carbohydrate-Binding Module and Leaf-Branch Compost Cutinase to Enhance the Degradation Efficiency of Polyethylene Terephthalate. International Journal of Molecular Sciences, 24(3), 2780. https://doi.org/10.3390/ijms24032780





