PTCHD1 Binds Cholesterol but Not Sonic Hedgehog, Suggesting a Distinct Cellular Function
Abstract
1. Introduction
2. Results
2.1. Expression and Purification of PTCHD1
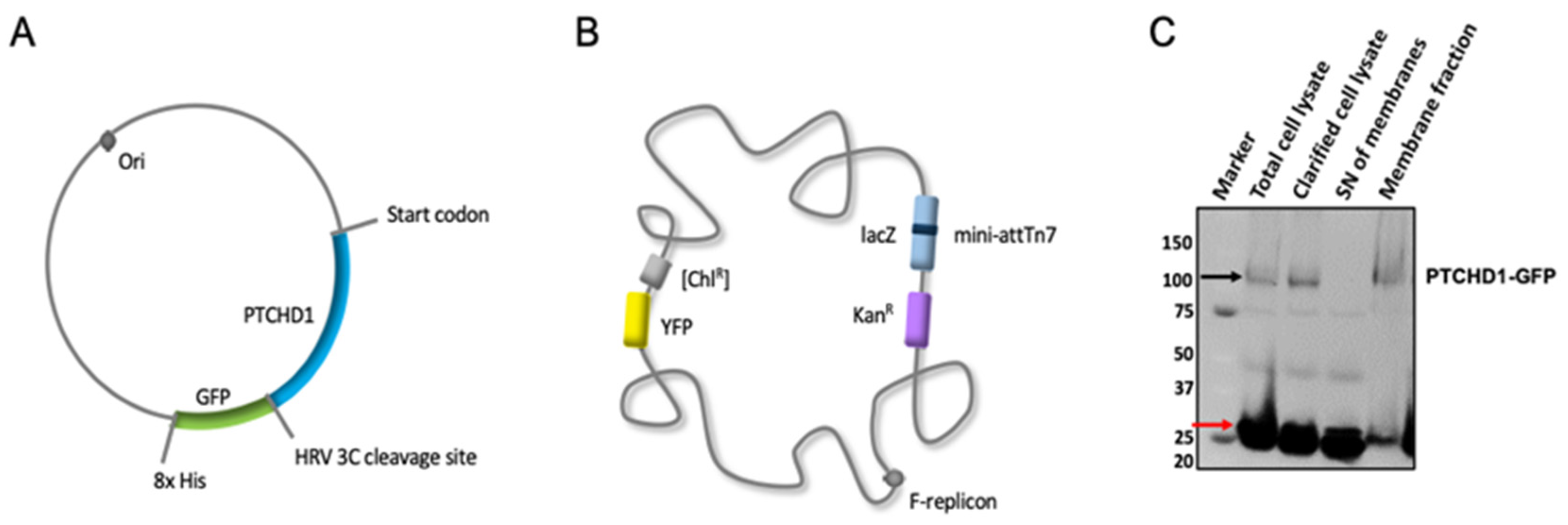
2.1.1. Cloning, Expression, and Membrane Preparation
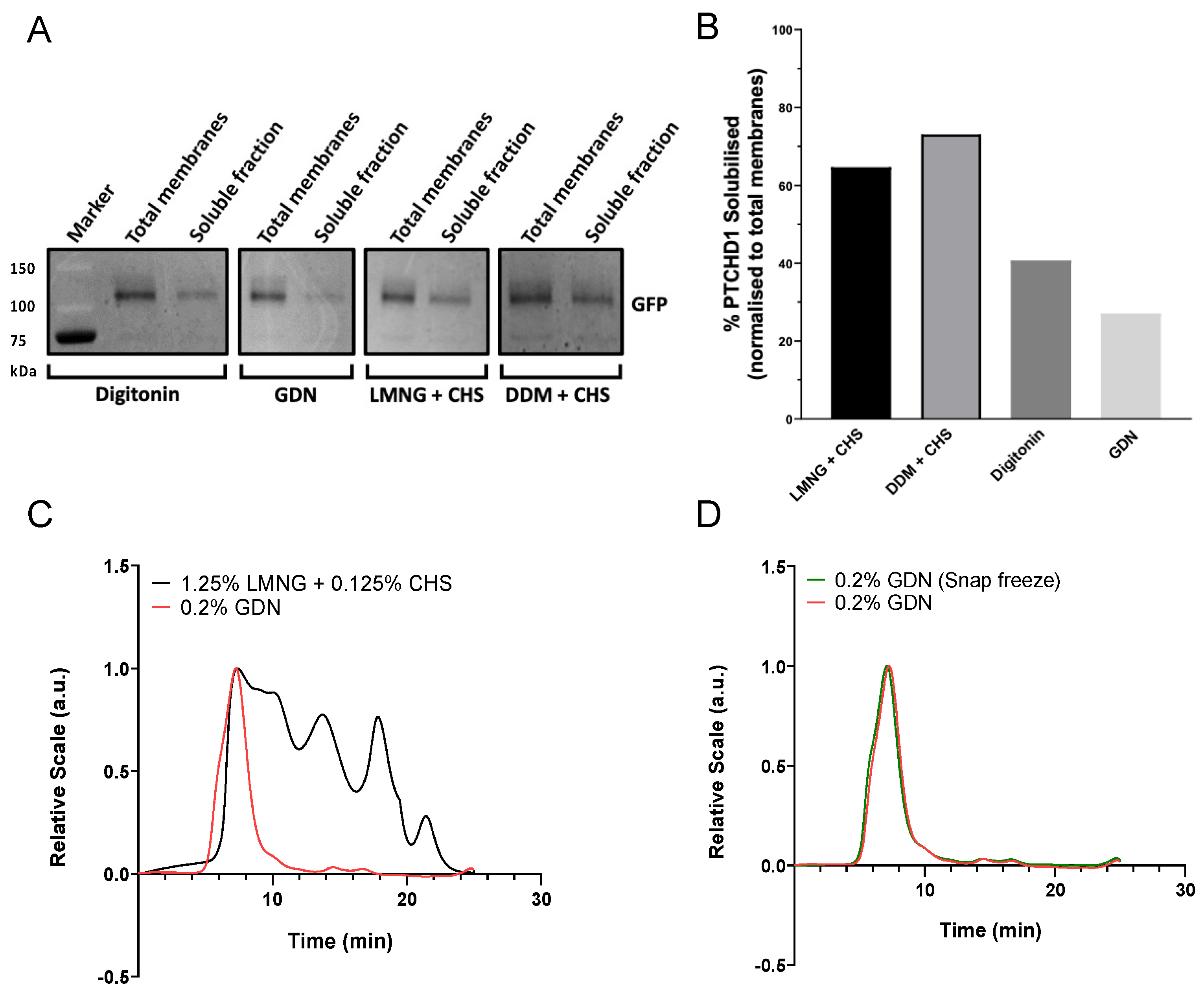
2.1.2. Solubilisation Detergent Screening
2.1.3. Purification of PTCHD1 with GDN Produces a Monodispersed Peak by SEC-MALS
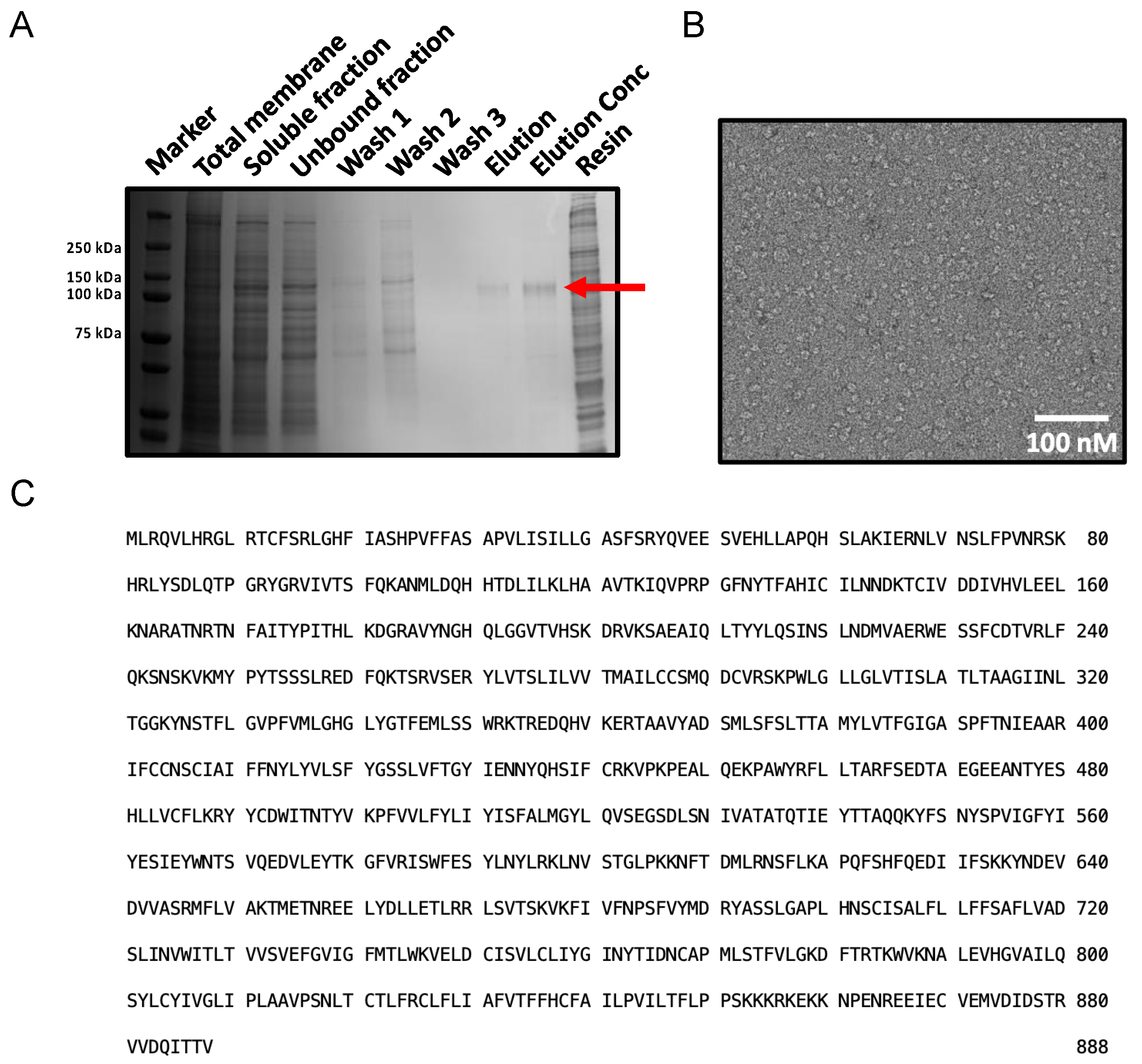
2.1.4. Optimised Large-Scale Purification of GDN-Solubilised PTCHD1
2.2. PTCHD1 Binds Cholesterol but Displays No Inhibitory Activity in Canonical Hedgehog Signalling
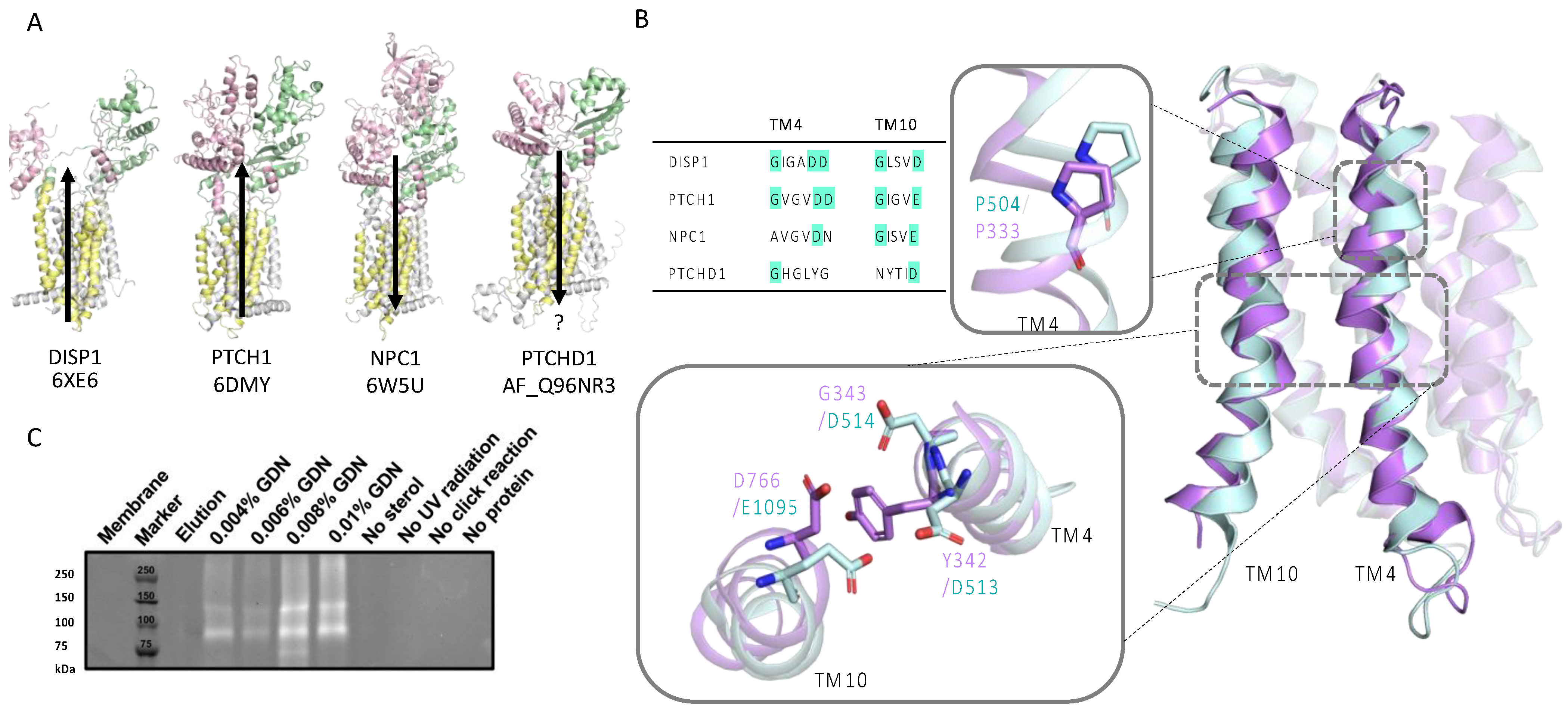
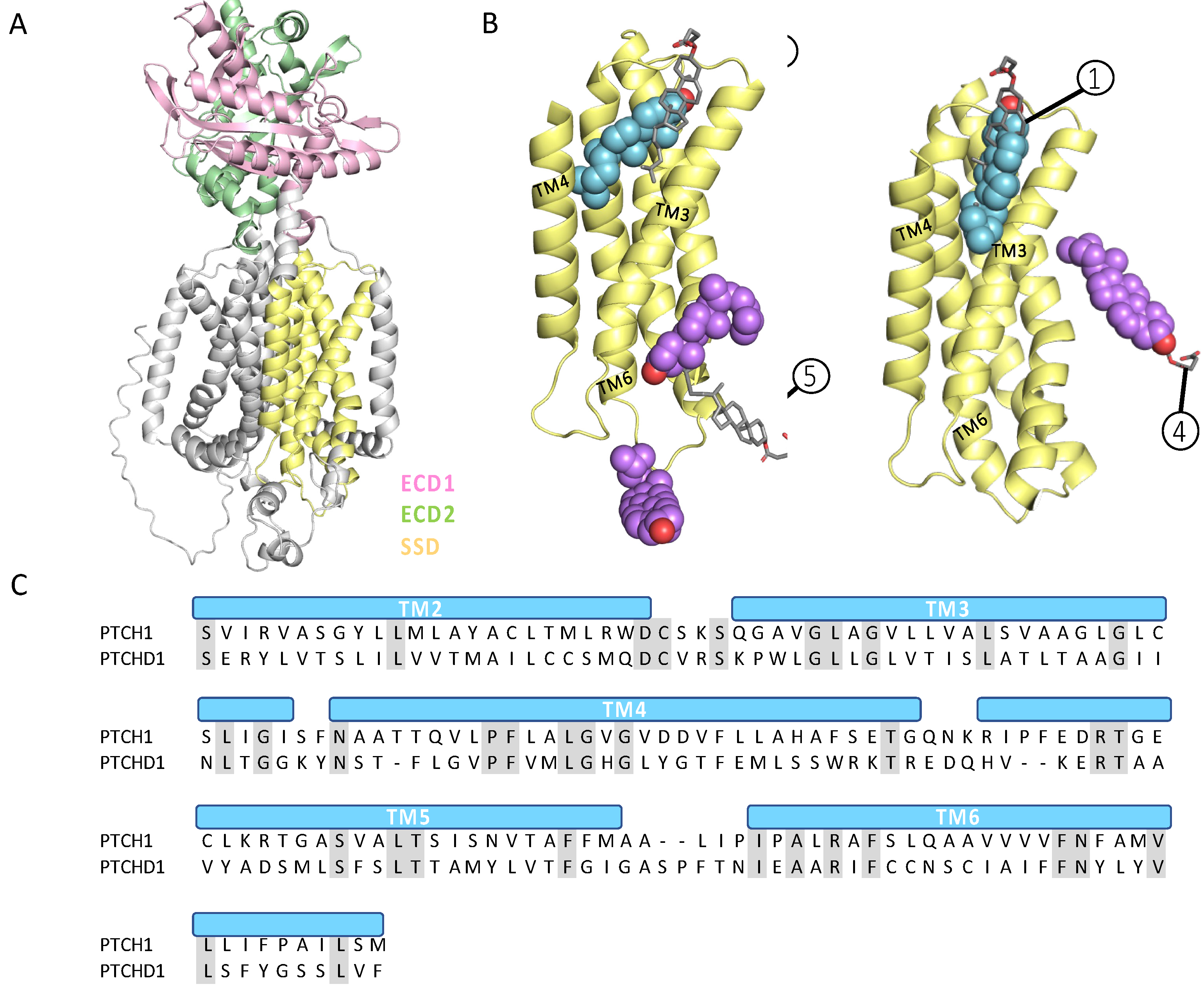
2.2.1. Alignment Reveals Conserved Cholesterol-Binding Elements and a Lack of Ion Flux Motifs
2.2.2. Click-Cholesterol Assay Shows PTCHD1 Binds Cholesterol
2.2.3. Docking Indicates PTCHD1 Can Bind Cholesterol Similarly to PTCH1
2.2.4. PTCHD1 Does Not Inhibit Canonical Hedgehog Signalling
2.2.5. Structural Analysis and Complex Prediction Showed That PTCHD1 Is Unlikely to Bind Shh
2.2.6. Structural Analysis Reveals Key Differences in PTCHD1 and PTCH1 Ectodomains
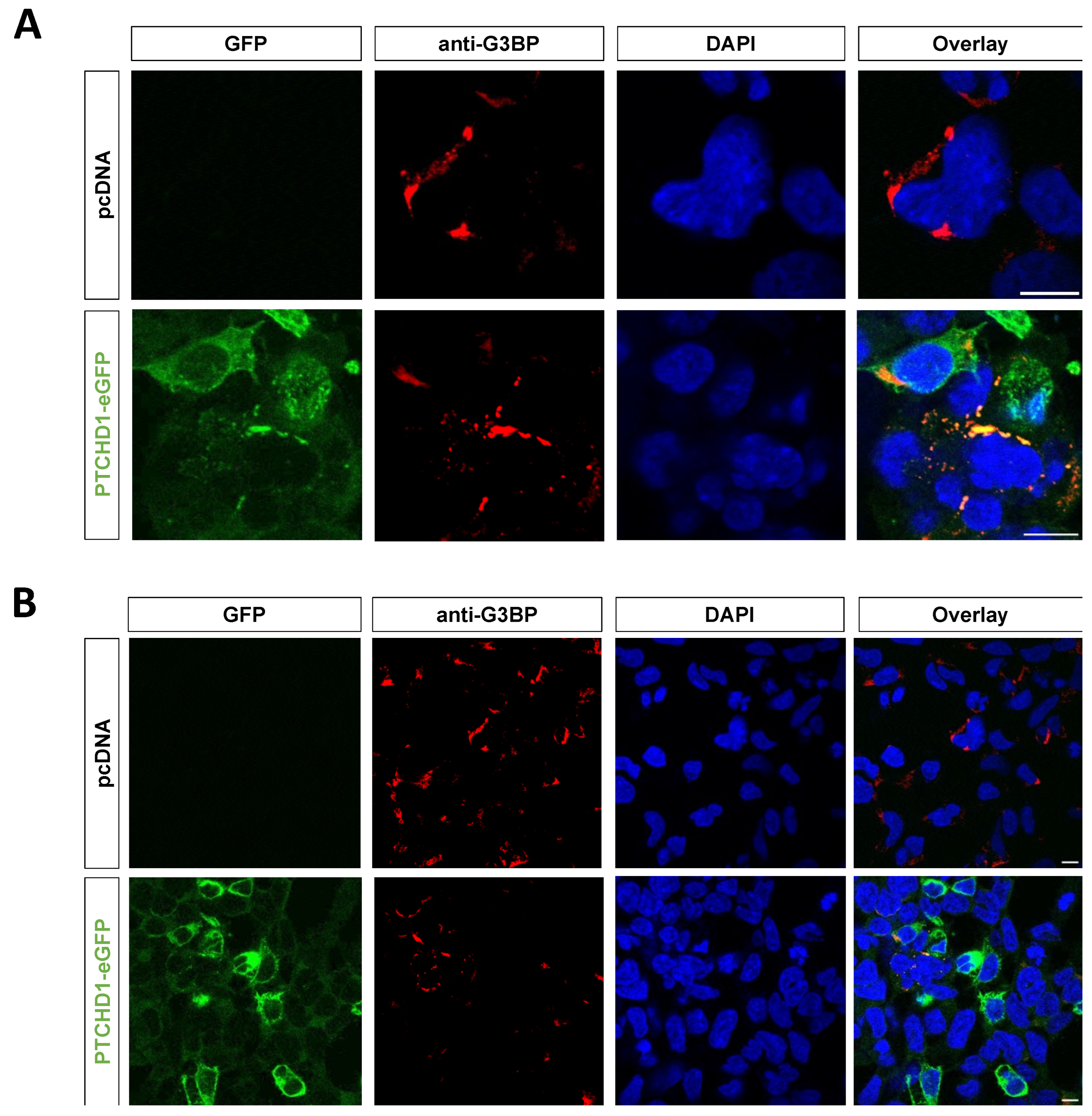
2.3. PTCHD1 Specific Interaction Network Linked to Ribonucleoprotein Granule Proteins and Stress Granules
Gene Ontology Analysis
3. Discussion
4. Materials and Methods
4.1. Cloning and Production of PTCHD1 Baculovirus-Infected Cells
4.2. Expression in Sf9 Insect Cells and Membrane Preparation
4.3. Small-Scale Detergent Screening
4.4. Optimisation of PTCHD1 Purification
4.5. PhotoClickable Cholesterol Assay
4.6. GLI Luciferase Assay in Mouse Embryonic Fibroblasts
4.7. Sequence Analysis
4.8. ShhN-ECD1-ECD2 Complex Prediction
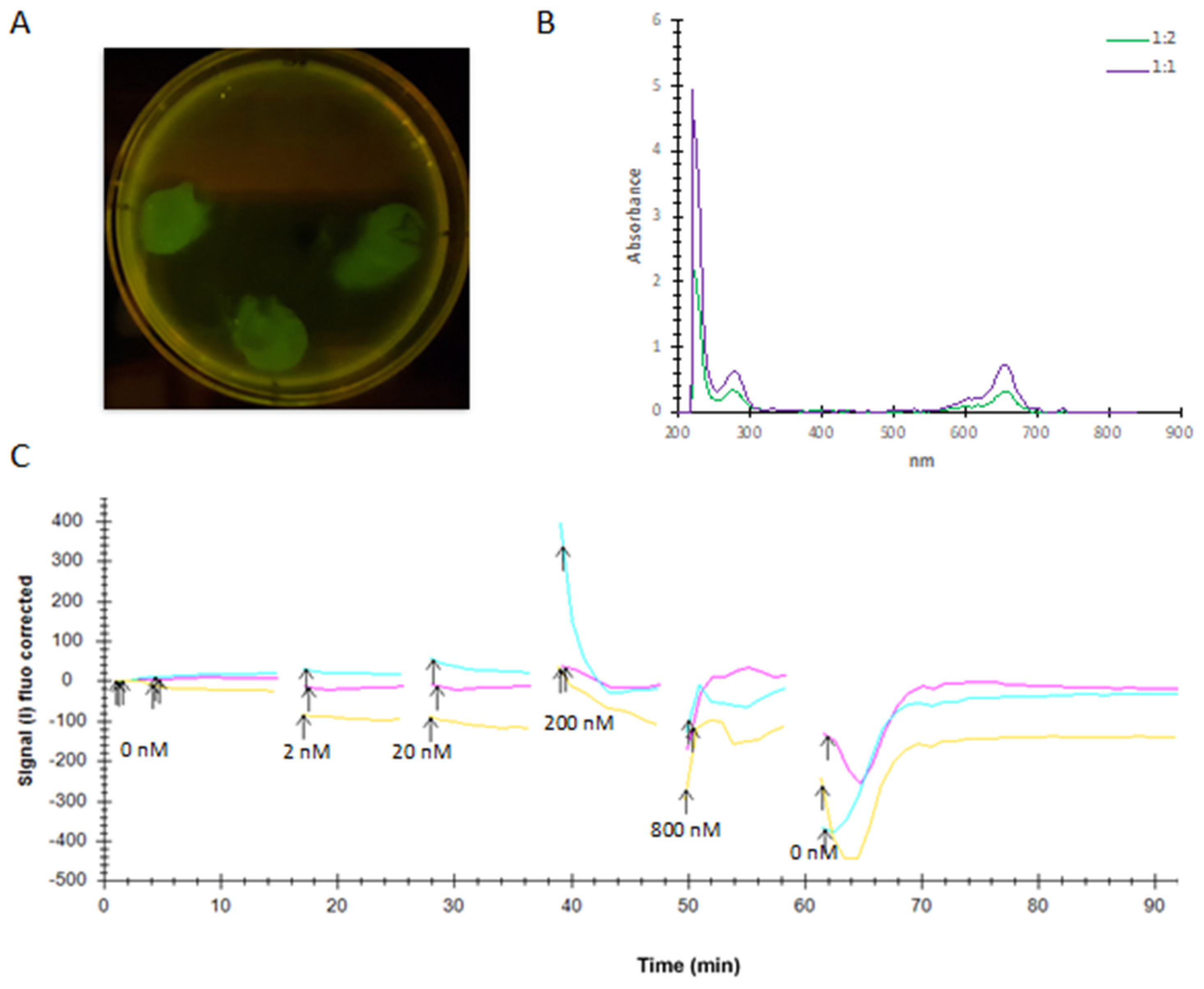
4.9. LigandTracerTM Experiment
4.10. Docking Analysis
4.11. HEK293 Cell Culture and Co-immunoprecipitation Assay
4.12. Mass Spectrometry
4.13. Gene ontology Analysis
4.14. Confocal Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilman, S.R.; Iossifov, I.; Levy, D.; Ronemus, M.; Wigler, M.; Vitkup, D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 2011, 70, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Girirajan, S.; Dennis, M.Y.; Baker, C.; Malig, M.; Coe, B.P.; Campbell, C.D.; Mark, K.; Vu, T.H.; Alkan, C.; Cheng, Z.; et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am. J. Hum. Genet. 2013, 92, 221–237. [Google Scholar] [CrossRef] [PubMed]
- Szatmari, P.; Paterson, A.D.; Zwaigenbaum, L.; Roberts, W.; Brian, J.; Liu, X.-Q.; Vincent, J.B.; Skaug, J.L.; Thompson, A.P.; Senman, L.; et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007, 39, 319–328. [Google Scholar] [PubMed]
- Wells, M.F.; Wimmer, R.D.; Schmitt, L.I.; Feng, G.; Halassa, M.M. Thalamic reticular impairment underlies attention deficit in Ptchd1(Y/-) mice. Nature 2016, 532, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Noor, A.; Whibley, A.; Marshall, C.R.; Gianakopoulos, P.J.; Piton, A.; Carson, A.R.; Orlic-Milacic, M.; Lionel, A.C.; Sato, D.; Pinto, D.; et al. Disruption at the PTCHD1 Locus on Xp22.11 in Autism spectrum disorder and intellectual disability. Sci. Transl. Med. 2010, 2, 49ra68. [Google Scholar] [CrossRef]
- Tora, D.; Gomez, A.M.; Michaud, J.F.; Yam, P.T.; Charron, F.; Scheiffele, P. Cellular Functions of the Autism Risk Factor PTCHD1 in Mice. J. Neurosci. 2017, 37, 11993–12005. [Google Scholar] [CrossRef]
- Ung, D.C.; Iacono, G.; Méziane, H.; Blanchard, E.; Papon, M.A.; Selten, M.; van Rhijn, J.R.; Montjean, R.; Rucci, J.; Martin, S.; et al. Ptchd1 deficiency induces excitatory synaptic and cognitive dysfunctions in mouse. Mol. Psychiatry. 2018, 23, 1356–1367. [Google Scholar] [CrossRef]
- Halewa, J.; Marouillat, S.; Dixneuf, M.; Thépault, R.A.; Ung, D.C.; Chatron, N.; Gérard, B.; Ghoumid, J.; Lesca, G.; Till, M.; et al. Novel missense mutations in PTCHD1 alter its plasma membrane subcellular localization and cause intellectual disability and autism spectrum disorder. Hum. Mutat. 2021, 42, 848–861. [Google Scholar] [CrossRef]
- Chaudhry, A.; Noor, A.; Degagne, B.; Baker, K.; Bok, L.A.; Brady, M.F.; Chitayat, D.; Chung, B.H.; Cytrynbaum, C.; Dyment, D.; et al. Phenotypic spectrum associated with PTCHD1 deletions and truncating mutations includes intellectual disability and autism spectrum disorder. Clin. Genet. 2015, 88, 224–233. [Google Scholar] [CrossRef]
- Marshall, C.R.; Noor, A.; Vincent, J.B.; Lionel, A.C.; Feuk, L.; Skaug, J.; Shago, M.; Moessner, R.; Pinto, D.; Ren, Y.; et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008, 82, 477–488. [Google Scholar] [CrossRef]
- Pastore, S.F.; Ko, S.Y.; Frankland, P.W.; Hamel, P.A.; Vincent, J.B. PTCHD1: Identification and neurodevelopmental contributions of an autism spectrum disorder and intellectual disability susceptibility gene. Genes 2022, 13, 527. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Pagnamenta, A.T.; Klei, L.; Anney, R.; Merico, D.; Regan, R.; Conroy, J.; Magalhaes, T.R.; Correia, C.; Abrahams, B.S.; et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010, 466, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Kinnebrew, M.; Luchetti, G.; Sircar, R.; Frigui, S.; Viti, L.V.; Naito, T.; Beckert, F.; Saheki, Y.; Siebold, C.; Radhakrishnan, A.; et al. Patched 1 reduces the accessibility of cholesterol in the outer leaflet of membranes. Elife 2021, 10, e70504. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Rohatgi, R.; Siebold, C. Cholesterol access in cellular membranes controls Hedgehog signaling. Nat. Chem. Biol. 2020, 16, 1303–1313. [Google Scholar] [CrossRef] [PubMed]
- Kinnebrew, M.; Iverson, E.J.; Patel, B.B.; Pusapati, G.V.; Kong, J.H.; Johnson, K.A.; Luchetti, G.; Eckert, K.M.; McDonald, J.G.; Covey, D.F.; et al. Cholesterol accessibility at the ciliary membrane controls hedgehog signaling. Elife 2019, 8, e50051. [Google Scholar] [CrossRef]
- Chung, J.H.; Larsen, A.R.; Chen, E.; Bunz, F. A PTCH1 homolog transcriptionally activated by p53 suppresses Hedgehog signaling. J. Biol. Chem. 2014, 289, 33020–33031. [Google Scholar] [CrossRef]
- Loftus, S.K.; Morris, J.A.; Carstea, E.D.; Gu, J.Z.; Cummings, C.; Brown, A.; Ellison, J.; Ohno, K.; Rosenfeld, M.A.; Tagle, D.A.; et al. Murine model of Niemann-Pick C disease: Mutation in a cholesterol homeostasis gene. Science 1997, 277, 232–235. [Google Scholar] [CrossRef]
- Carstea, E.D.; Morris, J.A.; Coleman, K.G.; Loftus, S.K.; Zhang, D.; Cummings, C.; Gu, J.; Rosenfeld, M.A.; Pavan, W.J.; Krizman, D.B.; et al. Niemann-Pick C1 disease gene: Homology to mediators of cholesterol homeostasis. Science 1997, 277, 228–231. [Google Scholar] [CrossRef]
- Karten, B.; Vance, D.E.; Campenot, R.B.; Vance, J.E. Trafficking of cholesterol from cell bodies to distal axons in Niemann Pick C1-deficient neurons. J. Biol. Chem. 2003, 278, 4168–4175. [Google Scholar] [CrossRef]
- Distl, R.; Treiber-Held, S.; Albert, F.; Meske, V.; Harzer, K.; Ohm, T.G. Cholesterol storage and tau pathology in Niemann-Pick type C disease in the brain. J. Pathol. 2003, 200, 104–111. [Google Scholar] [CrossRef]
- Depta, L.; Whitmarsh-Everiss, T.; Laraia, L. Structure, function and small molecule modulation of intracellular sterol transport proteins. Bioorg. Med. Chem. 2022, 68, 116856. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Bondza, S.; Foy, E.; Brooks, J.; Andersson, K.; Robinson, J.; Richalet, P.; Buijs, J. Real-time characterization of antibody binding to receptors on living immune cells. Front. Immunol. 2017, 8, 455. [Google Scholar] [CrossRef]
- Gong, X.; Qian, H.; Cao, P.; Zhao, X.; Zhou, Q.; Lei, J.; Yan, N. Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science 2018, 361, eaas8935. [Google Scholar] [CrossRef]
- Morita, K.; Lo Celso, C.; Spencer-Dene, B.; Zouboulis, C.C.; Watt, F.M. HAN11 binds mDia1 and controls GLI1 transcriptional activity. J. Dermatol. Sci. 2006, 44, 11–20. [Google Scholar] [CrossRef]
- Skurat, A.V.; Dietrich, A.D. Phosphorylation of Ser640 in muscle glycogen synthase by DYRK family protein kinases. J. Biol. Chem. 2004, 279, 2490–2498. [Google Scholar] [CrossRef]
- Qi, C.; Di Minin, G.; Vercellino, I.; Wutz, A.; Korkhov, V.M. Structural basis of sterol recognition by human hedgehog receptor PTCH1. Sci. Adv. 2019, 5, eaaw6490. [Google Scholar] [CrossRef]
- Qian, H.; Wu, X.; Du, X.; Yao, X.; Zhao, X.; Lee, J.; Yang, H.; Yan, N. Structural Basis of low-pH-dependent lysosomal cholesterol egress by NPC1 and NPC2. Cell 2020, 182, 98–111.e18. [Google Scholar] [CrossRef]
- Wang, Q.; Asarnow, D.E.; Ding, K.; Mann, R.K.; Hatakeyama, J.; Zhang, Y.; Ma, Y.; Cheng, Y.; Beachy, P.A. Dispatched uses Na+ flux to power release of lipid-modified Hedgehog. Nature 2021, 599, 320–324. [Google Scholar] [CrossRef]
- Zhang, Y.; Bulkley, D.P.; Xin, Y.; Roberts, K.J.; Asarnow, D.E.; Sharma, A.; Myers, B.R.; Cho, W.; Cheng, Y.; Beachy, P.A. Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched. Cell 2018, 175, 1352–1364.e14. [Google Scholar] [CrossRef] [PubMed]
- Petrov, K.; Wierbowski, B.M.; Liu, J.; Salic, A. Distinct cation gradients power cholesterol transport at different key points in the Hedgehog signaling pathway. Dev. Cell. 2020, 55, 314–327.e7. [Google Scholar] [CrossRef]
- Infante, R.E.; Wang, M.L.; Radhakrishnan, A.; Kwon, H.J.; Brown, M.S.; Goldstein, J.L. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. USA 2008, 105, 15287–15292. [Google Scholar] [CrossRef]
- Bryant, P.; Pozzati, G.; Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 2022, 13, 1265. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.; Kedersha, N.; Anderson, P.; Ivanov, P. Molecular mechanisms of stress granule assembly and disassembly. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118876. [Google Scholar] [CrossRef] [PubMed]
- Formicola, N.; Vijayakumar, J.; Besse, F. Neuronal ribonucleoprotein granules: Dynamic sensors of localized signals. Traffic 2019, 20, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Glauninger, H.; Wong Hickernell, C.J.; Bard, J.A.M.; Drummond, D.A. Stressful steps: Progress and challenges in understanding stress-induced mRNA condensation and accumulation in stress granules. Mol. Cell 2022, 82, 2544–2556. [Google Scholar] [CrossRef]
- Popper, B.; Scheidt, T.; Schieweck, R. RNA-binding protein dysfunction in neurodegeneration. Essays Biochem. 2021, 65, 975–986. [Google Scholar]
- Lennox, A.L.; Hoye, M.L.; Jiang, R.; Johnson-Kerner, B.L.; Suit, L.A.; Venkataramanan, S.; Sheehan, C.J.; Alsina, F.C.; Fregeau, B.; Aldinger, K.A.; et al. Pathogenic DDX3X mutations impair RNA metabolism and neurogenesis during fetal cortical development. Neuron 2020, 106, 404–420.e8. [Google Scholar] [CrossRef]
- Richter, J.D.; Zhao, X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021, 22, 209–222. [Google Scholar] [CrossRef]
- Lai, A.; Valdez-Sinon, A.N.; Bassell, G.J. Regulation of RNA granules by FMRP and implications for neurological diseases. Traffic 2020, 21, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.; Hollingworth, D.; Adinolfi, S.; Castets, M.; Kelly, G.; Frenkiel, T.A.; Bardoni, B.; Pastore, A. The structure of the N-terminal domain of the fragile X mental retardation protein: A platform for protein-protein interaction. Structure 2006, 14, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.S.; Haghighi, F.; Stefanski, A.; Nakhaei-Rad, S.; Kazemein Jasemi, N.S.; Al Kabbani, M.A.; Görg, B.; Fujii, M.; Lang, P.A.; Häussinger, D.; et al. Novel FMRP interaction networks linked to cellular stress. FEBS J. 2021, 288, 837–860. [Google Scholar] [CrossRef] [PubMed]
- Pasciuto, E.; Bagni, C. SnapShot: FMRP interacting proteins. Cell 2014, 159, 218.e1. [Google Scholar] [CrossRef]
- Trowitzsch, S.; Bieniossek, C.; Nie, Y.; Garzoni, F.; Berger, I. New baculovirus expression tools for recombinant protein complex production. J. Struct. Biol. 2010, 172, 45–54. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Jumper, J.; Hassabis, D. Protein structure predictions to atomic accuracy with AlphaFold. Nat. Methods 2022, 19, 11–12. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef]
- Mariani, V.; Biasini, M.; Barbato, A.; Schwede, T. lDDT: A local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 2013, 29, 2722–2728. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Kejariwal, A.; Guo, N.; Mi, H.; Campbell, M.J.; Muruganujan, A.; Lazareva-Ulitsky, B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006, 34, W645–W650. [Google Scholar] [CrossRef]
- Gene, O.C. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar]

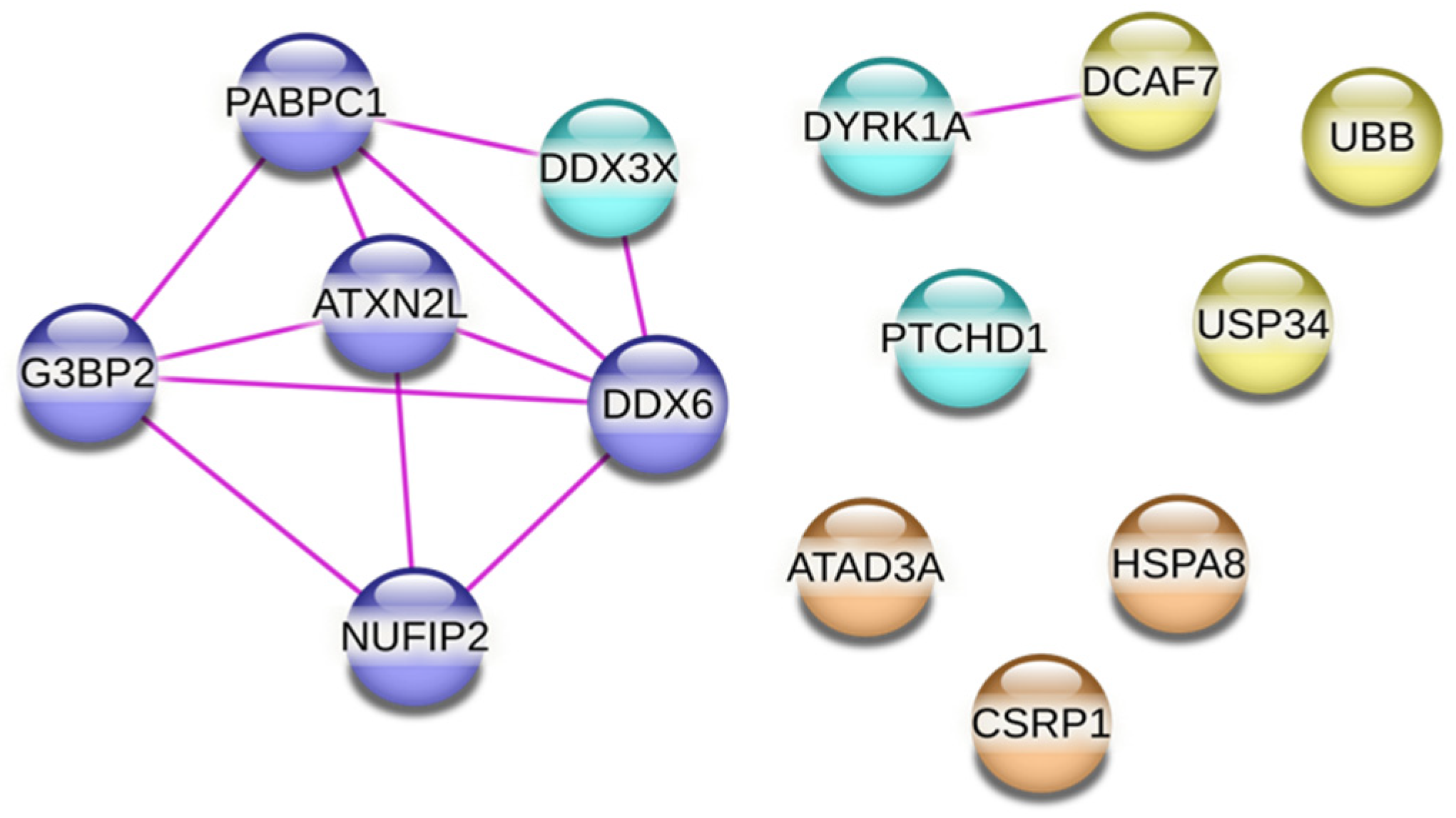
| Protein Name | No Unique Peptides | Reported Function |
|---|---|---|
| ATXN2L * | 39 | Regulation of stress granule and P-body formation. |
| NUFIP2 * | 25 | RNA binding protein found in stress granules. |
| PABP1 * | 23 | Binds the poly(A) tail of mRNA, regulates processes of mRNA metabolism such as pre-mRNA splicing and mRNA stability. |
| HSPA8 * | 12 | Molecular chaperone implicated in a wide variety of cellular processes, including protection of the proteome from stress, and acts as a repressor of transcriptional activation. |
| DCAF7 * | 10 | Associates with DIAPH1 and controls GLI1 transcriptional activity. Binds DYRK1A through its CTD. Inhibits DYRK1A dependent GLI activation and nuclear retention [26]. |
| DYRK1A A | 16 | Dual-specificity kinase which possesses both serine/threonine and tyrosine kinase activities. Plays an important role in double-strand break (DSB) repair following DNA damage. Phosphorylates GLI1 to promote nuclear localisation and transcriptional activity [27]. |
| DDX3X *A | 6 | ATPase/helicase activity. Required for efficient stress granule assembly via interaction with eIF4E. Direct interaction partner of PABP1. |
| UBB * | 4 | Involved in protein synthesis and degradation. Highly expressed in gonadotropin-releasing hormone neurons [26]. |
| CSRP1 * | 4 | Potential role in neuronal development. Cysteine-rich domain is highly conserved in steroid receptors. |
| G3BP2 * | 4 | Scaffold protein with essential role in cytoplasmic stress granule formation. |
| ATAD3A *° | 8 | Required for enhanced channelling of cholesterol for hormone- dependent steroidogenesis. |
| UBP34 | 4 | Ubiquitin hydrolase that removes conjugated ubiquitin from AXIN1 and AXIN2, regulating Wnt signalling pathway downstream of the beta-catenin destruction complex. |
| DDX6 * | 4 | Essential for the formation of P-bodies and ribonucleoprotein particles. |
| Cellular Component | Fold Enrichment | p-Value | False Discovery Rate | ||
|---|---|---|---|---|---|
| cytoplasmic stress granule | (GO:0010494) | >100 | 10 × 10−12 | 2 × 10−8 | |
| cytoplasmic ribonucleoprotein particle | (GO:0036464) | 38 | 5 × 10−9 | 5 × 10−6 | |
| ribonucleoprotein particle | (GO:0035770) | 36 | 8 × 10−9 | 5 × 10−6 | |
| ribonucleoprotein complex | (GO:1990904) | 14 | 2 × 10−6 | 8 × 10−4 | |
| supramolecular complex | (GO:0099080) | 9 | 4 × 10−7 | 2 × 10−4 | |
| intracellular non-membrane-bounded organelle | (GO:0043232) | 3 | 1 × 10−4 | 5 × 10−2 | |
| non-membrane-bounded organelle | (GO:0043228) | 3 | 1 × 10−4 | 4 × 10−2 | |
| Biological process | |||||
| stress granule assembly | (GO:0034063) | >100 | 2 × 10−9 | 3 × 10−5 | |
| non-membrane-bound organelle assembly | (GO:0140694) | 25 | 10 × 10−7 | 8 × 10−3 | |
| Molecular function | |||||
| RNA binding | (GO:0003723) | 8 | 2 × 10−6 | 8 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hiltunen, M.K.; Timmis, A.J.; Thomsen, M.; Gkotsi, D.S.; Iwaï, H.; Ribeiro, O.M.; Goldman, A.; Riobo-Del Galdo, N.A. PTCHD1 Binds Cholesterol but Not Sonic Hedgehog, Suggesting a Distinct Cellular Function. Int. J. Mol. Sci. 2023, 24, 2682. https://doi.org/10.3390/ijms24032682
Hiltunen MK, Timmis AJ, Thomsen M, Gkotsi DS, Iwaï H, Ribeiro OM, Goldman A, Riobo-Del Galdo NA. PTCHD1 Binds Cholesterol but Not Sonic Hedgehog, Suggesting a Distinct Cellular Function. International Journal of Molecular Sciences. 2023; 24(3):2682. https://doi.org/10.3390/ijms24032682
Chicago/Turabian StyleHiltunen, Mimmu K., Alex J. Timmis, Maren Thomsen, Danai S. Gkotsi, Hideo Iwaï, Orquidea M. Ribeiro, Adrian Goldman, and Natalia A. Riobo-Del Galdo. 2023. "PTCHD1 Binds Cholesterol but Not Sonic Hedgehog, Suggesting a Distinct Cellular Function" International Journal of Molecular Sciences 24, no. 3: 2682. https://doi.org/10.3390/ijms24032682
APA StyleHiltunen, M. K., Timmis, A. J., Thomsen, M., Gkotsi, D. S., Iwaï, H., Ribeiro, O. M., Goldman, A., & Riobo-Del Galdo, N. A. (2023). PTCHD1 Binds Cholesterol but Not Sonic Hedgehog, Suggesting a Distinct Cellular Function. International Journal of Molecular Sciences, 24(3), 2682. https://doi.org/10.3390/ijms24032682








