Theoretical Studies on Selectivity of HPK1/JAK1 Inhibitors by Molecular Dynamics Simulations and Free Energy Calculations
Abstract
1. Introduction
2. Results and Discussion
2.1. Sequence Alignment of HPK1 and JAK1
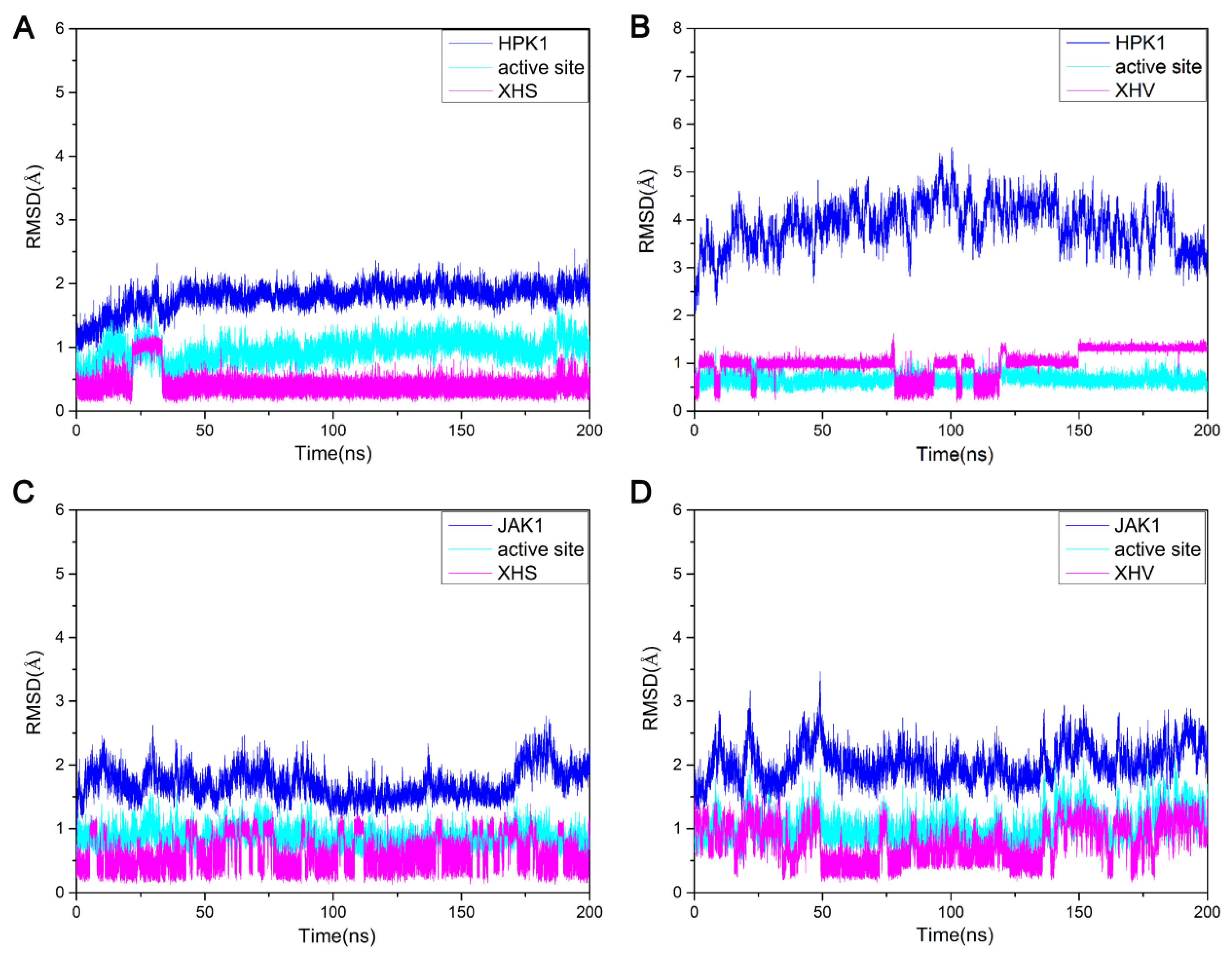
2.2. The Conformational Stabilities of the Four Systems
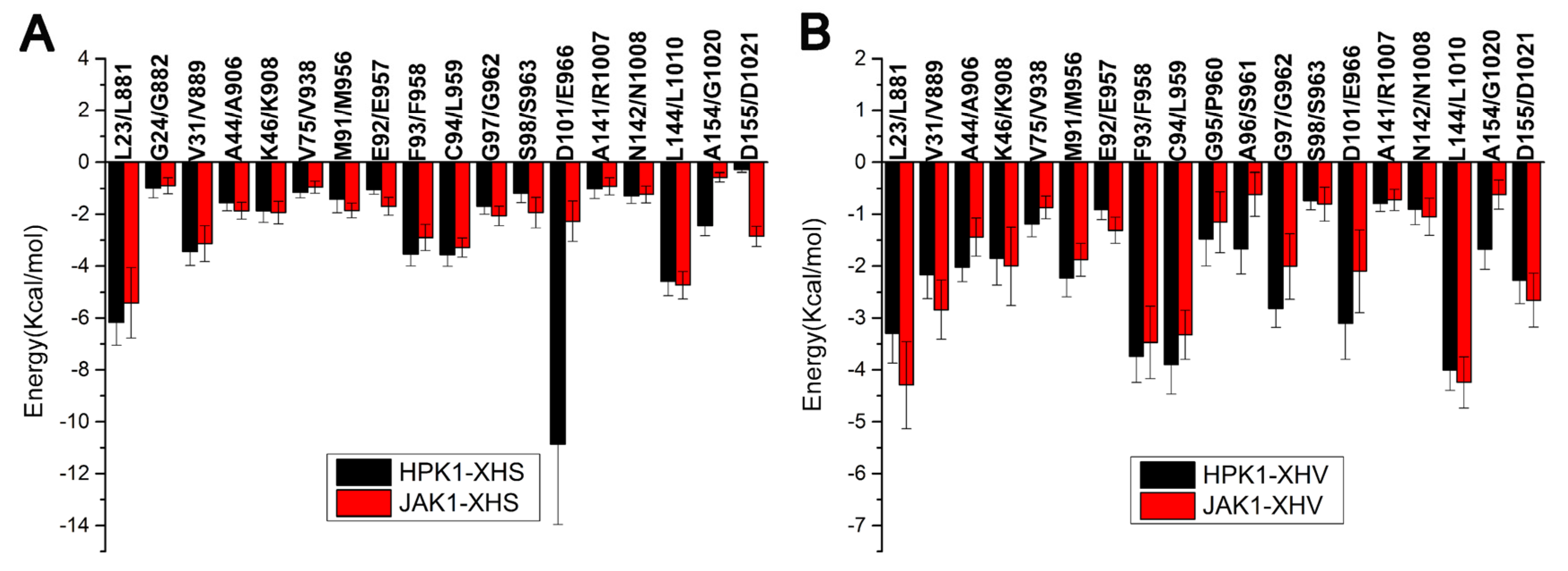
2.3. Binding Free Energy Calculations
2.4. Hydrogen Bond and Salt Bridge Analysis
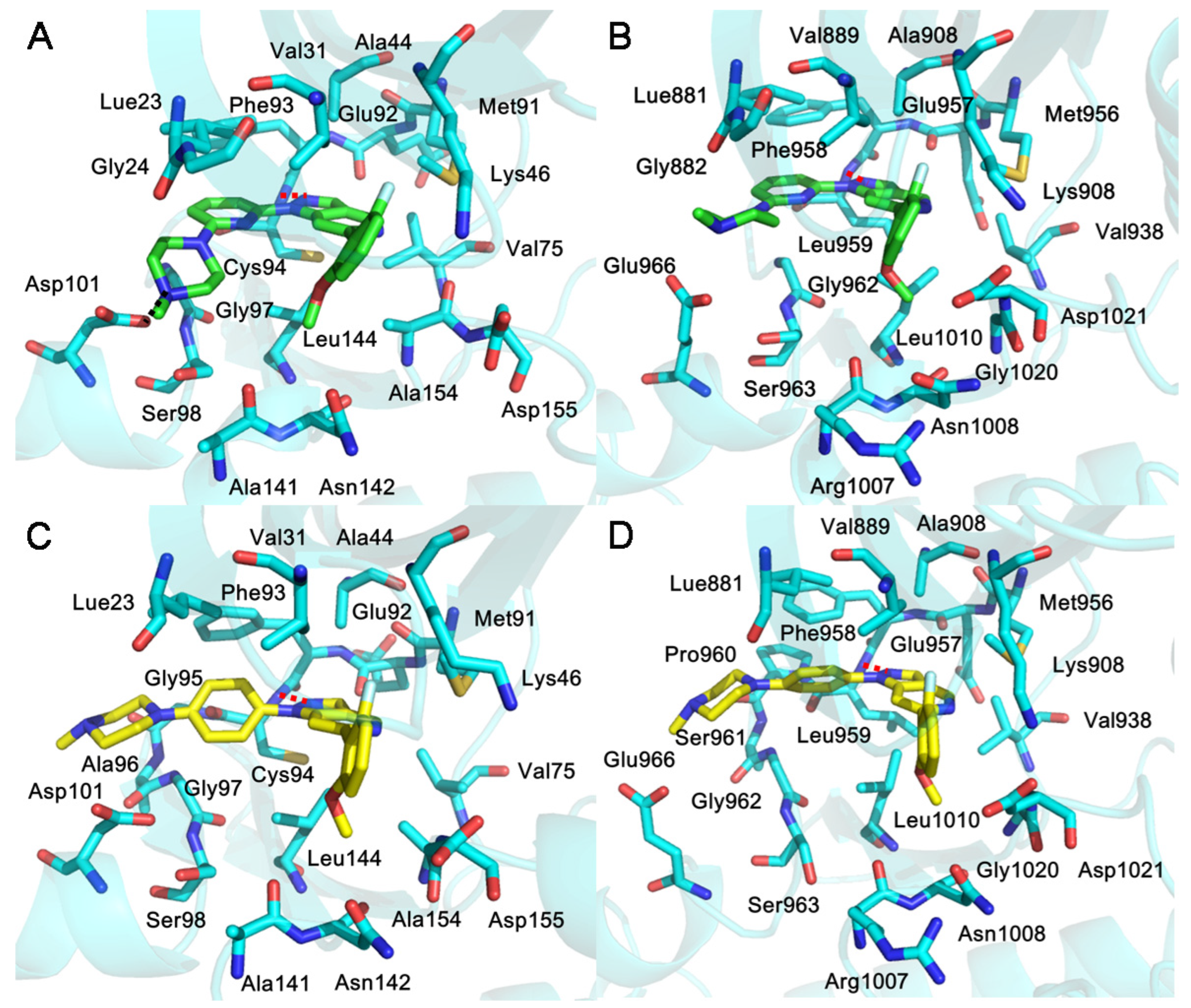
2.5. The Binding Mode between Kinases (HPK1/JAK1) and Inhibitors (XHS/XHV)
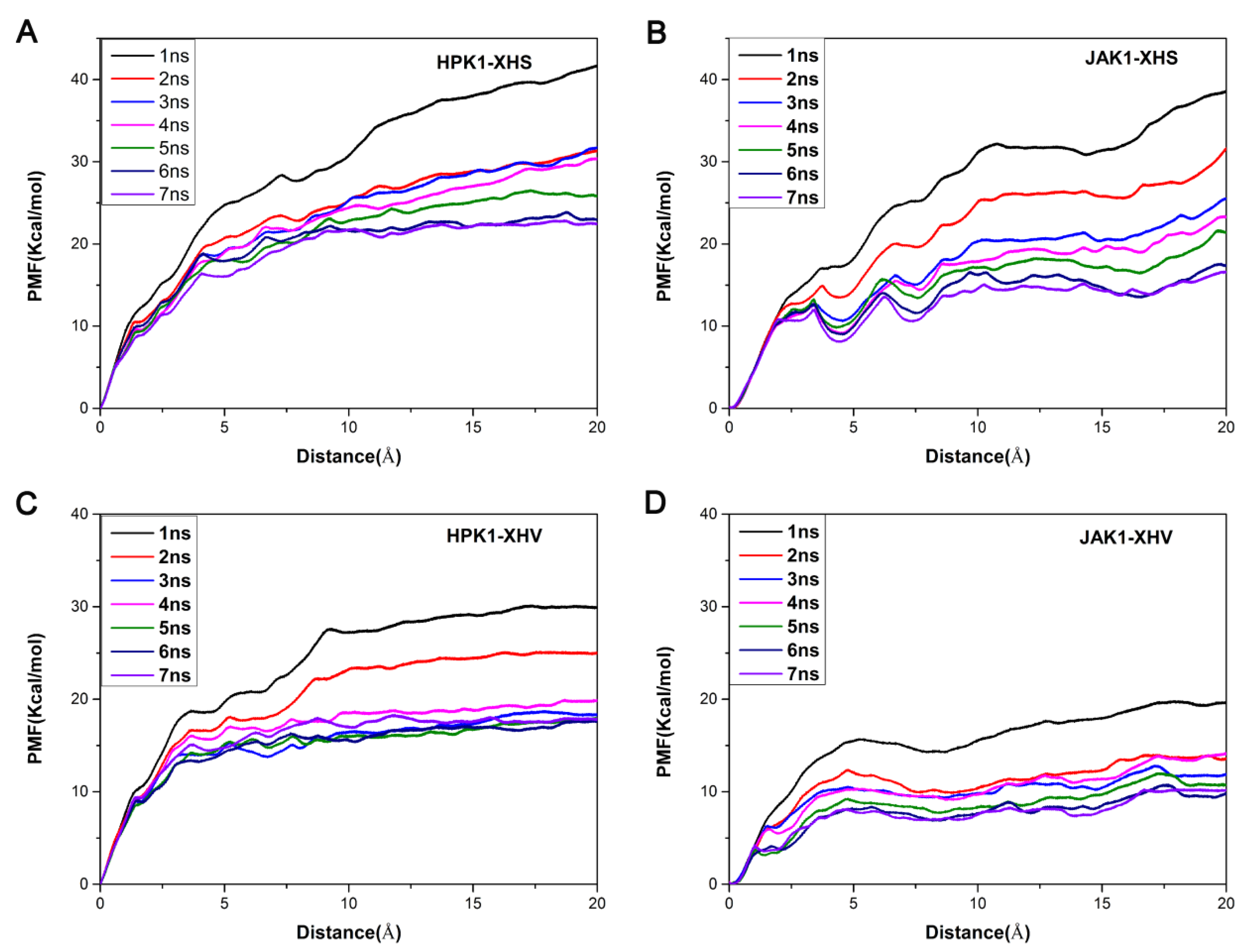
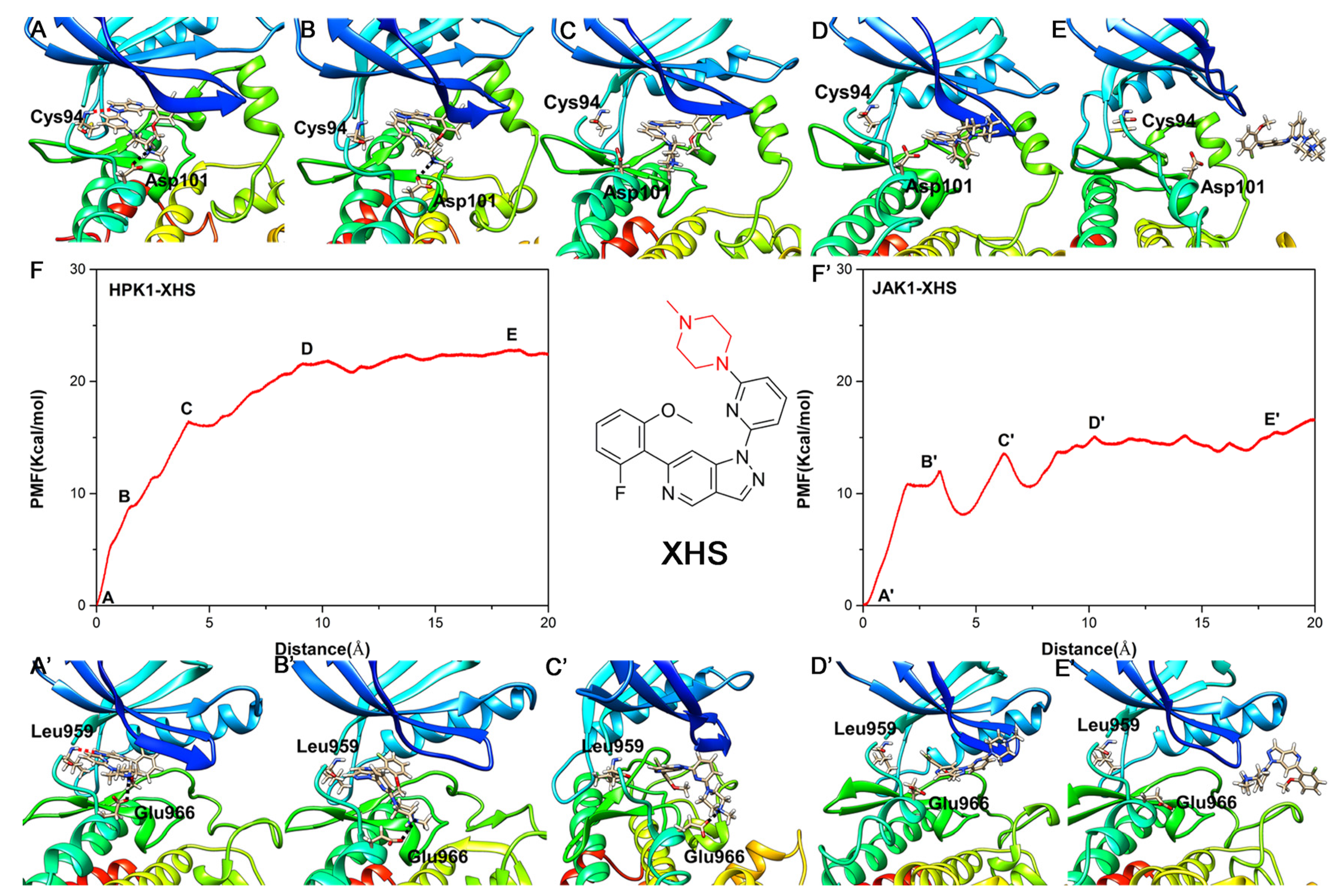
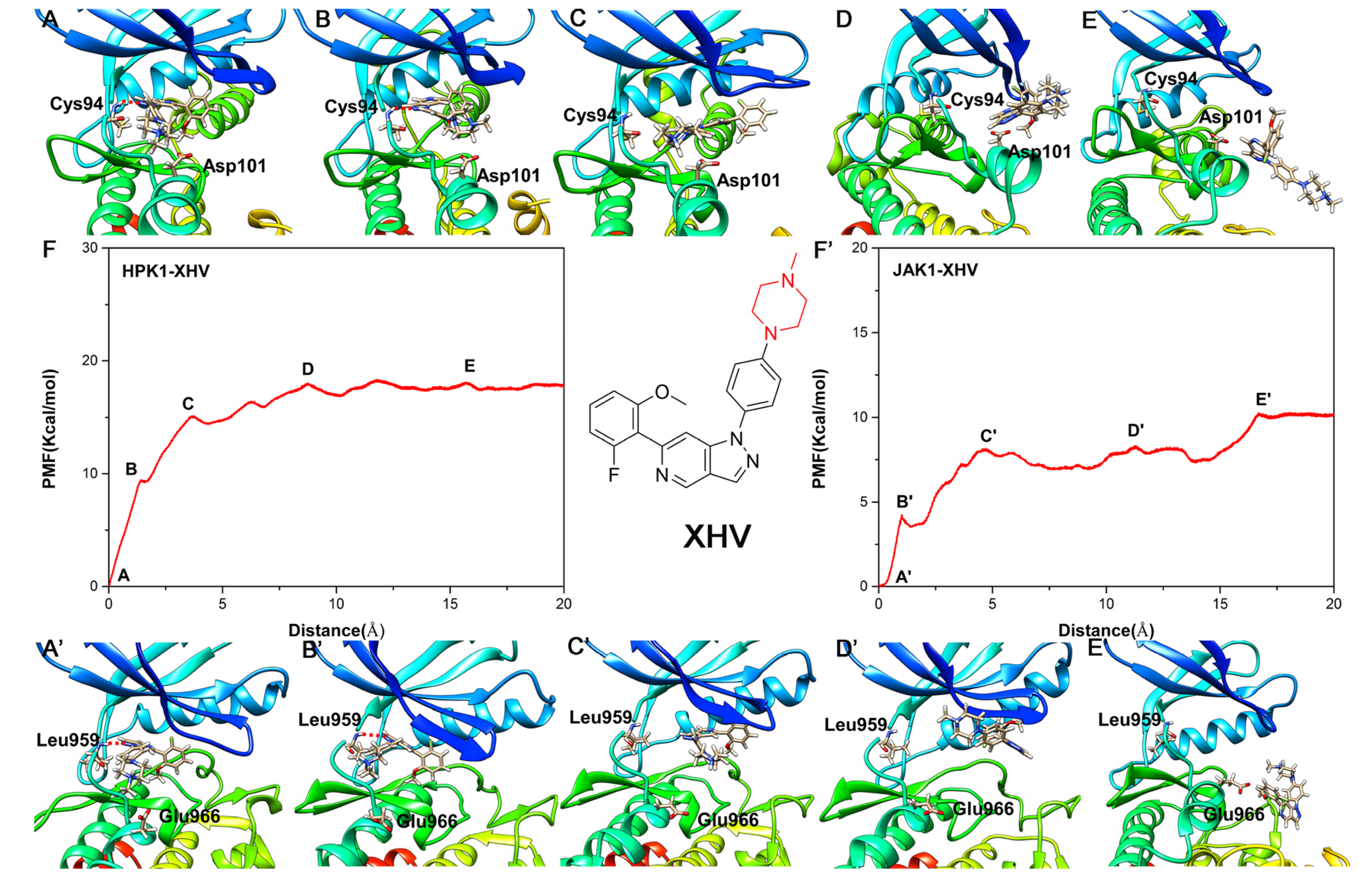
2.6. Unbinding Pathways of Inhibitors (XHS and XHV) Dissociating from Kinases (HPK1 and JAK1)
3. Materials and Methods
3.1. Sequence Alignment and Structure Superposition
3.2. Molecular Dynamics Simulation
3.3. Binding Free Energy Calculations
3.4. Free Energy Decomposition Analysis
3.5. Umbrella Sampling Simulations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sawasdikosol, S.; Zha, R.; Yang, B.; Burakoff, S. HPK1 as a novel target for cancer immunotherapy. Immunol Res. 2012, 54, 262–265. [Google Scholar] [PubMed]
- Si, J.; Shi, X.; Sun, S.; Zou, B.; Li, Y.; An, D.; Lin, X.; Gao, Y.; Long, F.; Pang, B.; et al. Hematopoietic Progenitor Kinase1 (HPK1) Mediates T Cell Dysfunction and Is a Druggable Target for T Cell-Based Immunotherapies. Cancer Cell 2020, 38, 551–566. [Google Scholar] [PubMed]
- Shui, J.W.; Boomer, J.S.; Han, J.; Xu, J.; Dement, G.A.; Zhou, G.; Tan, T.H. Hematopoietic progenitor kinase 1 negatively regulates T cell receptor signaling and T cell-mediated immune responses. Nat. Immunol. 2007, 8, 84–91. [Google Scholar]
- Sauer, K.; Liou, J.; Singh, S.B.; Yablonski, D.; Weiss, A.; Perlmutter, R.M. Hematopoietic progenitor kinase 1 associates physically and functionally with the adaptor proteins B cell linker protein and SLP-76 in lymphocytes. J. Biol. Chem. 2001, 276, 45207–45216. [Google Scholar] [CrossRef] [PubMed]
- Di Bartolo, V.; Montagne, B.; Salek, M.; Jungwirth, B.; Carrette, F.; Fourtane, J.; Sol-Foulon, N.; Michel, F.; Schwartz, O.; Lehmann, W.D.; et al. A novel pathway down-modulating T cell activation involves HPK-1-dependent recruitment of 14-3-3 proteins on SLP-76. J. Exp. Med. 2007, 204, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Ling, P.; Meyer, C.F.; Redmond, L.P.; Shui, J.W.; Davis, B.; Rich, R.R.; Hu, M.C.; Wange, R.L.; Tan, T.H. Involvement of hematopoietic progenitor kinase 1 in T cell receptor signaling. J. Biol. Chem. 2001, 276, 18908–18914. [Google Scholar] [PubMed]
- Liou, J.; Kiefer, F.; Dang, A.; Hashimoto, A.; Cobb, M.H.; Kurosaki, T.; Weiss, A. HPK1 Is Activated by Lymphocyte Antigen Receptors and Negatively Regulates AP-1. Immunity 2000, 12, 399–408. [Google Scholar]
- Wang, X.; Li, J.P.; Chiu, L.L.; Lan, J.L.; Chen, D.Y.; Boomer, J.; Tan, T.H. Attenuation of T cell receptor signaling by serine phosphorylation-mediated lysine 30 ubiquitination of SLP-76 protein. J. Biol. Chem. 2012, 287, 34091–34100. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, S.; Qing, J.; Thibodeau, R.H.; Du, X.; Park, S.; Lee, H.M.; Xu, M.; Oh, S.; Navarro, A.; Roose-Girma, M.; et al. The Kinase Activity of Hematopoietic Progenitor Kinase 1 Is Essential for the Regulation of T Cell Function. Cell Rep. 2018, 25, 80–94. [Google Scholar] [CrossRef]
- Alzabin, S.; Bhardwaj, N.; Kiefer, F.; Sawasdikosol, S.; Burakoff, S. Hematopoietic progenitor kinase 1 is a negative regulator of dendritic cell activation. J. Immunol. 2009, 182, 6187–6194. [Google Scholar] [CrossRef]
- Sawasdikosol, S.; Zha, R.; Fisher, T.S.; Alzabin, S.; Burakoff, S.J. HPK1 Influences Regulatory T Cell Functions. ImmunoHorizons 2020, 4, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Alzabin, S.; Pyarajan, S.; Yee, H.; Kiefer, F.; Suzuki, A.; Burakoff, S.; Sawasdikosol, S. Hematopoietic progenitor kinase 1 is a critical component of prostaglandin E2-mediated suppression of the anti-tumor immune response. Cancer Immunol. Immunother. 2010, 59, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, K.; Georgiev, P.; Wells, S.; Xu, H.; Lacey, B.M.; Xu, Z.; Laskey, J.; McLeod, R.; Methot, J.L.; et al. Pharmacological inhibition of hematopoietic progenitor kinase 1 positively regulates T-cell function. PLoS ONE 2020, 15, e0243145. [Google Scholar]
- You, D.; Hillerman, S.; Locke, G.; Chaudhry, C.; Stromko, C.; Murtaza, A.; Fan, Y.; Koenitzer, J.; Chen, Y.; Briceno, S.; et al. Enhanced antitumor immunity by a novel small molecule HPK1 inhibitor. J. Immunother. Cancer 2021, 9, e001402. [Google Scholar] [CrossRef]
- Yu, E.C.; Methot, J.L.; Fradera, X.; Lesburg, C.A.; Lacey, B.M.; Siliphaivanh, P.; Liu, P.; Smith, D.M.; Xu, Z.; Piesvaux, J.A.; et al. Identification of Potent Reverse Indazole Inhibitors for HPK1. ACS Med. Chem. Lett. 2021, 12, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, K.; Saharinen, P.; Pesu, M.; Holt, V.E.T.; Silvennoinen, O.; O’Shea, J.J. The Janus kinases (Jaks). Genome Biol. 2004, 5, 253. [Google Scholar]
- Banerjee, S.; Biehl, A.; Gadina, M.; Hasni, S.; Schwartz, D.M. JAK-STAT Signaling as a Target for Inflammatory and Autoimmune Diseases: Current and Future Prospects. Drugs 2017, 77, 521–546. [Google Scholar]
- Wang, Q.; Shao, X.; Leung, E.L.H.; Chen, Y.; Yao, X. Selectively targeting individual bromodomain: Drug discovery and molecular mechanisms. Pharmacol. Res. 2021, 172, 105804. [Google Scholar]
- Fu, W.; Chen, L.; Wang, Z.; Kang, Y.; Wu, C.; Xia, Q.; Liu, Z.; Zhou, J.; Liang, G.; Cai, Y.A.-O. Theoretical studies on FGFR isoform selectivity of FGFR1/FGFR4 inhibitors by molecular dynamics simulations and free energy calculations. Phys. Chem. Chem. Phys. 2017, 19, 3649–3659. [Google Scholar]
- Tian, Y.; Yu, Y.; Shen, Y.; Wan, H.; Chang, S.; Zhang, T.; Wan, S.; Zhang, J. Molecular Simulation Studies on the Binding Selectivity of Type-I Inhibitors in the Complexes with ROS1 versus ALK. J. Chem. Inf. Model. 2017, 57, 977–987. [Google Scholar]
- Wang, H.; Gao, Z.; Song, P.; Hu, B.; Wang, J.; Cheng, M. Molecular dynamics simulation and QM/MM calculation reveal the selectivity mechanism of type I 1/2 kinase inhibitors: The effect of intramolecular H-bonds and conformational restriction for improved selectivity. Phys. Chem. Chem. Phys. 2019, 21, 24147–24164. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Song, P.; Wang, H.; Hu, B.; Wang, J.; Cheng, M.S. Precise design of highly isoform-selective p21-activated kinase 4 inhibitors: Computational insights into the selectivity mechanism through molecular dynamics simulation and binding free energy calculation. J. Biomol. Struct. Dyn. 2020, 38, 3825–3837. [Google Scholar] [CrossRef]
- Chen, Q.; Cui, W.; Cheng, Y.; Zhang, F.; Ji, M. Studying the mechanism that enables paullones to selectively inhibit glycogen synthase kinase 3 rather than cyclin-dependent kinase 5 by molecular dynamics simulations and free-energy calculations. J. Mol. Model. 2011, 17, 795–803. [Google Scholar] [PubMed]
- Zhang, Z.; Xu, Y.; Wu, J.; Shen, Y.; Cheng, H.; Xiang, Y. Exploration of the selective binding mechanism of protein kinase Aurora A selectivity via a comprehensive molecular modeling study. PeerJ 2019, 7, e7832. [Google Scholar] [CrossRef] [PubMed]
- Keretsu, S.; Bhujbal, S.P.; Joo Cho, S. Computational study of paroxetine-like inhibitors reveals new molecular insight to inhibit GRK2 with selectivity over ROCK1. Sci. Rep. 2019, 9, 13053. [Google Scholar] [PubMed]
- Lin, Y.L.; Meng, Y.; Huang, L.; Roux, B. Computational study of Gleevec and G6G reveals molecular determinants of kinase inhibitor selectivity. J. Am. Chem. Soc. 2014, 136, 14753–14762. [Google Scholar]
- Shi, D.; Bai, Q.; Zhou, S.; Liu, X.; Liu, H.; Yao, X. Molecular dynamics simulation, binding free energy calculation and unbinding pathway analysis on selectivity difference between FKBP51 and FKBP52: Insight into the molecular mechanism of isoform selectivity. Proteins 2018, 86, 43–56. [Google Scholar]
- Wang, L.; Zheng, G.; Liu, X.; Ni, D.; He, X.; Cheng, J.; Lu, S. Molecular dynamics simulations provide insights into the origin of gleevec’s selectivity toward human tyrosine kinases. J. Biomol. Struct. Dyn. 2019, 37, 2733–2744. [Google Scholar] [CrossRef]
- Chohan, T.; Chen, J.; Qian, H.; Pan, Y.; Chen, J. Molecular modeling studies to characterize N-phenylpyrimidin-2-amine selectivity for CDK2 and CDK4 through 3D-QSAR and molecular dynamics simulations. Mol. Biosyst. 2016, 12, 1250–1268. [Google Scholar]
- Kong, X.; Sun, H.; Pan, P.; Tian, S.; Li, D.; Li, Y.; Hou, T. Molecular principle of the cyclin-dependent kinase selectivity of 4-(thiazol-5-yl)-2-(phenylamino) pyrimidine-5-carbonitrile derivatives revealed by molecular modeling studies. Phys. Chem. Chem. Phys. 2016, 18, 2034–2046. [Google Scholar]
- Chohan, T.; Qian, H.; Pan, Y.; Chen, J. Molecular simulation studies on the binding selectivity of 2-anilino-4-(thiazol-5-yl)-pyrimidines in complexes with CDK2 and CDK7. Mol. Biosyst. 2016, 12, 145–161. [Google Scholar] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic. Acids. Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Gilli, P.; Gilli, G. Hydrogen bond models and theories: The dual hydrogen bond model and its consequences. J. Mol. Struct. 2010, 972, 2–10. [Google Scholar] [CrossRef]
- Gilli, P.; Pretto, L.; Bertolasi, V.; Gilli, G. Predicting hydrogen-bond strengths from acid-base molecular properties. The pK(a) slide rule: Toward the solution of a long-lasting problem. Acc. Chem. Res. 2009, 42, 33–44. [Google Scholar] [CrossRef]
- Gilli, P.; Pretto, L.; Gilli, G. PA/pKa equalization and the prediction of the hydrogen-bond strength: A synergism of classical thermodynamics and structural crystallography. J. Mol. Struct. 2007, 844–845, 328–339. [Google Scholar]
- Kurczab, R.; Sliwa, P.; Rataj, K.; Kafel, R.; Bojarski, A.J. Salt Bridge in Ligand-Protein Complexes-Systematic Theoretical and Statistical Investigations. J. Chem. Inf. Model. 2018, 58, 2224–2238. [Google Scholar] [CrossRef]
- Su, Q.; Banks, E.; Bebernitz, G.; Bell, K.; Borenstein, C.F.; Chen, H.; Chuaqui, C.E.; Deng, N.; Ferguson, A.D.; Kawatkar, S.; et al. Discovery of (2R)-N-[3-[2-[(3-Methoxy-1-methyl-pyrazol-4-yl)amino]pyrimidin-4-yl]-1H-indol-7-y l]-2-(4-methylpiperazin-1-yl)propenamide (AZD4205) as a Potent and Selective Janus Kinase 1 Inhibitor. J. Med. Chem. 2020, 63, 4517–4527. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. AMBER 18; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Frisch, M.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Petersson, G. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Chen, Y.; Zheng, Y.; Fong, P.; Mao, S.; Wang, Q. The application of the MM/GBSA method in the binding pose prediction of FGFR inhibitors. Phys. Chem. Chem. Phys. 2020, 22, 9656–9663. [Google Scholar] [CrossRef] [PubMed]
- Luan, J.; Hu, B.; Wang, S.; Liu, H.; Lu, S.; Li, W.; Sun, X.; Shi, J.; Wang, J. Selectivity mechanism of BCL-XL/2 inhibition through in silico investigation. Phys. Chem. Chem. Phys. 2022, 24, 17105–17115. [Google Scholar]
- Sahakyan, H. Improving virtual screening results with MM/GBSA and MM/PBSA rescoring. J. Comput. Aided. Mol. Des. 2021, 35, 731–736. [Google Scholar] [PubMed]
- Sitkoff, D.; Sharp, K.A.; Honig, B. Accurate Calculation of Hydration Free Energies Using Macroscopic Solvent Models. J. Phys. Chem. 1994, 98, 1978–1988. [Google Scholar] [CrossRef]
- Kästner, J.; Thiel, W. Bridging the gap between thermodynamic integration and umbrella sampling provides a novel analysis method: “Umbrella integration”. J. Chem. Phys. 2005, 123, 144104. [Google Scholar] [CrossRef]
- Chovancova, E.; Pavelka, A.; Benes, P.; Strnad, O.; Brezovsky, J.; Kozlikova, B.; Gora, A.; Sustr, V.; Klvana, M.; Medek, P.; et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 2012, 8, e1002708. [Google Scholar]
- Pavelka, A.; Sebestova, E.; Kozlikova, B.; Brezovsky, J.; Sochor, J.; Damborsky, J. CAVER: Algorithms for Analyzing Dynamics of Tunnels in Macromolecules. IEEE/ACM. Trans. Comput. Biol. Bioinform. 2016, 13, 505–517. [Google Scholar]
- Yu, Z.; Liu, Y.; Zhu, J.; Han, J.; Tian, X.; Han, W.; Zhao, L. Insights from molecular dynamics simulations and steered molecular dynamics simulations to exploit new trends of the interaction between HIF-1α and p300. J. Biomol. Struct. Dyn. 2020, 38, 1–12. [Google Scholar] [CrossRef]
- Wang, D.; Jin, H.; Wang, J.; Guan, S.; Zhang, Z.; Han, W. Exploration of the chlorpyrifos escape pathway from acylpeptide hydrolases using steered molecular dynamics simulations. J. Biomol. Struct. Dyn. 2016, 34, 749–761. [Google Scholar] [CrossRef]
- Sun, H.; Tian, S.; Zhou, S.; Li, Y.; Li, D.; Xu, L.; Shen, M.; Pan, P.; Hou, T. Revealing the favorable dissociation pathway of type II kinase inhibitors via enhanced sampling simulations and two-end-state calculations. Sci. Rep. 2015, 5, 8457. [Google Scholar]
- Kim, I.; Allen, T.W. Bennett’s acceptance ratio and histogram analysis methods enhanced by umbrella sampling along a reaction coordinate in configurational space. J. Chem. Phys. 2012, 136, 164103. [Google Scholar] [CrossRef]

| Energy | HPK1-XHS | JAK1-XHS | HPK1-XHV | JAK1-XHV |
|---|---|---|---|---|
| ΔEele | −31.58 ± 13.41 | −53.77 ± 19.26 | −37.81 ± 9.73 | −41.81 ± 19.85 |
| ΔEvdw | −49.88 ± 2.80 | −48.37 ± 2.91 | −50.30 ± 2.67 | −43.62 ± 3.57 |
| ΔEgas | −81.45 ± 14.14 | −102.14 ± 19.47 | −88.11 ± 9.85 | −85.43 ± 20.40 |
| ΔGGB | 41.10 ± 11.98 | 71.24 ± 18.55 | 55.18 ± 9.55 | 57.67 ± 19.46 |
| ΔGnonpl,sol | −5.99 ± 0.25 | −5.99 ± 0.29 | −6.18 ± 0.23 | −5.54 ± 0.42 |
| ΔGsol | 35.11 ± 11.88 | 65.25 ± 18.42 | 49.00 ± 9.54 | 52.14 ± 52.14 |
| ΔGpl | 9.52 ± 1.43 | 17.47 ± 0.71 | 17.37 ± 0.18 | 15.86 ± 0.39 |
| ΔGnonpl | −55.87 ± 3.05 | −54.36 ± 3.20 | −56.48 ± 2.90 | −49.16 ± 3.99 |
| ΔH (GB) | −46.35 ± 3.91 | −36.89 ± 3.19 | −39.11 ± 2.89 | −33.30 ± 3.78 |
| −TΔS | 19.75 ± 5.85 | 18.80 ± 8.28 | 20.43 ± 6.32 | 22.82 ± 3.25 |
| ΔG | −26.6 ± 7.08 | −18.09 ± 8.87 | −18.68 ± 6.92 | −10.48 ± 4.98 |
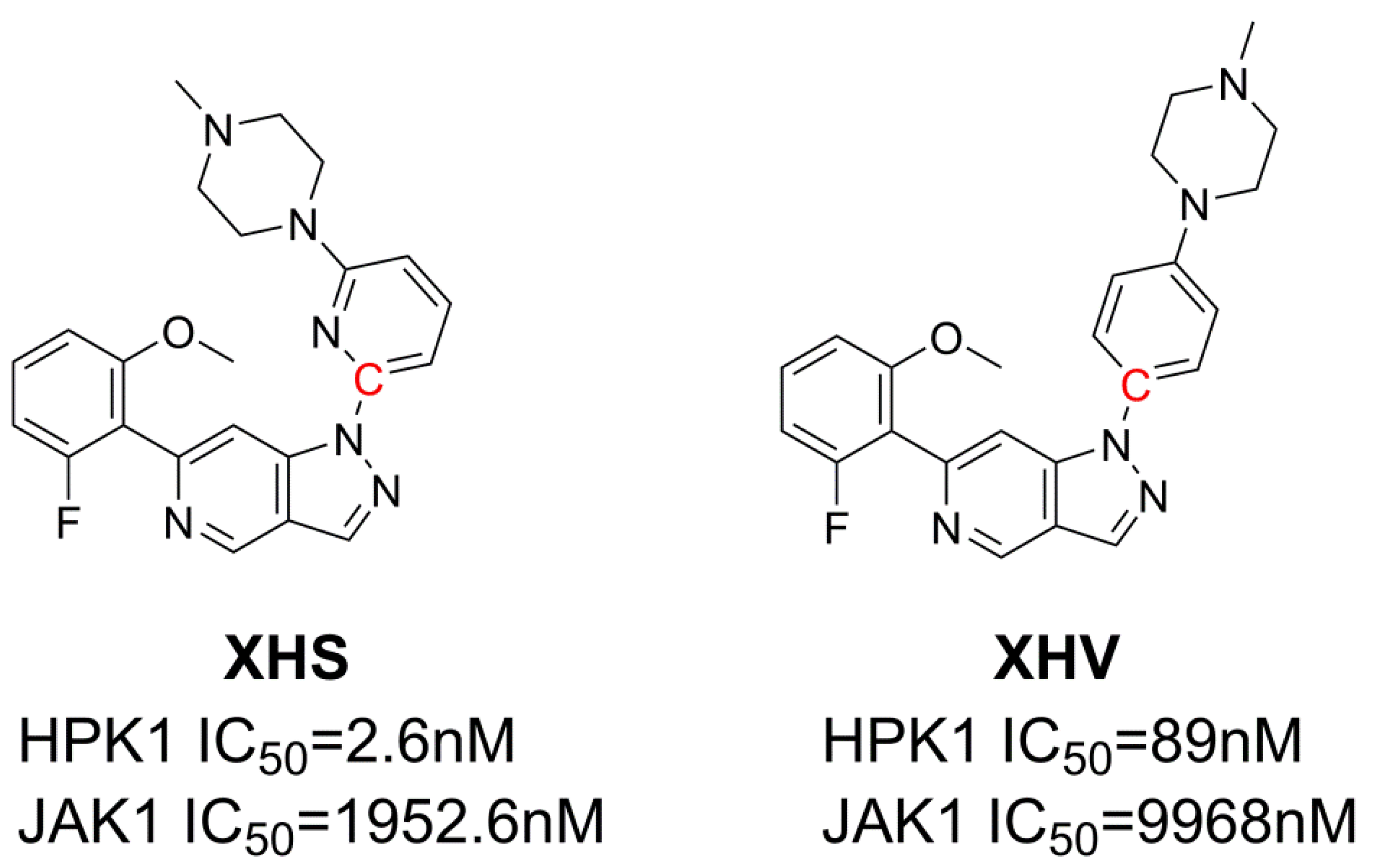
| IC50 | 2.6 nM | 1952.6 nM | 89 nM | 9968 nM |
| ΔGexp a | −11.71 | −7.79 | −9.61 | −6.82 |
| Acceptor | Donor | Occupancy (%) | Distance (Å) | Angle (°) |
|---|---|---|---|---|
| HPK1-XHS | ||||
| XHS@N5 | Cys94@HN | 90.80 | 3.17 | 160.46 |
| JAK1-XHS | ||||
| XHS@N5 | Leu959@HN | 86.64 | 3.20 | 150.60 |
| HPK1-XHV | ||||
| XHV@N4 | Cys94@HN | 82.36 | 3.14 | 152.55 |
| JAK1-XHV | ||||
| XHV@N4 | Leu959@HN | 65.76 | 3.25 | 153.23 |
| System | Positively Charged Residue | Negatively Charged Residue | Occupancy (%) | Diatance (Å) |
|---|---|---|---|---|
| HPK1-XHS | XHS@N1 | Asp101@OD | 62.03 | 2.86 |
| JAK1-XHS | XHS@N1 | Glu966@OE | 7.41 | 2.86 |
| Energy | HPK1-XHS | JAK1-XHS | HPK1-XHV | JAK1-XHV |
|---|---|---|---|---|
| PMF/kcal∙mol−1 | 22.43 ± 0.07 | 15.09 ± 0.86 | 17.68 ± 0.16 | 10.02 ± 0.29 |
| IC50/nM | 2.6 | 1952.6 | 89 | 9968 |
| ΔGexp/kcal∙mol−1 | −11.71 | −7.79 | −9.61 | −6.82 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, H.; Tang, C.; Pan, Y.; Yao, X. Theoretical Studies on Selectivity of HPK1/JAK1 Inhibitors by Molecular Dynamics Simulations and Free Energy Calculations. Int. J. Mol. Sci. 2023, 24, 2649. https://doi.org/10.3390/ijms24032649
Ge H, Tang C, Pan Y, Yao X. Theoretical Studies on Selectivity of HPK1/JAK1 Inhibitors by Molecular Dynamics Simulations and Free Energy Calculations. International Journal of Molecular Sciences. 2023; 24(3):2649. https://doi.org/10.3390/ijms24032649
Chicago/Turabian StyleGe, Huizhen, Chunchao Tang, Yiting Pan, and Xiaojun Yao. 2023. "Theoretical Studies on Selectivity of HPK1/JAK1 Inhibitors by Molecular Dynamics Simulations and Free Energy Calculations" International Journal of Molecular Sciences 24, no. 3: 2649. https://doi.org/10.3390/ijms24032649
APA StyleGe, H., Tang, C., Pan, Y., & Yao, X. (2023). Theoretical Studies on Selectivity of HPK1/JAK1 Inhibitors by Molecular Dynamics Simulations and Free Energy Calculations. International Journal of Molecular Sciences, 24(3), 2649. https://doi.org/10.3390/ijms24032649




