Transcriptional Insights of Oxidative Stress and Extracellular Traps in Lung Tissues of Fatal COVID-19 Cases
Abstract
1. Introduction
2. Results
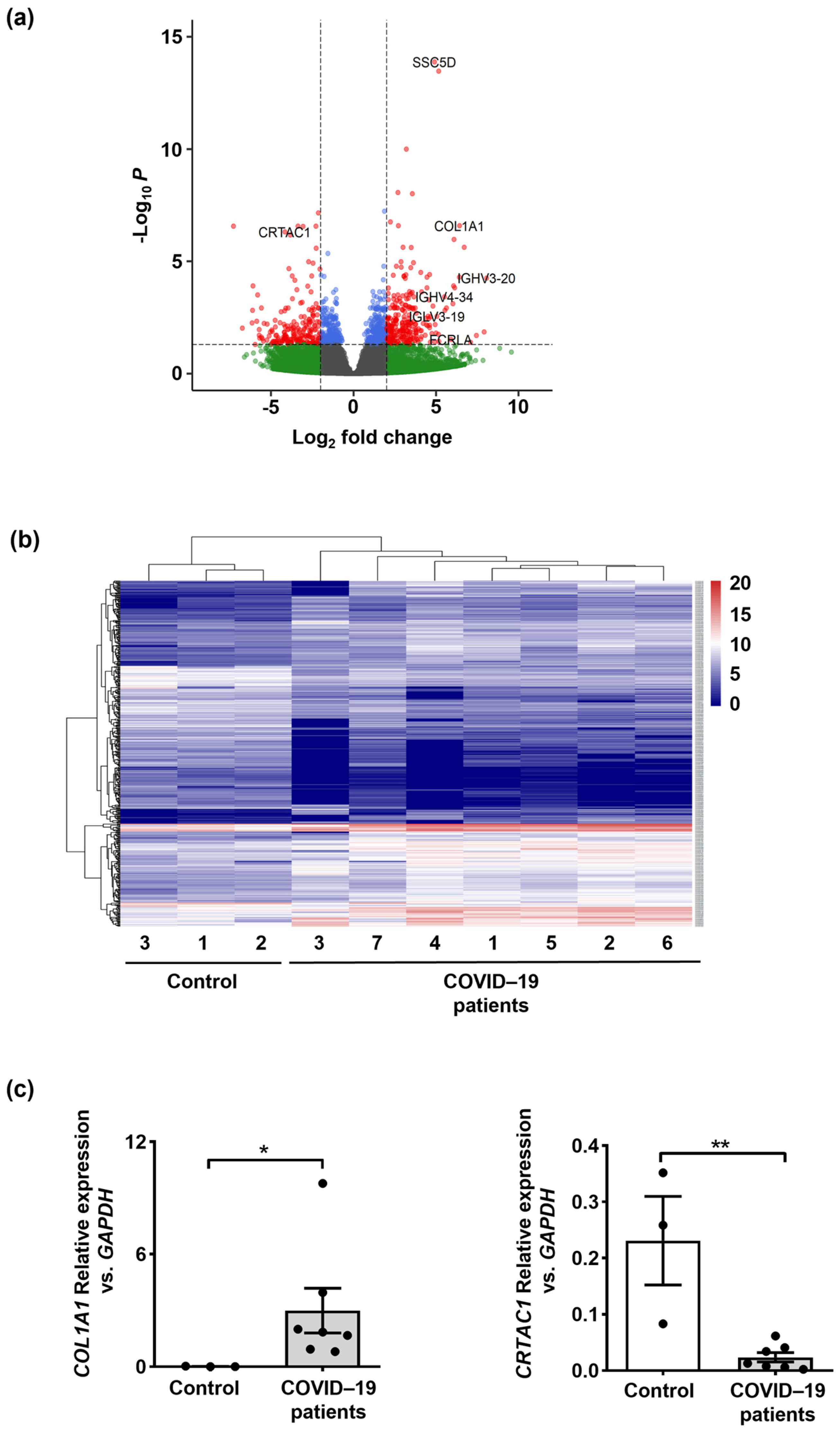
2.1. Lung Transcriptomes Reveal Upregulation of Inflammatory Immune Response Genes in Fatal Cases of COVID-19 Patients
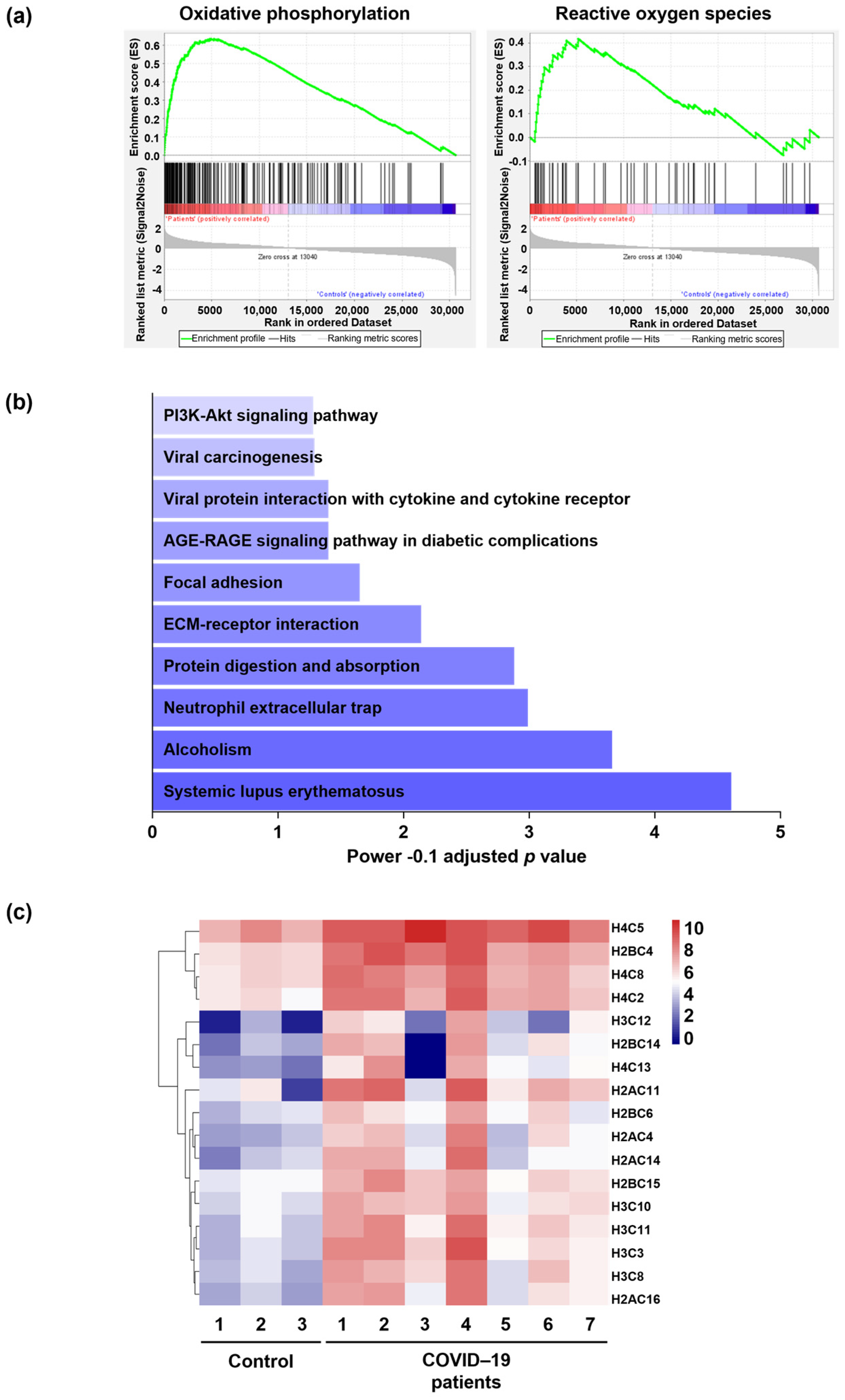
2.2. Molecular Signaling Pathway Enrichment Analysis Proposes Possible Inflammation and Neutrophil Extracellular Trap (NET) Formation in the Lungs of COVID-19 Patients
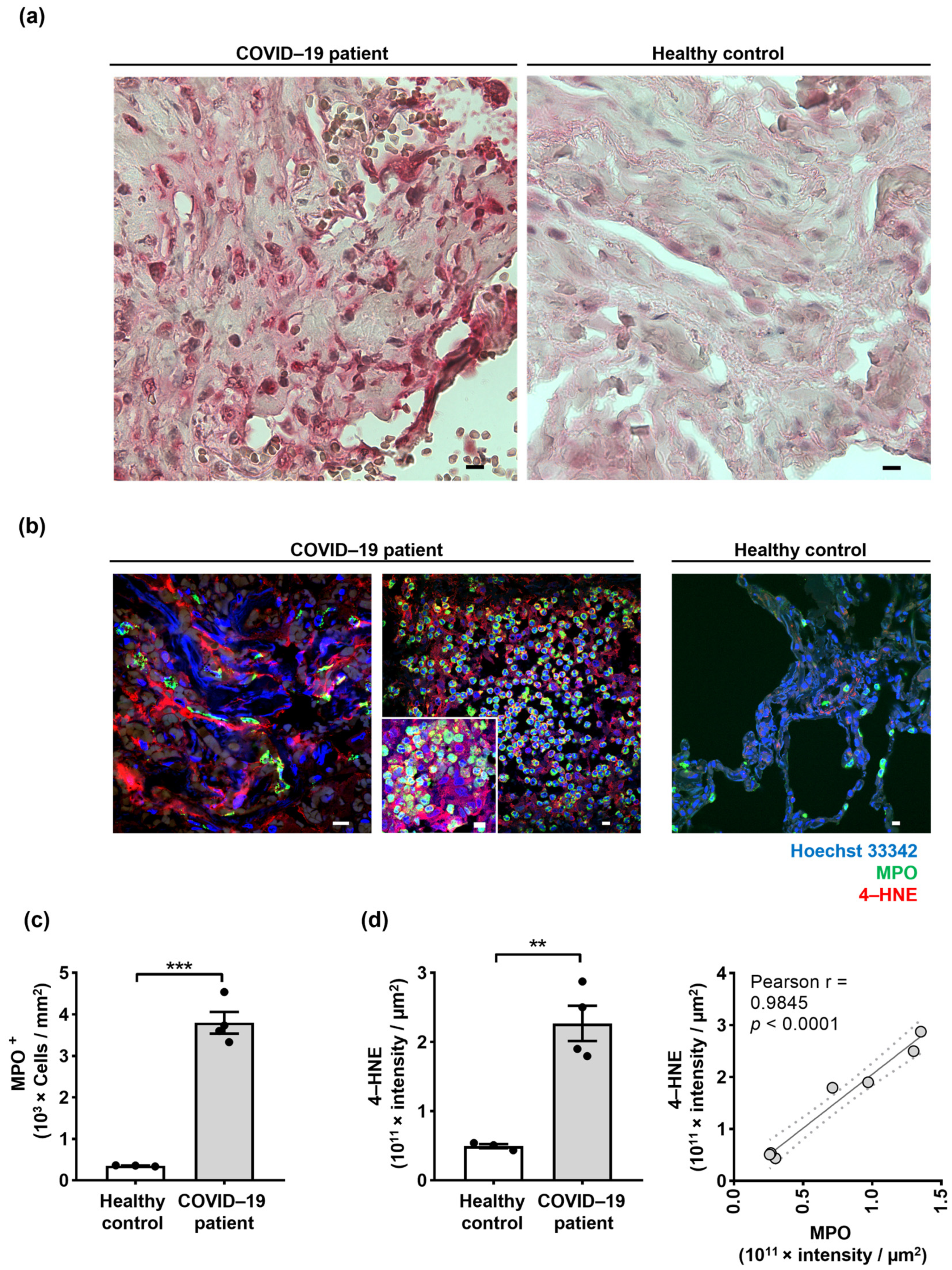
2.3. Neutrophil Extracellular Traps (NETs) in the Lungs of COVID-19 Patients Are Associated with Oxidized dsDNA
2.4. Lipid Oxidation Occurs in the Lungs of COVID-19 Patients, and 4-Hydroxynonenal (4-HNE) Co-Localizes with NETs
3. Discussion
4. Materials and Methods
4.1. RNA Isolation and RNA Sequencing
4.2. RNA-Sequencing Analysis
4.3. Complementary DNA Synthesis and Quantitative PCR
4.3.1. Immunohistochemistry
4.3.2. Immunofluorescence and Confocal Microscopy
4.3.3. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lynch, S.M.; Guo, G.; Gibson, D.S.; Bjourson, A.J.; Rai, T.S. Role of Senescence and Aging in SARS-CoV-2 Infection and COVID-19 Disease. Cells 2021, 10, 3367. [Google Scholar] [CrossRef] [PubMed]
- Rocha, B.C.; Marques, P.E.; Leoratti, F.M.S.; Junqueira, C.; Pereira, D.B.; Antonelli, L.; Menezes, G.B.; Golenbock, D.T.; Gazzinelli, R.T. Type I Interferon Transcriptional Signature in Neutrophils and Low-Density Granulocytes Are Associated with Tissue Damage in Malaria. Cell Rep. 2015, 13, 2829–2841. [Google Scholar] [CrossRef] [PubMed]
- McKenna, E.; Wubben, R.; Isaza-Correa, J.M.; Melo, A.M.; Mhaonaigh, A.U.; Conlon, N.; O’Donnell, J.S.; Ní Cheallaigh, C.; Hurley, T.; Stevenson, N.J. Neutrophils in COVID-19: Not Innocent Bystanders. Front. Immunol. 2022, 13, 2548. [Google Scholar] [CrossRef]
- Yousefi, S.; Stojkov, D.; Germic, N.; Simon, D.; Wang, X.; Benarafa, C.; Simon, H.U. Untangling “NETosis” from NETs. Eur. J. Immunol. 2019, 49, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Anders, H.-J.; Bilyy, R.; Bowlin, G.L.; Daniel, C.; De Lorenzo, R.; Egeblad, M.; Henneck, T.; Hidalgo, A.; Hoffmann, M. Patients with COVID-19: In the dark-NETs of neutrophils. Cell Death Differ. 2021, 28, 3125–3139. [Google Scholar] [CrossRef] [PubMed]
- Leppkes, M.; Knopf, J.; Naschberger, E.; Lindemann, A.; Singh, J.; Herrmann, I.; Stürzl, M.; Staats, L.; Mahajan, A.; Schauer, C. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine 2020, 58, 102925. [Google Scholar] [CrossRef] [PubMed]
- Middleton, E.A.; He, X.-Y.; Denorme, F.; Campbell, R.A.; Ng, D.; Salvatore, S.P.; Mostyka, M.; Baxter-Stoltzfus, A.; Borczuk, A.C.; Loda, M. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood 2020, 136, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Skendros, P.; Mitsios, A.; Chrysanthopoulou, A.; Mastellos, D.C.; Metallidis, S.; Rafailidis, P.; Ntinopoulou, M.; Sertaridou, E.; Tsironidou, V.; Tsigalou, C. Complement and tissue factor–enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Invest. 2020, 130, 6151–6157. [Google Scholar] [CrossRef]
- Uddin, M.; Watz, H.; Malmgren, A.; Pedersen, F. NETopathic inflammation in chronic obstructive pulmonary disease and severe asthma. Front. Immunol. 2019, 10, 47. [Google Scholar] [CrossRef]
- Dworski, R.; Simon, H.-U.; Hoskins, A.; Yousefi, S. Eosinophil and neutrophil extracellular DNA traps in human allergic asthmatic airways. J. Allergy Clin. Immunol. 2011, 127, 1260–1266. [Google Scholar] [CrossRef]
- Twaddell, S.H.; Baines, K.J.; Grainge, C.; Gibson, P.G. The emerging role of neutrophil extracellular traps in respiratory disease. Chest 2019, 156, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.D.; Hardisty, G.; Regan, K.H.; Smith, M.; Robb, C.T.; Duffin, R.; Mackellar, A.; Felton, J.M.; Paemka, L.; McCullagh, B.N.; et al. Delayed neutrophil apoptosis enhances NET formation in cystic fibrosis. Thorax 2018, 73, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5, e138999. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.; Havervall, S.; Rosell, A.; Aguilera, K.; Parv, K.; Von Meijenfeldt, F.A.; Lisman, T.; Mackman, N.; Thålin, C.; Phillipson, M. Circulating markers of neutrophil extracellular traps are of prognostic value in patients with COVID-19. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Burn, G.L.; Foti, A.; Marsman, G.; Patel, D.F.; Zychlinsky, A. The neutrophil. Immunity 2021, 54, 1377–1391. [Google Scholar] [CrossRef]
- Szturmowicz, M.; Demkow, U. Neutrophil extracellular traps (NETs) in severe SARS-CoV-2 lung disease. Int. J. Mol. Sci. 2021, 22, 8854. [Google Scholar] [CrossRef]
- Ouwendijk, W.J.; Raadsen, M.P.; Van Kampen, J.J.; Verdijk, R.M.; Von Der Thusen, J.H.; Guo, L.; Hoek, R.A.; Van Den Akker, J.P.; Endeman, H.; Langerak, T. High levels of neutrophil extracellular traps persist in the lower respiratory tract of critically ill patients with coronavirus disease 2019. J. Infect. Dis. 2021, 223, 1512–1521. [Google Scholar] [CrossRef]
- Kinnare, N.; Hook, J.S.; Patel, P.A.; Monson, N.L.; Moreland, J.G. Neutrophil extracellular trap formation potential correlates with lung disease severity in COVID-19 patients. Inflammation 2022, 45, 800–811. [Google Scholar] [CrossRef]
- Ombrello, M.J.; Schulert, G.S. COVID-19 and cytokine storm syndrome: Are there lessons from macrophage activation syndrome? Transl. Res. 2021, 232, 1–12. [Google Scholar] [CrossRef]
- Al-Kuraishy, H.M.; Al-Gareeb, A.I.; Al-Hussaniy, H.A.; Al-Harcan, N.A.H.; Alexiou, A.; Batiha, G.E.-S. Neutrophil Extracellular Traps (NETs) and COVID-19: A new frontiers for therapeutic modality. Int. Immunopharmacol. 2022, 104, 108516. [Google Scholar] [CrossRef]
- Block, H.; Zarbock, A. A fragile balance: Does neutrophil extracellular trap formation drive pulmonary disease progression? Cells 2021, 10, 1932. [Google Scholar] [CrossRef]
- Zuo, Y.; Zuo, M.; Yalavarthi, S.; Gockman, K.; Madison, J.A.; Shi, H.; Woodard, W.; Lezak, S.P.; Lugogo, N.L.; Knight, J.S. Neutrophil extracellular traps and thrombosis in COVID-19. J. Thromb. Thrombolysis. 2021, 51, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A.; Borczuk, A.; Cools-Lartigue, J.; Crawford, J.M.; Daßler-Plenker, J.; Guerci, P.; Huynh, C.; Knight, J.S. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020, 217, e20200652. [Google Scholar] [CrossRef] [PubMed]
- Arcanjo, A.; Logullo, J.; Menezes, C.C.B.; de Souza Carvalho Giangiarulo, T.C.; Dos Reis, M.C.; de Castro, G.M.M.; da Silva Fontes, Y.; Todeschini, A.R.; Freire-de-Lima, L.; Decoté-Ricardo, D. The emerging role of neutrophil extracellular traps in severe acute respiratory syndrome coronavirus 2 (COVID-19). Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Teluguakula, N. Neutrophils set extracellular traps to injure lungs in coronavirus disease 2019. J. Infect. Dis. 2021, 223, 1503–1505. [Google Scholar] [CrossRef]
- Schönrich, G.; Raftery, M.J.; Samstag, Y. Devilishly radical NETwork in COVID-19: Oxidative stress, neutrophil extracellular traps (NETs), and T cell suppression. Adv. Biol. Regul. 2020, 77, 100741. [Google Scholar] [CrossRef]
- Vajdovich, P. Free radicals and antioxidants in inflammatory processes and ischemia-reperfusion injury. Vet. Clin. North Am. Small Anim. Pract. 2008, 38, 31–123. [Google Scholar] [CrossRef]
- Mittal, M.; Siddiqui, M.R.; Tran, K.; Reddy, S.P.; Malik, A.B. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox. Signal. 2014, 20, 1126–1167. [Google Scholar] [CrossRef]
- Kuwano, K.; Nakashima, N.; Inoshima, I.; Hagimoto, N.; Fujita, M.; Yoshimi, M.; Maeyama, T.; Hamada, N.; Watanabe, K.; Hara, N. Oxidative stress in lung epithelial cells from patients with idiopathic interstitial pneumonias. Eur. Respir. J. 2003, 21, 232–240. [Google Scholar] [CrossRef]
- Valavanidis, A.; Vlachogianni, T.; Fiotakis, C. 8-hydroxy-2′-deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2009, 27, 120–139. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, Y.; Li, L.; Liu, S.; Wang, C.; Yuan, Y.; Yang, G.; Chen, Y.; Cheng, J.; Lu, Y. Mitochondrial ROS promote mitochondrial dysfunction and inflammation in ischemic acute kidney injury by disrupting TFAM-mediated mtDNA maintenance. Theranostics 2021, 11, 1845. [Google Scholar] [CrossRef] [PubMed]
- Amini, P.; Stojkov, D.; Felser, A.; Jackson, C.B.; Courage, C.; Schaller, A.; Gelman, L.; Soriano, M.E.; Nuoffer, J.M.; Scorrano, L.; et al. Neutrophil extracellular trap formation requires OPA1-dependent glycolytic ATP production. Nat. Commun. 2018, 9, 2958. [Google Scholar] [CrossRef] [PubMed]
- Lood, C.; Blanco, L.P.; Purmalek, M.M.; Carmona-Rivera, C.; De Ravin, S.S.; Smith, C.K.; Malech, H.L.; Ledbetter, J.A.; Elkon, K.B.; Kaplan, M.J. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 2016, 22, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Piantadosi, C.A. Mitochondrial DNA, oxidants, and innate immunity. Free Radic. Biol. Med. 2020, 152, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Laforge, M.; Elbim, C.; Frère, C.; Hémadi, M.; Massaad, C.; Nuss, P.; Benoliel, J.-J.; Becker, C. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat. Rev. Immunol. 2020, 20, 515–516. [Google Scholar] [CrossRef]
- Barciszewska, A.-M. Elucidating of oxidative distress in COVID-19 and methods of its prevention. Chem. Biol. Interact. 2021, 344, 109501. [Google Scholar] [CrossRef]
- Bakadia, B.M.; Boni, B.O.O.; Ahmed, A.A.Q.; Yang, G. The impact of oxidative stress damage induced by the environmental stressors on COVID-19. Life Sci. 2021, 264, 118653. [Google Scholar] [CrossRef]
- Suhail, S.; Zajac, J.; Fossum, C.; Lowater, H.; McCracken, C.; Severson, N.; Laatsch, B.; Narkiewicz-Jodko, A.; Johnson, B.; Liebau, J. Role of oxidative stress on SARS-CoV (SARS) and SARS-CoV-2 (COVID-19) infection: A review. Protein J. 2020, 39, 644–656. [Google Scholar] [CrossRef]
- Qin, M.; Cao, Z.; Wen, J.; Yu, Q.; Liu, C.; Wang, F.; Zhang, J.; Yang, F.; Li, Y.; Fishbein, G. An antioxidant enzyme therapeutic for COVID-19. Adv. Mater. 2020, 32, 2004901. [Google Scholar] [CrossRef]
- Muhoberac, B.B. What can cellular redox, iron, and reactive oxygen species suggest about the mechanisms and potential therapy of COVID-19? Front. Cell. Infect. Microbiol. 2020, 10, 569709. [Google Scholar] [CrossRef]
- Miripour, Z.S.; Sarrami-Forooshani, R.; Sanati, H.; Makarem, J.; Taheri, M.S.; Shojaeian, F.; Eskafi, A.H.; Abbasvandi, F.; Namdar, N.; Ghafari, H. Real-time diagnosis of reactive oxygen species (ROS) in fresh sputum by electrochemical tracing; correlation between COVID-19 and viral-induced ROS in lung/respiratory epithelium during this pandemic. Biosens. Bioelectron. 2020, 165, 112435. [Google Scholar] [CrossRef] [PubMed]
- Ramana, K.V.; Srivastava, S.; Singhal, S.S. Lipid peroxidation products in human health and disease 2019. Oxid. Med. Cell. Longev. 2019, 2019, 7147235. [Google Scholar] [CrossRef] [PubMed]
- Zarkovic, N. 4-Hydroxynonenal as a bioactive marker of pathophysiological processes. Mol. Aspects Med. 2003, 24, 281–291. [Google Scholar] [CrossRef]
- Zarkovic, N.; Orehovec, B.; Milkovic, L.; Barsic, B.; Tatzber, F.; Wonisch, W.; Tarle, M.; Kmet, M.; Mataic, A.; Jakovcevic, A.; et al. Preliminary Findings on the Association of the Lipid Peroxidation Product 4-Hydroxynonenal with the Lethal Outcome of Aggressive COVID-19. Antioxidants 2021, 10, 1341. [Google Scholar] [CrossRef]
- Gimpel, A.-K.; Maccataio, A.; Unterweger, H.; Sokolova, M.V.; Schett, G.; Steffen, U. IgA Complexes Induce Neutrophil Extracellular Trap Formation More Potently Than IgG Complexes. Front. Immunol. 2022, 12, 761816. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.G.; Zilhão, R.; Thorsteinsdóttir, S.; Carlos, A.R. Linking oxidative stress and DNA damage to changes in the expression of ECM components. Front. Genet. 2021, 12, 1279. [Google Scholar] [CrossRef] [PubMed]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst. 2021, 12, 23–40.e27. [Google Scholar] [CrossRef] [PubMed]
- Katerji, M.; Filippova, M.; Duerksen-Hughes, P. Approaches and Methods to Measure Oxidative Stress in Clinical Samples: Research Applications in the Cancer Field. Oxid. Med. Cell. Longev. 2019, 2019, 1279250. [Google Scholar] [CrossRef]
- Murphy, M.P.; Bayir, H.; Belousov, V.; Chang, C.J.; Davies, K.J.A.; Davies, M.J.; Dick, T.P.; Finkel, T.; Forman, H.J.; Janssen-Heininger, Y.; et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 2022, 4, 651–662. [Google Scholar] [CrossRef]
- Su, L.J.; Zhang, J.H.; Gomez, H.; Murugan, R.; Hong, X.; Xu, D.; Jiang, F.; Peng, Z.Y. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxid. Med. Cell. Longev. 2019, 2019, 5080843. [Google Scholar] [CrossRef]
- Liou, G.Y.; Storz, P. Detecting reactive oxygen species by immunohistochemistry. Methods Mol. Biol. 2015, 1292, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Milkovic, L.; Cipak Gasparovic, A.; Zarkovic, N. Overview on major lipid peroxidation bioactive factor 4-hydroxynonenal as pluripotent growth-regulating factor. Free Radic. Res. 2015, 49, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Kosanovic, T.; Sagic, D.; Djukic, V.; Pljesa-Ercegovac, M.; Savic-Radojevic, A.; Bukumiric, Z.; Lalosevic, M.; Djordjevic, M.; Coric, V.; Simic, T. Time Course of Redox Biomarkers in COVID-19 Pneumonia: Relation with Inflammatory, Multiorgan Impairment Biomarkers and CT Findings. Antioxidants 2021, 10, 1126. [Google Scholar] [CrossRef] [PubMed]
- Tantry, U.S.; Bliden, K.P.; Cho, A.; Walia, N.; Dahlen, J.R.; Ens, G.; Traianova, M.; Jerjian, C.; Usman, A.; Gurbel, P.A. First Experience Addressing the Prognostic Utility of Novel Urinary Biomarkers in Patients With COVID-19. Open Forum. Infect. Dis. 2021, 8, ofab274. [Google Scholar] [CrossRef]
- Nadhan, R.; Patra, D.; Krishnan, N.; Rajan, A.; Gopala, S.; Ravi, D.; Srinivas, P. Perspectives on mechanistic implications of ROS inducers for targeting viral infections. Eur. J. Pharmacol. 2021, 890, 173621. [Google Scholar] [CrossRef]
- Wu, M.; Chen, Y.; Xia, H.; Wang, C.; Tan, C.Y.; Cai, X.; Liu, Y.; Ji, F.; Xiong, P.; Liu, R. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc. Natl. Acad. Sci. USA 2020, 117, 28336–28343. [Google Scholar] [CrossRef]
- Martín-Fernández, M.; Aller, R.; Heredia-Rodríguez, M.; Gómez-Sánchez, E.; Martínez-Paz, P.; Gonzalo-Benito, H.; Sánchez-de Prada, L.; Gorgojo, Ó.; Carnicero-Frutos, I.; Tamayo, E. Lipid peroxidation as a hallmark of severity in COVID-19 patients. Redox Biol. 2021, 48, 102181. [Google Scholar] [CrossRef]
- LaSalle, T.J.; Gonye, A.L.; Freeman, S.S.; Kaplonek, P.; Gushterova, I.; Kays, K.R.; Manakongtreecheep, K.; Tantivit, J.; Rojas-Lopez, M.; Russo, B.C. Longitudinal characterization of circulating neutrophils uncovers distinct phenotypes associated with disease severity in hospitalized COVID-19 patients. Cell Rep. Med. 2021, 3, 100779. [Google Scholar] [CrossRef]
- McElvaney, O.J.; McEvoy, N.L.; McElvaney, O.F.; Carroll, T.P.; Murphy, M.P.; Dunlea, D.M.; Ní Choileáin, O.; Clarke, J.; O’Connor, E.; Hogan, G. Characterization of the inflammatory response to severe COVID-19 illness. Am. J. Respir. Crit. Care Med. 2020, 202, 812–821. [Google Scholar] [CrossRef]
- Panasenko, O.M.; Ivanov, V.A.; Mikhalchik, E.V.; Gorudko, I.V.; Grigorieva, D.V.; Basyreva, L.Y.; Shmeleva, E.V.; Gusev, S.A.; Kostevich, V.A.; Gorbunov, N.P. Methylglyoxal-modified human serum albumin binds to leukocyte myeloperoxidase and inhibits its enzymatic activity. Antioxidants 2022, 11, 2263. [Google Scholar] [CrossRef]
- Goud, P.T.; Bai, D.; Abu-Soud, H.M. A multiple-hit hypothesis involving reactive oxygen species and myeloperoxidase explains clinical deterioration and fatality in COVID-19. Int. J. Biol. Sci. 2021, 17, 62. [Google Scholar] [CrossRef] [PubMed]
- Camp, O.G.; Bai, D.; Gonullu, D.C.; Nayak, N.; Abu-Soud, H.M. Melatonin interferes with COVID-19 at several distinct ROS-related steps. J. Inorg. Biochem. 2021, 223, 111546. [Google Scholar] [CrossRef] [PubMed]
- Dowey, R.; Iqbal, A.; Heller, S.R.; Sabroe, I.; Prince, L.R. A bittersweet response to infection in diabetes; targeting neutrophils to modify inflammation and improve host immunity. Front. Immunol. 2021, 12, 678771. [Google Scholar] [CrossRef] [PubMed]
- Bardaweel, S.K.; Gul, M.; Alzweiri, M.; Ishaqat, A.; ALSalamat, H.A.; Bashatwah, R.M. Reactive oxygen species: The dual role in physiological and pathological conditions of the human body. Eurasian J. Med. 2018, 50, 193. [Google Scholar] [CrossRef]
- Wingler, K.; Hermans, J.; Schiffers, P.; Moens, A.; Paul, M.; Schmidt, H. NOX1, 2, 4, 5: Counting out oxidative stress. Br. J. Pharmacol. 2011, 164, 866–883. [Google Scholar] [CrossRef]
- Alexeyev, M.F. Is there more to aging than mitochondrial DNA and reactive oxygen species? FEBS J. 2009, 276, 5768–5787. [Google Scholar] [CrossRef]
- Sharif-Askari, N.S.; Sharif-Askari, F.S.; Mdkhana, B.; Alsayed, H.A.H.; Alsafar, H.; Alrais, Z.F.; Hamid, Q.; Halwani, R. Upregulation of oxidative stress gene markers during SARS-COV-2 viral infection. Free Radic. Biol. Med. 2021, 172, 688–698. [Google Scholar] [CrossRef]
- Yu, G.; Liang, Y.; Huang, Z.; Jones, D.W.; Pritchard, K.A.; Zhang, H. Inhibition of myeloperoxidase oxidant production by N-acetyl lysyltyrosylcysteine amide reduces brain damage in a murine model of stroke. J. Neuroinflamm. 2016, 13, 1–13. [Google Scholar]
- Matsubara, T.; Soh, J.; Morita, M.; Uwabo, T.; Tomida, S.; Fujiwara, T.; Kanazawa, S.; Toyooka, S.; Hirasawa, A. DV200 index for assessing RNA integrity in next-generation sequencing. Biomed. Res. Int. 2020, 2020, 9349132. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. R Package Version 3.10. 2019. Available online: https://bioconductor.org/packages/devel/bioc/vignettes/EnhancedVolcano/inst/doc/EnhancedVolcano.html (accessed on 24 December 2022).
- Kolde, R. Pheatmap: Pretty heatmaps. R package version 1.0. 12. 2019. Available online: https://rdrr.io/cran/pheatmap/ (accessed on 24 December 2022).
- Wickham, H. Programming with ggplot2. In Ggplot2; Springer: Berlin/Heidelberg, Germany, 2016; pp. 241–253. [Google Scholar]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.; Clarke, D.; Evangelista, J.; Jenkins, S.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Greuter, T.; Straumann, A.; Fernandez-Marrero, Y.; Germic, N.; Hosseini, A.; Yousefi, S.; Simon, D.; Collins, M.H.; Bussmann, C.; Chehade, M.; et al. Characterization of eosinophilic esophagitis variants by clinical, histological, and molecular analyses: A cross-sectional multi-center study. Allergy 2022, 77, 2520–2533. [Google Scholar] [CrossRef] [PubMed]

| Primer | Sequence (5′–3′) | Amplicon Size (bp) | T (°C) |
|---|---|---|---|
| GAPDH F | CAA CAG CCT CAA GAT CAT CAG CAA | 103 | 60 |
| GAPDH R | CAT GAG TCC TTC CAC GAT ACC | ||
| CRTAC1 F | ATC TTC TTC AAC AAC ATT GCC TAC | 80 | 60 |
| CRTAC1 R | GGG TCT CCG TGC TCT CTA | ||
| COL1A1 F | GTT CGG AGG AGA GTC AGG AAG G | 128 | 60 |
| COL1A1 R | CAG CAA CAC AGT TAC ACA AGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hosseini, A.; Stojkov, D.; Fettrelet, T.; Bilyy, R.; Yousefi, S.; Simon, H.-U. Transcriptional Insights of Oxidative Stress and Extracellular Traps in Lung Tissues of Fatal COVID-19 Cases. Int. J. Mol. Sci. 2023, 24, 2646. https://doi.org/10.3390/ijms24032646
Hosseini A, Stojkov D, Fettrelet T, Bilyy R, Yousefi S, Simon H-U. Transcriptional Insights of Oxidative Stress and Extracellular Traps in Lung Tissues of Fatal COVID-19 Cases. International Journal of Molecular Sciences. 2023; 24(3):2646. https://doi.org/10.3390/ijms24032646
Chicago/Turabian StyleHosseini, Aref, Darko Stojkov, Timothée Fettrelet, Rostyslav Bilyy, Shida Yousefi, and Hans-Uwe Simon. 2023. "Transcriptional Insights of Oxidative Stress and Extracellular Traps in Lung Tissues of Fatal COVID-19 Cases" International Journal of Molecular Sciences 24, no. 3: 2646. https://doi.org/10.3390/ijms24032646
APA StyleHosseini, A., Stojkov, D., Fettrelet, T., Bilyy, R., Yousefi, S., & Simon, H.-U. (2023). Transcriptional Insights of Oxidative Stress and Extracellular Traps in Lung Tissues of Fatal COVID-19 Cases. International Journal of Molecular Sciences, 24(3), 2646. https://doi.org/10.3390/ijms24032646







