Genome-Wide Identification and Posttranscriptional Regulation Analyses Elucidate Roles of Key Argonautes and Their miRNA Triggers in Regulating Complex Yield Traits in Rapeseed
Abstract
1. Introduction
2. Results
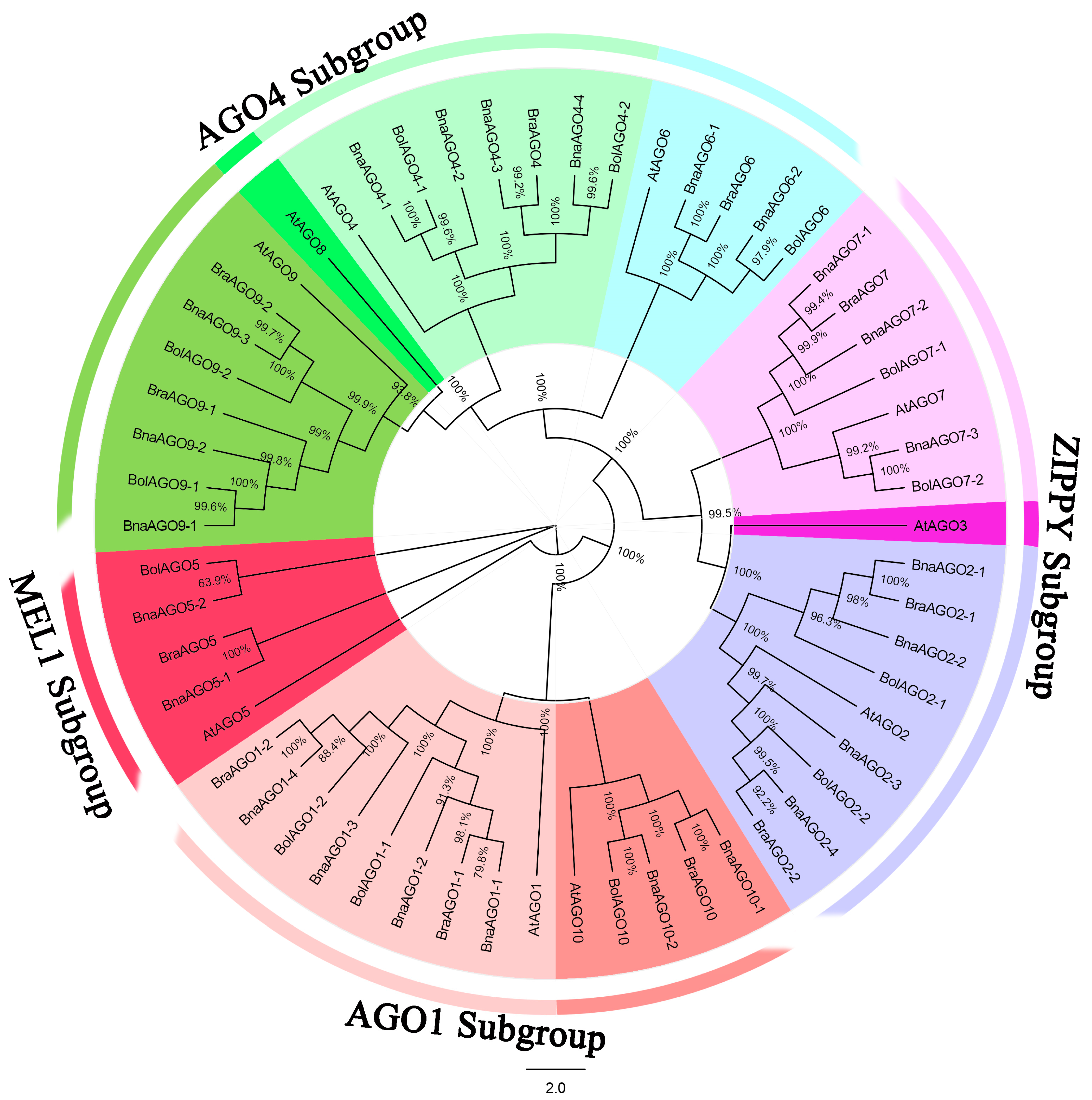
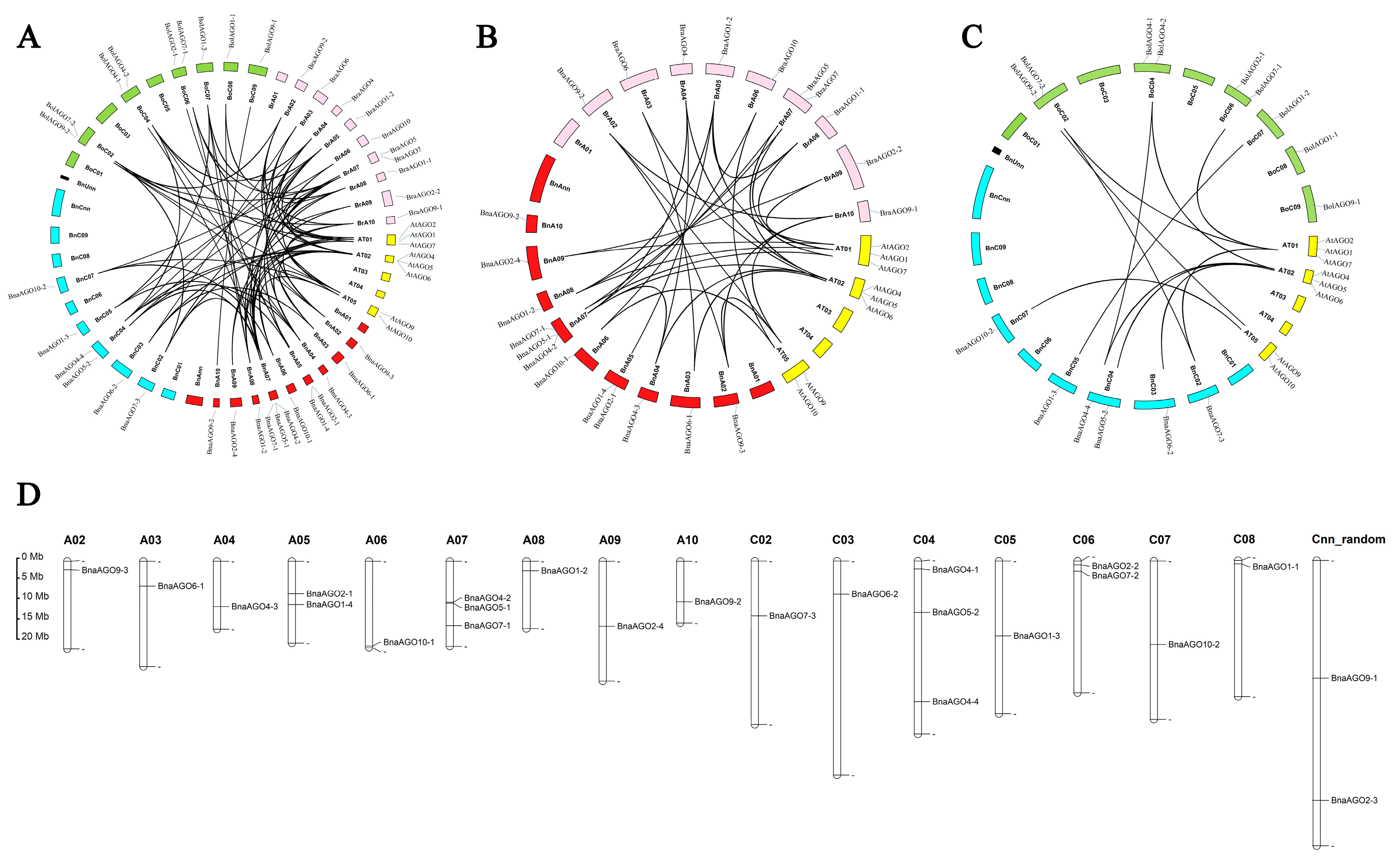
2.1. Genome-Wide Prediction, Phylogenetic Analysis, and Nomenclature of AGO Genes
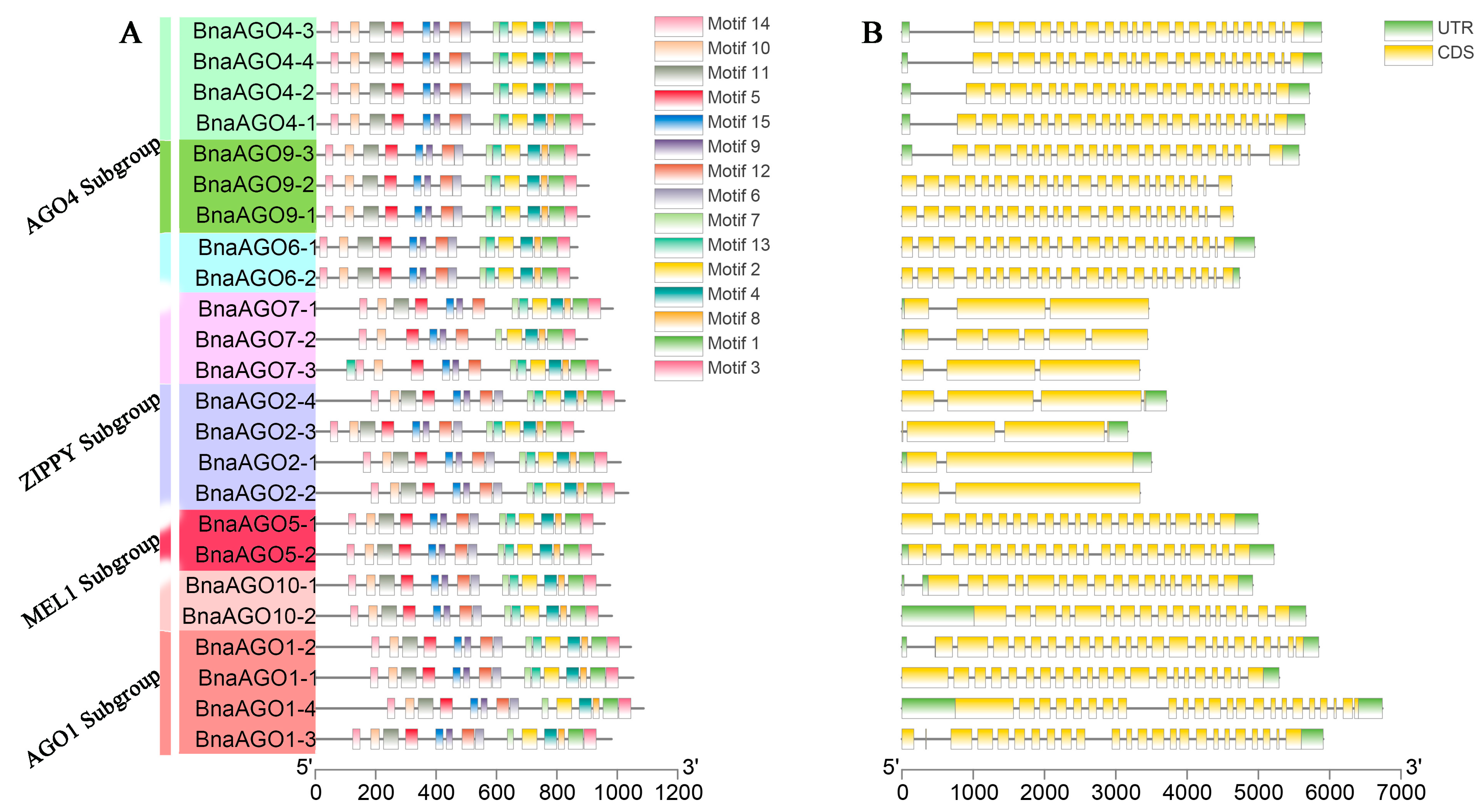
2.2. Protein Profiles, Gene Structure and Conserved Motif Analysis of the 24 BnaAGOs
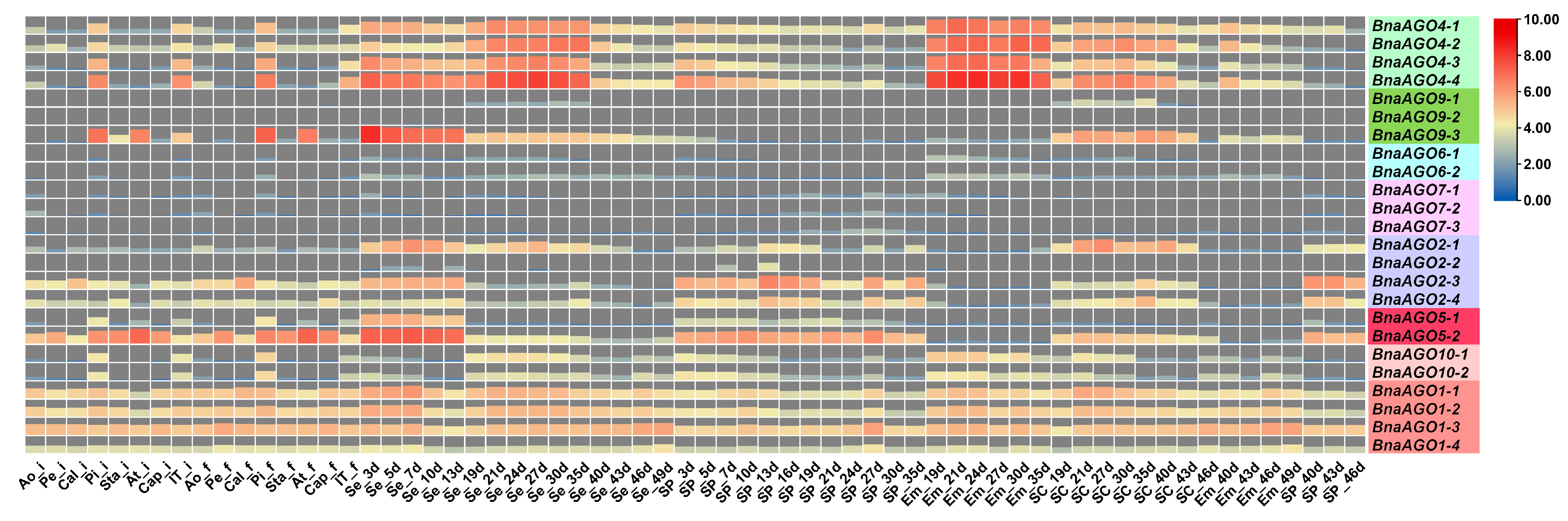
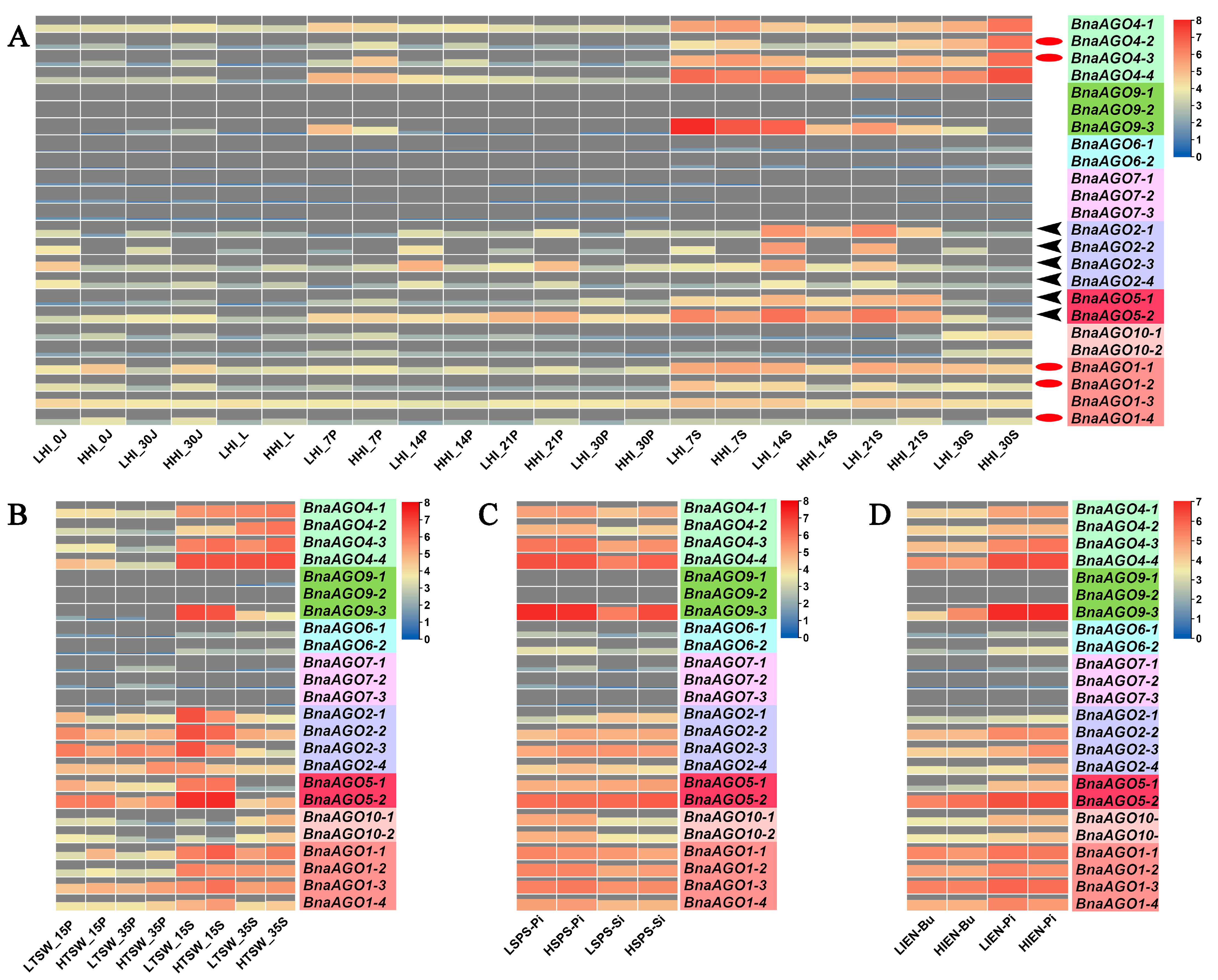
2.3. Differential Expression of 24 BnaAGOs among Differently Yielding B. napus Cultivars
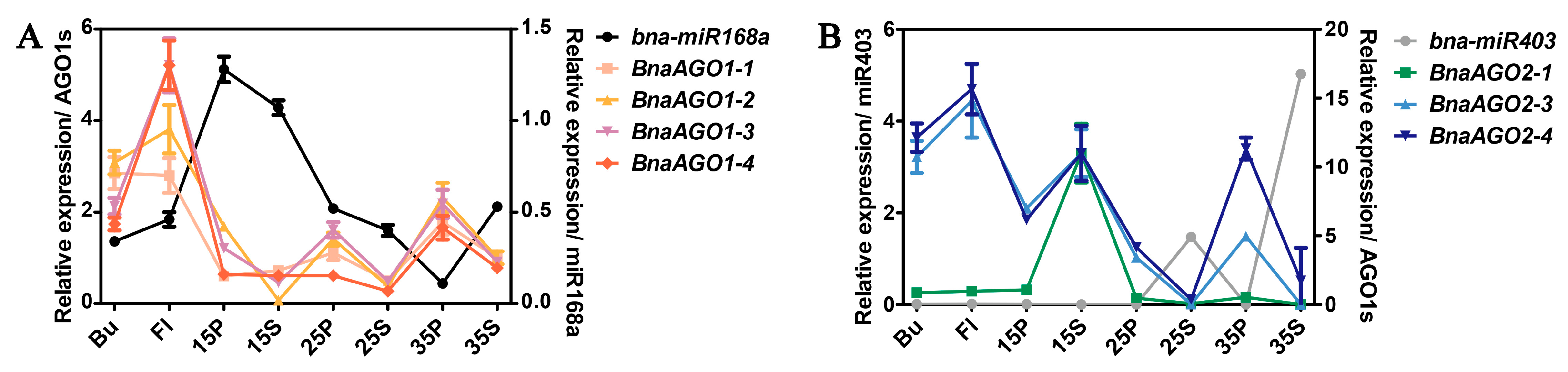
2.4. MiRNA-Mediated Posttranscriptional Regulation of the BnaAGOs
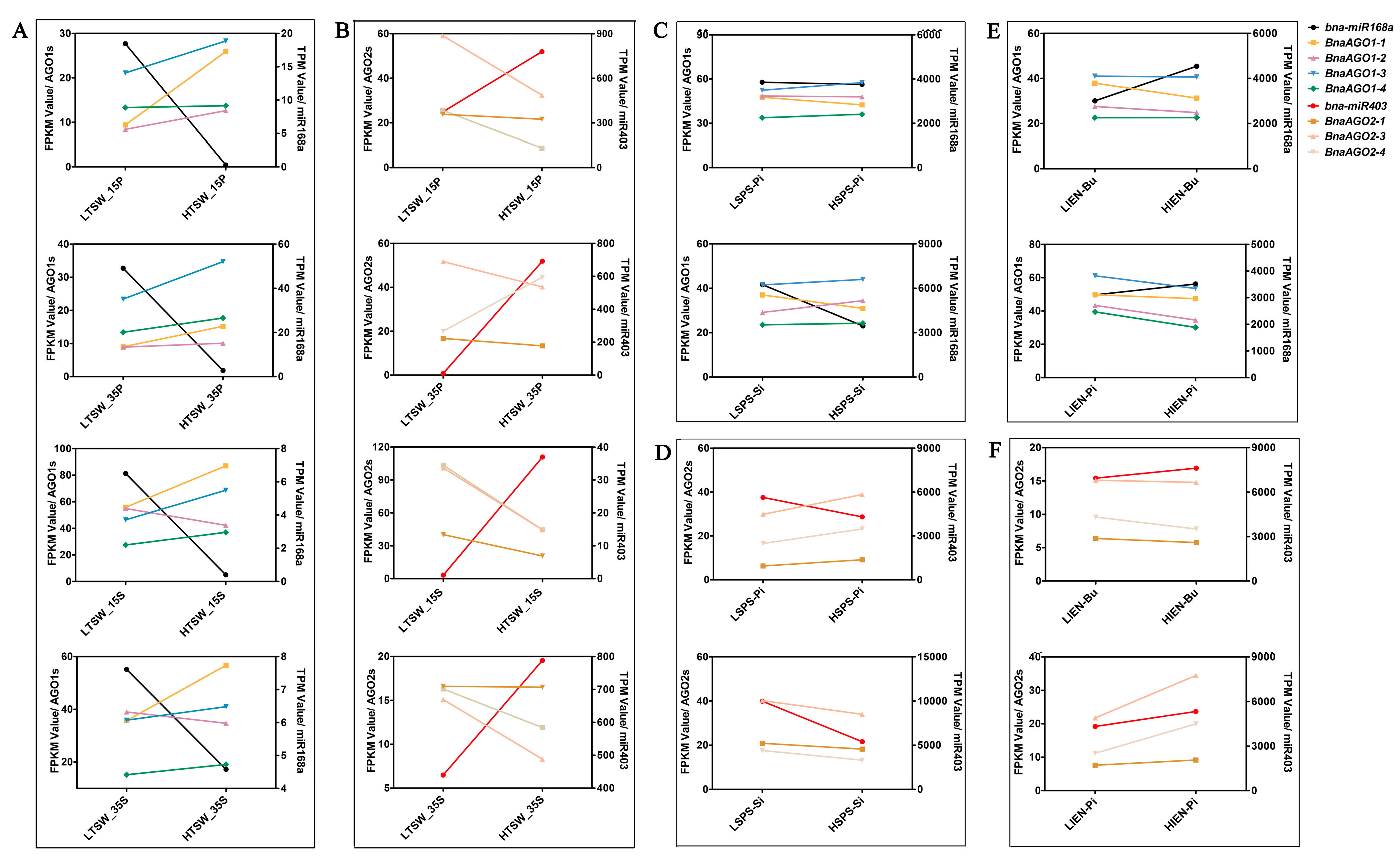
2.5. miRNA-Seq and mRNA-Seq Analysis of miR168a–AGO1s and miR403–AGO2s in Differentially Yielding B. napus Materials
3. Discussion
3.1. Genome-Wide Identification, Phylogenetic Analysis of AGOs, and Their miRNA Triggers
3.2. Diversity and Conservation of the 24 BnaAGOs
3.3. Posttranscriptional Regulation by the B. napus miRNA–AGOs and the Future Regulation of Yield-Related Traits
4. Materials and Methods
4.1. Genome-Wide Prediction of AGOs and Their miRNA Triggers
4.2. Chromosomal Location, Gene Structure and Protein Properties
4.3. Identification of B. napus miRNAs with Perfect Complementarity to the BnaAGOs
4.4. Plant Materials
4.5. miRNA-Seq and mRNA-Seq Analysis among Multiple Yield-Related Materials
4.6. RNA Extraction and qRT-PCR Verification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and functional diversification of MIRNA genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Z.; Bai, J.; Tao, X.; Wang, L.; Zhang, H.; Zhu, J.K. Disruption of MIR396e and MIR396f improves rice yield under nitrogen-deficient conditions. Natl. Sci. Rev. 2020, 7, 102–112. [Google Scholar] [CrossRef]
- Miao, C.; Wang, D.; He, R.; Liu, S.; Zhu, J.K. Mutations in MIR396e and MIR396f increase grain size and modulate shoot architecture in rice. Plant Biotechnol. J. 2020, 18, 491–501. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Yu, Y.; Wang, C.Y.; Li, Z.Y.; Liu, Q.; Xu, J.; Liao, J.Y.; Wang, X.J.; Qu, L.H.; Chen, F.; et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013, 31, 848–852. [Google Scholar] [CrossRef]
- Zhang, J.P.; Yu, Y.; Feng, Y.Z.; Zhou, Y.F.; Zhang, F.; Yang, Y.W.; Lei, M.Q.; Zhang, Y.C.; Chen, Y.Q. MiR408 Regulates Grain Yield and Photosynthesis via a Phytocyanin Protein. Plant Physiol. 2017, 175, 1175–1185. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, H.; Guo, S.; Wang, B.; Li, Z.; Chong, K.; Xu, Y. OsmiR396d Affects Gibberellin and Brassinosteroid Signaling to Regulate Plant Architecture in Rice. Plant Physiol. 2018, 176, 946–959. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, J.; Wang, K.; Li, D.; Yang, R.; Luo, H.; Zhang, W. MiR396-GRF module associates with switchgrass biomass yield and feedstock quality. Plant Biotechnol. J. 2021, 19, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, Y.P.; Zhou, X.H.; Yang, X.M.; He, X.R.; Feng, Q.; Zhu, Y.; Li, G.B.; Wang, H.; Zhao, J.H.; et al. Rice miR1432 Fine-Tunes the Balance of Yield and Blast Disease Resistance via Different Modules. Rice 2021, 14, 87. [Google Scholar] [CrossRef] [PubMed]
- Niaz, S. The AGO proteins: An overview. Biol. Chem. 2018, 399, 525–547. [Google Scholar] [CrossRef] [PubMed]
- Azlan, A.; Dzaki, N.; Azzam, G. Argonaute: The executor of small RNA function. J. Genet Genom. 2016, 43, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Barford, D. Argonaute: A scaffold for the function of short regulatory RNAs. Trends Biochem. Sci. 2006, 31, 622–630. [Google Scholar] [CrossRef]
- Carbonell, A. Plant ARGONAUTEs: Features, Functions, and Unknowns. Methods Mol. Biol. 2017, 1640, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Arora, R.; Lama, T.; Nijhawan, A.; Khurana, J.P.; Tyagi, A.K.; Kapoor, S. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genom. 2008, 9, 45. [Google Scholar] [CrossRef]
- Qian, Y.X.; Cheng, Y.; Cheng, X.; Jiang, H.Y.; Zhu, S.W.; Cheng, B.J. Identification and characterization of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in maize. Plant Cell Rep. 2011, 30, 1347–1363. [Google Scholar] [CrossRef]
- Cui, D.L.; Meng, J.Y.; Ren, X.Y.; Yue, J.J.; Fu, H.Y.; Huang, M.T.; Zhang, Q.Q.; Gao, S.J. Genome-wide identification and characterization of DCL, AGO and RDR gene families in Saccharum spontaneum. Sci. Rep. 2020, 10, 13202. [Google Scholar] [CrossRef]
- Bai, M.; Yang, G.S.; Chen, W.T.; Mao, Z.C.; Kang, H.X.; Chen, G.H.; Yang, Y.H.; Xie, B.Y. Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 2012, 501, 52–62. [Google Scholar] [CrossRef]
- Dalmadi, A.; Miloro, F.; Balint, J.; Varallyay, E.; Havelda, Z. Controlled RISC loading efficiency of miR168 defined by miRNA duplex structure adjusts ARGONAUTE1 homeostasis. Nucleic Acids Res. 2021, 49, 12912–12928. [Google Scholar] [CrossRef]
- Carbonell, A.; Fahlgren, N.; Garcia-Ruiz, H.; Gilbert, K.B.; Montgomery, T.A.; Nguyen, T.; Cuperus, J.T.; Carrington, J.C. Functional Analysis of Three Arabidopsis ARGONAUTES Using Slicer-Defective Mutants. Plant Cell 2012, 24, 3613–3629. [Google Scholar] [CrossRef]
- Manavella, P.A.; Weigel, D.; Wu, L.A. Argonaute10 as a miRNA Locker. Cell 2011, 145, 173–174. [Google Scholar] [CrossRef]
- Ji, L.J.; Liu, X.G.; Yan, J.; Wang, W.M.; Yumul, R.E.; Kim, Y.J.; Dinh, T.T.; Liu, J.; Cui, X.; Zheng, B.L.; et al. ARGONAUTE10 and ARGONAUTE1 Regulate the Termination of Floral Stem Cells through Two MicroRNAs in Arabidopsis. PloS Genet. 2011, 7, e1001358. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.J.; Avrutsky, M.I.; Sifuentes, C.J.; Pereira, L.; Wierzbicki, A.T. Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing. PloS Genet. 2011, 7, e1002120. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.Q.; Wang, W.; Iki, T.; Liu, C.; Wu, Y.; Ishikawa, M.; Zhou, X.P.; Qi, Y.J. Cytoplasmic Assembly and Selective Nuclear Import of Arabidopsis ARGONAUTE4/siRNA Complexes. Mol. Cell 2012, 46, 859–870. [Google Scholar] [CrossRef] [PubMed]
- Vaucheret, H.; Vazquez, F.; Crete, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Chern, M.; Zhu, Y.; Zhang, L.L.; Lu, J.H.; Li, X.P.; Dang, W.Q.; Ma, X.C.; Yang, Z.R.; et al. Suppression of rice miR168 improves yield, flowering time and immunity. Nat. Plants 2021, 7, 129–136. [Google Scholar] [CrossRef]
- Shao, F.; Lu, S. Genome-wide identification, molecular cloning, expression profiling and posttranscriptional regulation analysis of the Argonaute gene family in Salvia miltiorrhiza, an emerging model medicinal plant. BMC Genom. 2013, 14, 512. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.X.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Sunitha, S.; Loyola, R.; Alcalde, J.A.; Arce-Johnson, P.; Matus, J.T.; Rock, C.D. The Role of UV-B light on Small RNA Activity During Grapevine Berry Development. G3 Genes Genomes Genet. 2019, 9, 769–787. [Google Scholar] [CrossRef]
- Harvey, J.J.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstadt, S.; Carr, J.P.; Baulcombe, D.C. An antiviral defense role of AGO2 in plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.J.; Lu, K.; Yang, B.; Wang, T.Y.; Zhang, L.; Zhang, A.X.; Wang, J.; Liu, L.Z.; Qu, C.M.; Li, J.N. Genome-Wide Analysis and Expression Profiling of the SUC and SWEET Gene Families of Sucrose Transporters in Oilseed Rape (Brassica napus L.). Front. Plant Sci. 2016, 7, 1464. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Peng, L.; Zhang, C.; Lu, J.H.; Yang, B.; Xiao, Z.C.; Liang, Y.; Xu, X.F.; Qu, C.M.; Zhang, K.; et al. Genome-Wide Association and Transcriptome Analyses Reveal Candidate Genes Underlying Yield-determining Traits in Brassica napus. Front. Plant Sci. 2017, 8, 206. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Zhang, C.; Yang, B.; Xiao, Z.C.; Ma, J.Q.; Liu, J.S.; Jian, H.J.; Qu, C.M.; Lu, K.; Li, J.N. Genome-Wide Identification and Expression Profiling of Monosaccharide Transporter Genes Associated with High Harvest Index Values in Rapeseed (Brassica napus L.). Genes 2020, 11, 653. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, C.; Ma, J.Q.; Zhang, L.Y.; Yang, B.; Tang, X.Y.; Huang, L.; Zhou, X.T.; Lu, K.; Li, J.N. Genome-Wide Identification and Expression Profiling of Cytokinin Oxidase/Dehydrogenase (CKX) Genes Reveal Likely Roles in Pod Development and Stress Responses in Oilseed Rape (Brassica napus L.). Genes 2018, 9, 168. [Google Scholar] [CrossRef]
- Ma, J.Q.; Jian, H.J.; Yang, B.; Lu, K.; Zhang, A.X.; Liu, P.; Li, J.N. Genome-wide analysis and expression profiling of the GRF gene family in oilseed rape (Brassica napus L.). Gene 2017, 620, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Ma, C.Z.; Yue, Y.; Hu, K.N.; Li, Y.Y.; Duan, Z.Q.; Wu, M.; Tu, J.X.; Shen, J.X.; Yi, B.; et al. Unravelling the complex trait of harvest index in rapeseed (Brassica napus L.) with association mapping. BMC Genom. 2015, 16, 379. [Google Scholar] [CrossRef]
- Dong, H.L.; Tan, C.D.; Li, Y.Z.; He, Y.; Wei, S.; Cui, Y.X.; Chen, Y.G.; Wei, D.Y.; Fu, Y.; He, Y.J.; et al. Genome-Wide Association Study Reveals Both Overlapping and Independent Genetic Loci to Control Seed Weight and Silique Length in Brassica napus. Front. Plant Sci. 2018, 9, 921. [Google Scholar] [CrossRef]
- Shen, W.; Qin, P.; Yan, M.; Li, B.; Wu, Z.; Wen, J.; Yi, B.; Ma, C.; Shen, J.; Fu, T.D.; et al. Fine mapping of a silique length- and seed weight-related gene in Brassica napus. Theor. Appl. Genet. 2019, 132, 2985–2996. [Google Scholar] [CrossRef]
- Nonomura, K.I.; Morohoshi, A.; Nakano, M.; Eiguchi, M.; Miyao, A.; Hirochika, H.; Kurata, N. A germ cell-specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 2007, 19, 2583–2594. [Google Scholar] [CrossRef]
- Chao, H.Y.; Li, T.; Luo, C.Y.; Huang, H.L.; Ruan, Y.F.; Li, X.D.; Niu, Y.; Fan, Y.H.; Sun, W.; Zhang, K.; et al. BrassicaEDB: A Gene Expression Database for Brassica Crops. Int. J. Mol. Sci. 2020, 21, 5831. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chang, W.; Li, X.D.; Yang, B.; Zhang, L.Y.; Xiao, Z.C.; Li, J.A.; Lu, K. Transcriptome and Small RNA Sequencing Reveal the Mechanisms Regulating Harvest Index in Brassica napus. Front. Plant Sci. 2022, 13, 855486. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.Y.; Parkin, I.A.P.; Tang, H.B.; Wang, X.Y.; Chiquet, J.; Belcram, H.; Tong, C.B.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Pfaff, J.; Meister, G. Argonaute and GW182 proteins: An effective alliance in gene silencing. Biochem. Soc. Trans. 2013, 41, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Hoden, K.P.; Singh, R.K.; Dixelius, C. Genome-wide identification of Argonautes in Solanaceae with emphasis on potato. Sci. Rep. 2020, 10, 20577. [Google Scholar] [CrossRef]
- Cao, J.Y.; Xu, Y.P.; Li, W.; Li, S.S.; Rahman, H.; Cai, X.Z. Genome-Wide Identification of Dicer-Like, Argonaute, and RNA-Dependent RNA Polymerase Gene Families in Brassica Species and Functional Analyses of Their Arabidopsis Homologs in Resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 2016, 7, 1614. [Google Scholar] [CrossRef]
- Willmann, M.R.; Poethig, R.S. Conservation and evolution of miRNA regulatory programs in plant development. Curr. Opin. Plant Biol. 2007, 10, 503–511. [Google Scholar] [CrossRef]
- Pietrykowska, H.; Sierocka, I.; Zielezinski, A.; Alisha, A.; Carrasco-Sanchez, J.C.; Jarmolowski, A.; Karlowski, W.M.; Szweykowska-Kulinska, Z. Biogenesis, conservation, and function of miRNA in liverworts. J. Exp. Bot. 2022, 73, 4528–4545. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Xian, Z.Q.; Huang, W.; Li, Z.G.; Zhang, C. Evidence for the biological function of miR403 in tomato development. Sci. Hortic. 2015, 197, 619–626. [Google Scholar] [CrossRef]
- Vaucheret, H.; Mallory, A.C.; Bartel, D.P. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 2006, 22, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Hereditas 2007, 29, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comp. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. Biotechniques 1998, 24, 318–319. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | A. thaliana | B. rapa | B. naus (A) | B. oleracea | B. naus (C) |
|---|---|---|---|---|---|
| Zero-copy in B. napus | |||||
| AGO3 | 1 | 0 | 0 | 0 | 0 |
| AGO8 | 1 | 0 | 0 | 0 | 0 |
| Two-copy in B. napus | |||||
| AGO5 | 1 | 1 | 1 | 1 | 1 |
| AGO10 | 1 | 1 | 1 | 1 | 1 |
| AGO6 | 1 | 1 | 1 | 1 | 1 |
| Three-copy in B. napus | |||||
| AGO7 | 1 | 1 | 1 | 2 | 2 |
| AGO9 | 1 | 2 | 2 | 2 | 1 |
| Four-copy in B. napus | |||||
| AGO1 | 1 | 2 | 2 | 2 | 2 |
| AGO2 | 1 | 2 | 2 | 2 | 2 |
| AGO4 | 1 | 1 | 2 | 2 | 2 |
| Gene Name | Transcript Name | gDNA Size (bp) | CDS Size (nts) | Protein | No. of Introns | Genomic Location | ||
|---|---|---|---|---|---|---|---|---|
| Length (aa) | Mw (kDa) | pI | ||||||
| BnaAGO1-1 | BnaC08g46720D | 5291 | 3159 | 1052 | 116.78 | 9.45 | 20 | chrC08_random:953598-958888 |
| BnaAGO1-2 | BnaA08g03260D | 5847 | 3135 | 1044 | 115.85 | 9.4 | 22 | chrA08:2681072-2686918 |
| BnaAGO1-3 | BnaC05g25730D | 5910 | 2943 | 980 | 100.15 | 9.17 | 22 | chrC05:21116236-21122145 |
| BnaAGO1-4 | BnaA05g17460D | 6740 | 3261 | 1086 | 120.33 | 9.38 | 22 | chrA05:12290150-12296889 |
| BnaAGO2-1 | BnaA05g14760D | 3501 | 3033 | 1010 | 112.74 | 9.51 | 1 | chrA05:9228612-9232112 |
| BnaAGO2-2 | BnaC06g41790D | 3340 | 3108 | 1035 | 114.35 | 9.54 | 1 | chrC06_random:1066401-1069740 |
| BnaAGO2-3 | BnaCnng68320D | 3169 | 2664 | 887 | 100.7 | 9.44 | 2 | chrCnn_random:67905947-67909115 |
| BnaAGO2-4 | BnaA09g25290D | 3712 | 3072 | 1023 | 113.91 | 9.66 | 3 | chrA09:18324937-18328648 |
| BnaAGO4-1 | BnaC04g54830D | 5652 | 2772 | 923 | 103.36 | 8.87 | 21 | chrC04_random:2230442-2236093 |
| BnaAGO4-2 | BnaA07g13010D | 5716 | 2772 | 923 | 103.36 | 8.82 | 21 | chrA07:11653377-11659092 |
| BnaAGO4-3 | BnaA04g15560D | 5887 | 2769 | 922 | 103.06 | 8.91 | 21 | chrA04:12884285-12890171 |
| BnaAGO4-4 | BnaC04g38560D | 5890 | 2769 | 922 | 103.11 | 8.87 | 21 | chrC04:39739228-39745117 |
| BnaAGO5-1 | BnaA07g13430D | 4997 | 2874 | 957 | 106.71 | 9.52 | 19 | chrA07:11907998-11912994 |
| BnaAGO5-2 | BnaC04g16450D | 5220 | 2859 | 952 | 106.03 | 9.62 | 20 | chrC04:14487678-14492897 |
| BnaAGO6-1 | BnaA03g15180D | 4948 | 2604 | 867 | 97.2 | 9.03 | 21 | chrA03:7005357-7010304 |
| BnaAGO6-2 | BnaC03g18310D | 4736 | 2604 | 867 | 97.37 | 8.99 | 21 | chrC03:9391663-9396398 |
| BnaAGO7-1 | BnaA07g24280D | 3461 | 2955 | 984 | 112.47 | 9.37 | 2 | chrA07:18160385-18163845 |
| BnaAGO7-2 | BnaC06g43420D | 3447 | 2700 | 899 | 102.67 | 9.38 | 5 | chrC06_random:2865213-2868659 |
| BnaAGO7-3 | BnaC02g19190D | 3334 | 2931 | 976 | 111.58 | 9.32 | 2 | chrC02:15451981-15455314 |
| BnaAGO9-1 | BnaCnng35060D | 4647 | 2721 | 906 | 102.57 | 9.42 | 21 | chrCnn_random:33265084-33269730 |
| BnaAGO9-2 | BnaA10g14450D | 4627 | 2715 | 904 | 102.04 | 9.31 | 21 | chrA10:11492941-11497567 |
| BnaAGO9-3 | BnaA02g05290D | 5571 | 2721 | 906 | 101.34 | 9.31 | 20 | chrA02:2403187-2408757 |
| BnaAGO10-1 | BnaA06g36540D | 4921 | 2928 | 975 | 109.27 | 9.38 | 16 | chrA06:23915363-23920283 |
| BnaAGO10-2 | BnaC07g17330D | 5667 | 2946 | 981 | 109.75 | 9.38 | 16 | chrC07:23533982-23539648 |
| miRNA_ID | Target_Name | Expection | miRNA_aligned_fragment | Target_aligned_fragment | Alignment | Inhibition |
|---|---|---|---|---|---|---|
| bna-miR403 | BnaAGO2-4 | 0 | UUAGAUUCACGCACAAACUCG | GGAGUUUGUGCGUGAAUCUAA | :::::::::::::::::::: | Cleavage |
| bna-miR403 | BnaAGO2-1 | 0 | UUAGAUUCACGCACAAACUCG | GGAGUUUGUGCGUGAAUCUAA | :::::::::::::::::::: | Cleavage |
| bna-miR403 | BnaAGO2-3 | 0 | UUAGAUUCACGCACAAACUCG | GGAGUUUGUGCGUGAAUCUAA | :::::::::::::::::::: | Cleavage |
| bna-miR168a | BnaAGO1-2 | 3 | UCGCUUGGUGCAGGUCGGGAA | UUCCCGAGCUGCAUCAAGCUA | ::::::: :::::.::::: : | Cleavage |
| bna-miR168a | BnaAGO1-1 | 3 | UCGCUUGGUGCAGGUCGGGAA | UUCCCGAGCUGCAUCAAGCUA | ::::::: :::::.::::: : | Cleavage |
| bna-miR168a | BnaAGO1-4 | 3 | UCGCUUGGUGCAGGUCGGGAA | UUCCCGAGCUGCAUCAAGCUA | ::::::: :::::.::::: : | Cleavage |
| bna-miR168a | BnaAGO1-3 | 3 | UCGCUUGGUGCAGGUCGGGAA | UUCCCGAGCUGCAUCAAGCUA | ::::::: :::::.::::: : | Cleavage |
| Trait | Material | 2016 | 2017 | 2018 | 2019 | 2021 | 2022 | Mean Value | SEM | p_Value |
|---|---|---|---|---|---|---|---|---|---|---|
| Thousand Seed Weight/g (TSW) | LTSW | - | 4.27 | 3.54 | 4.08 | 3.15 | 2.78 | 3.56 | 0.56 | |
| HTSW | - | 6.03 | 6.15 | 7.58 | 6.55 | 6.99 | 6.66 | 0.57 | 0.000 | |
| Seed number Per Silique (SPS) | LSPS-1 | - | 13.47 | 14.74 | 14.99 | - | 21.50 | 16.17 | 3.13 | |
| LSPS-2 | - | 15.30 | 13.97 | 8.98 | 15.59 | 11.50 | 13.07 | 2.51 | ||
| HSPS-1 | - | 31.43 | 33.53 | 25.23 | 34.60 | 31.00 | 31.16 | 3.25 | 0.00 | |
| HSPS-2 | - | 29.40 | 29.40 | - | 24.36 | 29.70 | 28.22 | 2.23 | ||
| Initial embryonic number (IEN) | LIEN-1 | - | - | - | - | 25.67 | 24.33 | 25.00 | 0.67 | |
| LIEN-2 | - | - | - | - | 24.00 | 23.00 | 23.50 | 0.50 | ||
| HIEN-1 | - | - | - | - | 38.33 | 34.33 | 36.33 | 2.00 | 0.00 | |
| HIEN-2 | - | - | - | - | 35.67 | 33.00 | 34.34 | 1.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Yang, B.; Zhang, C.; Chen, H.; Xu, J.; Qu, C.; Lu, K.; Li, J. Genome-Wide Identification and Posttranscriptional Regulation Analyses Elucidate Roles of Key Argonautes and Their miRNA Triggers in Regulating Complex Yield Traits in Rapeseed. Int. J. Mol. Sci. 2023, 24, 2543. https://doi.org/10.3390/ijms24032543
Zhang L, Yang B, Zhang C, Chen H, Xu J, Qu C, Lu K, Li J. Genome-Wide Identification and Posttranscriptional Regulation Analyses Elucidate Roles of Key Argonautes and Their miRNA Triggers in Regulating Complex Yield Traits in Rapeseed. International Journal of Molecular Sciences. 2023; 24(3):2543. https://doi.org/10.3390/ijms24032543
Chicago/Turabian StyleZhang, Liyuan, Bo Yang, Chao Zhang, Huan Chen, Jinxiong Xu, Cunmin Qu, Kun Lu, and Jiana Li. 2023. "Genome-Wide Identification and Posttranscriptional Regulation Analyses Elucidate Roles of Key Argonautes and Their miRNA Triggers in Regulating Complex Yield Traits in Rapeseed" International Journal of Molecular Sciences 24, no. 3: 2543. https://doi.org/10.3390/ijms24032543
APA StyleZhang, L., Yang, B., Zhang, C., Chen, H., Xu, J., Qu, C., Lu, K., & Li, J. (2023). Genome-Wide Identification and Posttranscriptional Regulation Analyses Elucidate Roles of Key Argonautes and Their miRNA Triggers in Regulating Complex Yield Traits in Rapeseed. International Journal of Molecular Sciences, 24(3), 2543. https://doi.org/10.3390/ijms24032543









