From 2D to 3D: Development of Monolayer Dopaminergic Neuronal and Midbrain Organoid Cultures for Parkinson’s Disease Modeling and Regenerative Therapy
Abstract
1. Introduction
2. The Three Stages of Midbrain DA (mDA) Neuronal Development
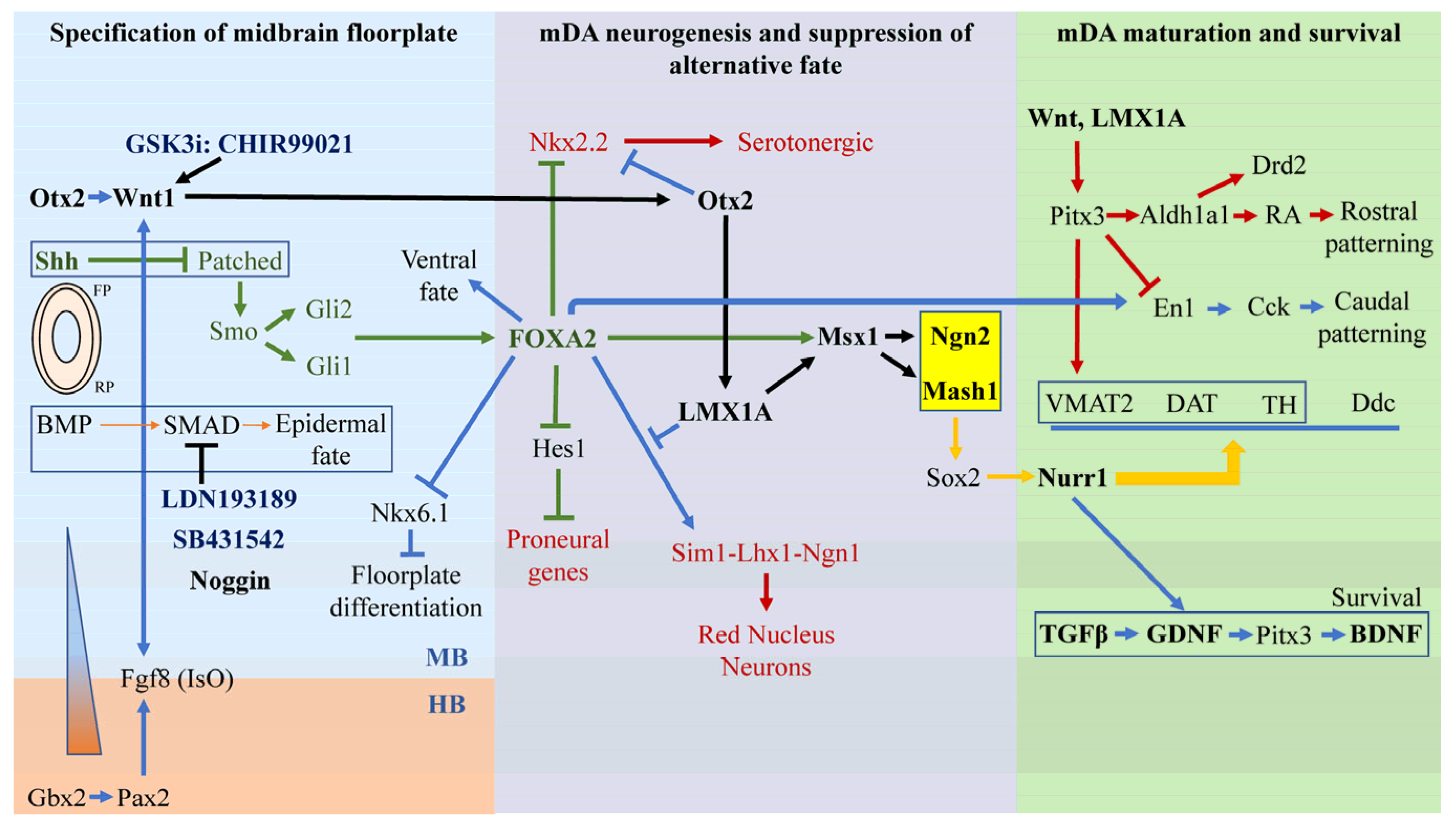
2.1. Stage 1—Specification of Ventral Midbrain Fate Driven by Otx2-Wnt1, Gbx2-Pax2-FGF8, and Shh-FoxA2
2.2. Stage 2—Specification of mDA Neurogenesis and Suppression of Alternative Fates
2.3. Stage 3—mDA Maturation Requires Nurr1, Pitx3, En1 and Survival Requires Gdnf, Bdnf, and Tgfβ
2.4. A9 SNpc DA Neurons and Their Selective Vulnerability to Degeneration in PD
2.5. Mimicking DA Neuronal Development In Vitro—A Quick Survey of Current Protocols
2.6. From 2D to 3D—Generating Midbrain Organoid Models of PD
2.7. Using MOs in PD Disease Modeling and Drug Discovery
2.8. The Future of Organoid Research—State-Of-The-Art Assembloids, Organ-On-A-Chip, and Vascularization
2.9. Use of mDA Precursor Cells for Neural Transplantation
2.10. From Bench to Bedside: Ongoing Human Trials Involving hPSC-Derived DA Precursors
3. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Parkinson Disease. Available online: https://www.who.int/news-room/fact-sheets/detail/parkinson-disease (accessed on 20 November 2022).
- Mackenzie, I.R.A. The pathology of Parkinson’s disease. Br. Columbia Med. J. 2001, 43, 142–147. [Google Scholar]
- Kelly, E.A.; Contreras, J.; Duan, A.; Vassell, R.; Fudge, J.L. Unbiased Stereological Estimates of Dopaminergic and GABAergic Neurons in the A10, A9, and A8 Subregions in the Young Male Macaque. Neuroscience 2022, 496, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Fiorenzano, A.; Sozzi, E.; Parmar, M.; Storm, P. Dopamine Neuron Diversity: Recent Advances and Current Challenges in Human Stem Cell Models and Single Cell Sequencing. Cells 2021, 10, 1366. [Google Scholar] [CrossRef] [PubMed]
- Sonne, J.R.V.; Beato, M.R. Neuroanatomy, Substantia Nigra; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Langley, J.; Huddleston, D.; Hu, X. Detecting parkinsonian degeneration in lateroventral tier of substantia nigra pars compacta with MRI. In Genetics, Neurology, Behavior, and Diet in Parkinson’s Disease; Elsevier: Amsterdam, The Netherlands, 2020; pp. 313–325. [Google Scholar]
- Hirsch, E.; Graybiel, A.M.; Agid, Y.A. Melanized dopaminergic neurons are differentially susceptible to degeneration in Parkinson’s disease. Nature 1988, 334, 345–348. [Google Scholar] [CrossRef]
- Lees, A.J.; Hardy, J.; Revesz, T. Parkinson’s disease. Lancet 2009, 373, 2055–2066. [Google Scholar] [CrossRef]
- Moustafa, A.A.; Chakravarthy, S.; Phillips, J.R.; Gupta, A.; Keri, S.; Polner, B.; Frank, M.J.; Jahanshahi, M. Motor symptoms in Parkinson’s disease: A unified framework. Neurosci. Biobehav. Rev. 2016, 68, 727–740. [Google Scholar] [CrossRef]
- Choudhury, S.P.; Bano, S.; Sen, S.; Suchal, K.; Kumar, S.; Nikolajeff, F.; Dey, S.K.; Sharma, V. Altered neural cell junctions and ion-channels leading to disrupted neuron communication in Parkinson’s disease. NPJ Park. Dis. 2022, 8, 66. [Google Scholar] [CrossRef]
- Zagare, A.; Barmpa, K.; Smajic, S.; Smits, L.M.; Grzyb, K.; Grünewald, A.; Skupin, A.; Nickels, S.L.; Schwamborn, J.C. Midbrain organoids mimic early embryonic neurodevelopment and recapitulate LRRK2-p.Gly2019Ser-associated gene expression. Am. J. Hum. Genet. 2022, 109, 311–327. [Google Scholar] [CrossRef]
- Marton, R.M.; Ioannidis, J.P.A. A Comprehensive Analysis of Protocols for Deriving Dopaminergic Neurons from Human Pluripotent Stem Cells. Stem Cells Transl. Med. 2019, 8, 366–374. [Google Scholar] [CrossRef]
- Mahajani, S.; Raina, A.; Fokken, C.; Kügler, S.; Bähr, M. Homogenous generation of dopaminergic neurons from multiple hiPSC lines by transient expression of transcription factors. Cell Death Dis. 2019, 10, 898. [Google Scholar] [CrossRef]
- Gantner, C.W.; Cota-Coronado, A.; Thompson, L.H.; Parish, C.L. An Optimized Protocol for the Generation of Midbrain Dopamine Neurons under Defined Conditions. STAR Protoc. 2020, 1, 100065. [Google Scholar] [CrossRef] [PubMed]
- SF, G. Formation of the Neural Tube. In Developmental Biology, 6th ed.; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Purves, D.; Augustine, G.J.; Fitzpatrick, D.; Hall, W.C.; LaMantia, A.S. Neuroscience; Oxford University Press: Oxford, UK, 2018. [Google Scholar]
- Kittappa, R.; Chang, W.W.; Awatramani, R.B.; McKay, R.D.G. The foxa2 Gene Controls the Birth and Spontaneous Degeneration of Dopamine Neurons in Old Age. PLoS Biol. 2007, 5, e325. [Google Scholar] [CrossRef] [PubMed]
- Arenas, E.; Denham, M.; Villaescusa, J.C. How to make a midbrain dopaminergic neuron. Development 2015, 142, 1918–1936. [Google Scholar] [CrossRef] [PubMed]
- Veenvliet, J.V.; Smidt, M.P. Molecular mechanisms of dopaminergic subset specification: Fundamental aspects and clinical perspectives. Cell Mol. Life Sci. 2014, 71, 4703–4727. [Google Scholar] [CrossRef]
- Sunmonu, N.A.; Li, K.; Guo, Q.; Li, J.Y. Gbx2 and Fgf8 are sequentially required for formation of the midbrain-hindbrain compartment boundary. Development 2011, 138, 725–734. [Google Scholar] [CrossRef]
- Joyner, A.L.; Liu, A.; Millet, S. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid–hindbrain organizer. Curr. Opin. Cell Biol. 2000, 12, 736–741. [Google Scholar] [CrossRef]
- Ye, W.; Shimamura, K.; Rubenstein, J.L.R.; Hynes, M.A.; Rosenthal, A. FGF and Shh Signals Control Dopaminergic and Serotonergic Cell Fate in the Anterior Neural Plate. Cell 1998, 93, 755–766. [Google Scholar] [CrossRef]
- Gibbs, H.C.; Chang-Gonzalez, A.; Hwang, W.; Yeh, A.T.; Lekven, A.C. Midbrain-Hindbrain Boundary Morphogenesis: At the Intersection of Wnt and Fgf Signaling. Front. Neuroanat. 2017, 11, 64. [Google Scholar] [CrossRef]
- Martinez, S.; Crossley, P.H.; Cobos, I.; Rubenstein, J.L.; Martin, G.R. FGF8 induces formation of an ectopic isthmic organizer and isthmocerebellar development via a repressive effect on Otx2 expression. Development 1999, 126, 1189–1200. [Google Scholar] [CrossRef]
- Dyer, C.; Blanc, E.; Hanisch, A.; Roehl, H.; Otto, G.W.; Yu, T.; Basson, M.A.; Knight, R. A bi-modal function of Wnt signalling directs an FGF activity gradient to spatially regulate neuronal differentiation in the midbrain. Development 2014, 141, 63–72. [Google Scholar] [CrossRef]
- Basson, M.A.; Echevarria, D.; Petersen Ahn, C.; Sudarov, A.; Joyner, A.L.; Mason, I.J.; Martinez, S.; Martin, G.R. Specific regions within the embryonic midbrain and cerebellum require different levels of FGF signaling during development. Development 2008, 135, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Carballo, G.B.; Honorato, J.R.; de Lopes, G.P.F.; Spohr, T.C.L.d.S.e. A highlight on Sonic hedgehog pathway. Cell Commun. Signal. 2018, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Cho, G.; Lim, Y.; Cho, I.T.; Simonet, J.C.; Golden, J.A. Arx together with FoxA2, regulates Shh floor plate expression. Dev. Biol. 2014, 393, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Norton, W.H.; Mangoli, M.; Lele, Z.; Pogoda, H.M.; Diamond, B.; Mercurio, S.; Russell, C.; Teraoka, H.; Stickney, H.L.; Rauch, G.J.; et al. Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphé neurones and cranial motoneurones. Development 2005, 132, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Prakash, N.; Brodski, C.; Naserke, T.; Puelles, E.; Gogoi, R.; Hall, A.; Panhuysen, M.; Echevarria, D.; Sussel, L.; Weisenhorn, D.M.V.; et al. A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development 2006, 133, 89–98. [Google Scholar] [CrossRef]
- Cheng, L.; Chen, C.L.; Luo, P.; Tan, M.; Qiu, M.; Johnson, R.; Ma, Q. Lmx1b, Pet-1, and Nkx2.2 coordinately specify serotonergic neurotransmitter phenotype. J. Neurosci. 2003, 23, 9961–9967. [Google Scholar] [CrossRef] [PubMed]
- Ono, Y.; Nakatani, T.; Sakamoto, Y.; Mizuhara, E.; Minaki, Y.; Kumai, M.; Hamaguchi, A.; Nishimura, M.; Inoue, Y.; Hayashi, H.; et al. Differences in neurogenic potential in floor plate cells along an anteroposterior location: Midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development 2007, 134, 3213–3225. [Google Scholar] [CrossRef]
- Nakatani, T.; Kumai, M.; Mizuhara, E.; Minaki, Y.; Ono, Y. Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev. Biol. 2010, 339, 101–113. [Google Scholar] [CrossRef]
- Andersson, E.; Tryggvason, U.; Deng, Q.; Friling, S.; Alekseenko, Z.; Robert, B.; Perlmann, T.; Ericson, J. Identification of Intrinsic Determinants of Midbrain Dopamine Neurons. Cell 2006, 124, 393–405. [Google Scholar] [CrossRef]
- Volpicelli, F.; Perrone-Capano, C.; Bellenchi, G.C.; Colucci-D’Amato, L.; di Porzio, U. Molecular Regulation in Dopaminergic Neuron Development. Cues to Unveil Molecular Pathogenesis and Pharmacological Targets of Neurodegeneration. Int. J. Mol. Sci. 2020, 21, 3995. [Google Scholar] [CrossRef]
- Kele, J.; Simplicio, N.; Ferri, A.L.M.; Mira, H.; Guillemot, F.O.; Arenas, E.; Ang, S.-L. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development 2006, 133, 495–505. [Google Scholar] [CrossRef]
- Ferri, A.L.M.; Lin, W.; Mavromatakis, Y.E.; Wang, J.C.; Sasaki, H.; Whitsett, J.A.; Ang, S.-L. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development 2007, 134, 2761–2769. [Google Scholar] [CrossRef]
- Caiazzo, M.; Dell’Anno, M.T.; Dvoretskova, E.; Lazarevic, D.; Taverna, S.; Leo, D.; Sotnikova, T.D.; Menegon, A.; Roncaglia, P.; Colciago, G.; et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 2011, 476, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Hulme, A.J.; Maksour, S.; St-Clair Glover, M.; Miellet, S.; Dottori, M. Making neurons, made easy: The use of Neurogenin-2 in neuronal differentiation. Stem Cell Rep. 2022, 17, 14–34. [Google Scholar] [CrossRef] [PubMed]
- Prakash, N.; Wurst, W. Genetic networks controlling the development of midbrain dopaminergic neurons. J. Physiol. 2006, 575, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Kouwenhoven, W.M.; von Oerthel, L.; Smidt, M.P. Pitx3 and En1 determine the size and molecular programming of the dopaminergic neuronal pool. PLoS ONE 2017, 12, e0182421. [Google Scholar] [CrossRef]
- Gale, E.; Li, M. Midbrain dopaminergic neuron fate specification: Of mice and embryonic stem cells. Mol. Brain 2008, 1, 8. [Google Scholar] [CrossRef]
- Chung, S.; Leung, A.; Han, B.-S.; Chang, M.-Y.; Moon, J.-I.; Kim, C.-H.; Hong, S.; Pruszak, J.; Isacson, O.; Kim, K.-S. Wnt1-lmx1a Forms a Novel Autoregulatory Loop and Controls Midbrain Dopaminergic Differentiation Synergistically with the SHH-FoxA2 Pathway. Cell Stem Cell 2009, 5, 646–658. [Google Scholar] [CrossRef]
- Stott, S.R.W.; Metzakopian, E.; Lin, W.; Kaestner, K.H.; Hen, R.; Ang, S.-L. Foxa1 and Foxa2 Are Required for the Maintenance of Dopaminergic Properties in Ventral Midbrain Neurons at Late Embryonic Stages. J. Neurosci. 2013, 33, 8022. [Google Scholar] [CrossRef]
- Smits, S.M.; Ponnio, T.; Conneely, O.M.; Burbach, J.P.H.; Smidt, M.P. Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur. J. Neurosci. 2003, 18, 1731–1738. [Google Scholar] [CrossRef]
- Saucedo-Cardenas, O.; Quintana-Hau, J.D.; Le, W.-D.; Smidt, M.P.; Cox, J.J.; De Mayo, F.; Burbach, J.P.H.; Conneely, O.M. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc. Natl. Acad. Sci. USA 1998, 95, 4013–4018. [Google Scholar] [CrossRef]
- Wang, M.; Ling, K.H.; Tan, J.J.; Lu, C.B. Development and Differentiation of Midbrain Dopaminergic Neuron: From Bench to Bedside. Cells 2020, 9, 1489. [Google Scholar] [CrossRef] [PubMed]
- Simon, H.H.; Saueressig, H.; Wurst, W.; Goulding, M.D.; O’Leary, D.D. Fate of midbrain dopaminergic neurons controlled by the engrailed genes. J. Neurosci. 2001, 21, 3126–3134. [Google Scholar] [CrossRef]
- Bachar-Dahan, L.; Goltzmann, J.; Yaniv, A.; Gazit, A. Engrailed-1 Negatively Regulates β-Catenin Transcriptional Activity by Destabilizing β-Catenin via a Glycogen Synthase Kinase-3β–independent Pathway. Mol. Biol. Cell 2006, 17, 2572–2580. [Google Scholar] [CrossRef]
- Hwang, D.-Y.; Ardayfio, P.; Kang, U.J.; Semina, E.V.; Kim, K.-S. Selective loss of dopaminergic neurons in the substantia nigra of Pitx3-deficient aphakia mice. Mol. Brain Res. 2003, 114, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, F.M.J.; Veenvliet, J.V.; Almirza, W.H.; Hoekstra, E.J.; von Oerthel, L.; van der Linden, A.J.A.; Neijts, R.; Koerkamp, M.G.; van Leenen, D.; Holstege, F.C.P.; et al. Retinoic acid-dependent and -independent gene-regulatory pathways of Pitx3 in meso-diencephalic dopaminergic neurons. Development 2011, 138, 5213–5222. [Google Scholar] [CrossRef] [PubMed]
- Veenvliet, J.V.; Dos Santos, M.T.A.; Kouwenhoven, W.M.; Von Oerthel, L.; Lim, J.L.; Van Der Linden, A.J.; Koerkamp, M.J.G.; Holstege, F.C.; Smidt, M.P. Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development 2013, 140, 3373–3384. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, F.M.; Smits, S.M.; Noorlander, C.W.; von Oerthel, L.; van der Linden, A.J.; Burbach, J.P.H.; Smidt, M.P. Retinoic acid counteracts developmental defects in the substantia nigra caused by Pitx3 deficiency. Development 2007, 134, 2673–2684. [Google Scholar] [CrossRef]
- Vaswani, A.R.; Weykopf, B.; Hagemann, C.; Fried, H.-U.; Brüstle, O.; Blaess, S. Correct setup of the substantia nigra requires Reelin-mediated fast, laterally-directed migration of dopaminergic neurons. eLife 2019, 8, e41623. [Google Scholar] [CrossRef]
- Tian, L.; Al-Nusaif, M.; Chen, X.; Li, S.; Le, W. Roles of Transcription Factors in the Development and Reprogramming of the Dopaminergic Neurons. Int. J. Mol. Sci. 2022, 23, 845. [Google Scholar] [CrossRef]
- Chalazonitis, A.; Rao, M.; Sulzer, D. Similarities and differences between nigral and enteric dopaminergic neurons unravel distinctive involvement in Parkinson’s disease. NPJ Park. Dis. 2022, 8, 50. [Google Scholar] [CrossRef]
- Roussa, E.; von Bohlen und Halback, O.; Krieglstein, K. TGF-β in Dopamine Neuron Development, Maintenance and Neuroprotection. In Development and Engineering of Dopamine Neurons; Pasterkamp, R.J., Smidt, M.P., Burbach, J.P.H., Eds.; Springer: New York, NY, USA, 2009; pp. 81–90. [Google Scholar]
- Arenas, E.; Trupp, M.; Akerud, P.; Ibáñez, C.F. GDNF prevents degeneration and promotes the phenotype of brain noradrenergic neurons in vivo. Neuron 1995, 15, 1465–1473. [Google Scholar] [CrossRef]
- Frim, D.M.; Uhler, T.A.; Galpern, W.R.; Beal, M.F.; Breakefield, X.O.; Isacson, O. Implanted fibroblasts genetically engineered to produce brain-derived neurotrophic factor prevent 1-methyl-4-phenylpyridinium toxicity to dopaminergic neurons in the rat. Proc. Natl. Acad. Sci. USA 1994, 91, 5104–5108. [Google Scholar] [CrossRef]
- Tesseur, I.; Nguyen, A.; Chang, B.; Li, L.; Woodling, N.S.; Wyss-Coray, T.; Luo, J. Deficiency in Neuronal TGF-β Signaling Leads to Nigrostriatal Degeneration and Activation of TGF-β Signaling Protects against MPTP Neurotoxicity in Mice. J. Neurosci. 2017, 37, 4584–4592. [Google Scholar] [CrossRef]
- Pascual, A.; Hidalgo-Figueroa, M.; Piruat, J.I.; Pintado, C.O.; Gómez-Díaz, R.; López-Barneo, J. Absolute requirement of GDNF for adult catecholaminergic neuron survival. Nat. Neurosci. 2008, 11, 755–761. [Google Scholar] [CrossRef]
- Baquet, Z.C.; Bickford, P.C.; Jones, K.R. Brain-derived neurotrophic factor is required for the establishment of the proper number of dopaminergic neurons in the substantia nigra pars compacta. J. Neurosci. 2005, 25, 6251–6259. [Google Scholar] [CrossRef]
- Krieglstein, K.; Farkas, L.; Unsicker, K. TGF-β regulates the survival of ciliary ganglionic neurons synergistically with ciliary neurotrophic factor and neurotrophins. J. Neurobiol. 1998, 37, 563–572. [Google Scholar] [CrossRef]
- Peng, C.; Aron, L.; Klein, R.; Li, M.; Wurst, W.; Prakash, N.; Le, W. Pitx3 is a critical mediator of GDNF-induced BDNF expression in nigrostriatal dopaminergic neurons. J. Neurosci. 2011, 31, 12802–12815. [Google Scholar] [CrossRef]
- Galleguillos, D.; Fuentealba, J.A.; Gómez, L.M.; Saver, M.; Gómez, A.; Nash, K.; Burger, C.; Gysling, K.; Andrés, M.E. Nurr1 regulates RET expression in dopamine neurons of adult rat midbrain. J. Neurochem. 2010, 114, 1158–1167. [Google Scholar] [CrossRef]
- Decressac, M.; Kadkhodaei, B.; Mattsson, B.; Laguna, A.; Perlmann, T.; Björklund, A. Alpha-Synuclein-Induced Down-Regulation of Nurr1 Disrupts GDNF Signaling in Nigral Dopamine Neurons. Sci. Transl. Med. 2012, 4, ra156–ra163. [Google Scholar] [CrossRef]
- Brichta, L.; Greengard, P. Molecular determinants of selective dopaminergic vulnerability in Parkinson’s disease: An update. Front. Neuroanat. 2014, 8, 152. [Google Scholar] [CrossRef] [PubMed]
- German, D.C.; Manaye, K.; Smith, W.K.; Woodward, D.J.; Saper, C.B. Midbrain dopaminergic cell loss in Parkinson’s disease: Computer visualization. Ann. Neurol. 1989, 26, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Damier, P.; Hirsch, E.C.; Agid, Y.; Graybiel, A.M. The substantia nigra of the human brain. II. Patterns of loss of dopamine-containing neurons in Parkinson’s disease. Brain 1999, 122, 1437–1448. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.L.; Sinton, C.M.; German, D.C. Midbrain dopaminergic neurons in the mouse: Co-localization with Calbindin-D28K and calretinin. Neuroscience 1996, 75, 523–533. [Google Scholar] [CrossRef]
- Surmeier, D.J.; Halliday, G.M.; Simuni, T. Calcium, mitochondrial dysfunction and slowing the progression of Parkinson’s disease. Exp. Neurol. 2017, 298, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.Y.; Wang, S.C.; Lei, M.; Wang, Z.; Xiong, K. Regulatory role of calpain in neuronal death. Neural Regen. Res. 2018, 13, 556–562. [Google Scholar]
- Nagley, P.; Higgins, G.C.; Atkin, J.D.; Beart, P.M. Multifaceted deaths orchestrated by mitochondria in neurones. Biochim. Biophys. Acta 2010, 1802, 167–185. [Google Scholar] [CrossRef]
- Goldberg, J.A.; Guzman, J.N.; Estep, C.M.; Ilijic, E.; Kondapalli, J.; Sanchez-Padilla, J.; Surmeier, D.J. Calcium entry induces mitochondrial oxidant stress in vagal neurons at risk in Parkinson’s disease. Nat. Neurosci. 2012, 15, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, P.; Ben Jelloun, N.; Febvret, A. Sparing of the dopaminergic neurons containing calbindin-D28k and of the dopaminergic mesocortical projections in weaver mutant mice. Neuroscience 1994, 61, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Meiser, J.; Weindl, D.; Hiller, K. Complexity of dopamine metabolism. Cell Commun. Signal. 2013, 11, 34. [Google Scholar] [CrossRef]
- Bisaglia, M.; Mammi, S.; Bubacco, L. Kinetic and structural analysis of the early oxidation products of dopamine: Analysis of the interactions with alpha-synuclein. J. Biol. Chem. 2007, 282, 15597–15605. [Google Scholar] [CrossRef] [PubMed]
- Penning, T.M. Genotoxicity of ortho-quinones: Reactive oxygen species versus covalent modification. Toxicol. Res. 2017, 6, 740–754. [Google Scholar] [CrossRef] [PubMed]
- Lohr, K.M.; Masoud, S.T.; Salahpour, A.; Miller, G.W. Membrane transporters as mediators of synaptic dopamine dynamics: Implications for disease. Eur. J. Neurosci. 2017, 45, 20–33. [Google Scholar] [CrossRef]
- Giros, B.; Jaber, M.; Jones, S.R.; Wightman, R.M.; Caron, M.G. Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature 1996, 379, 606–612. [Google Scholar] [CrossRef]
- Masoud, S.T.; Vecchio, L.M.; Bergeron, Y.; Hossain, M.M.; Nguyen, L.T.; Bermejo, M.K.; Kile, B.; Sotnikova, T.D.; Siesser, W.B.; Gainetdinov, R.R.; et al. Increased expression of the dopamine transporter leads to loss of dopamine neurons, oxidative stress and l-DOPA reversible motor deficits. Neurobiol. Dis. 2015, 74, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.W.; Erickson, J.D.; Perez, J.T.; Penland, S.N.; Mash, D.C.; Rye, D.B.; Levey, A.I. Immunochemical analysis of vesicular monoamine transporter (VMAT2) protein in Parkinson’s disease. Exp. Neurol. 1999, 156, 138–148. [Google Scholar] [CrossRef]
- Goldstein, D.S.; Sullivan, P.; Holmes, C.; Miller, G.W.; Alter, S.; Strong, R.; Mash, D.C.; Kopin, I.J.; Sharabi, Y. Determinants of buildup of the toxic dopamine metabolite DOPAL in Parkinson’s disease. J. Neurochem. 2013, 126, 591–603. [Google Scholar] [CrossRef]
- Bucher, M.L.; Barrett, C.W.; Moon, C.J.; Mortimer, A.D.; Burton, E.A.; Greenamyre, J.T.; Hastings, T.G. Acquired dysregulation of dopamine homeostasis reproduces features of Parkinson’s disease. NPJ Park. Dis. 2020, 6, 34. [Google Scholar] [CrossRef]
- Pereira Luppi, M.; Azcorra, M.; Caronia-Brown, G.; Poulin, J.F.; Gaertner, Z.; Gatica, S.; Moreno-Ramos, O.A.; Nouri, N.; Dubois, M.; Ma, Y.C.; et al. Sox6 expression distinguishes dorsally and ventrally biased dopamine neurons in the substantia nigra with distinctive properties and embryonic origins. Cell Rep. 2021, 37, 109975. [Google Scholar] [CrossRef]
- Friling, S.; Andersson, E.; Thompson, L.H.; Jönsson, M.E.; Hebsgaard, J.B.; Nanou, E.; Alekseenko, Z.; Marklund, U.; Kjellander, S.; Volakakis, N.; et al. Efficient production of mesencephalic dopamine neurons by Lmx1a expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA 2009, 106, 7613–7618. [Google Scholar] [CrossRef] [PubMed]
- Kriks, S.; Shim, J.W.; Piao, J.; Ganat, Y.M.; Wakeman, D.R.; Xie, Z.; Carrillo-Reid, L.; Auyeung, G.; Antonacci, C.; Buch, A.; et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 2011, 480, 547–551. [Google Scholar] [CrossRef]
- Niclis, J.C.; Gantner, C.W.; Alsanie, W.F.; McDougall, S.J.; Bye, C.R.; Elefanty, A.G.; Stanley, E.G.; Haynes, J.M.; Pouton, C.W.; Thompson, L.H.; et al. Efficiently Specified Ventral Midbrain Dopamine Neurons from Human Pluripotent Stem Cells Under Xeno-Free Conditions Restore Motor Deficits in Parkinsonian Rodents. Stem Cells Transl. Med. 2017, 6, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Chi, L.; Fan, B.; Zhang, K.; Du, Y.; Liu, Z.; Fang, Y.; Chen, Z.; Ren, X.; Xu, X.; Jiang, C.; et al. Targeted Differentiation of Regional Ventral Neuroprogenitors and Related Neuronal Subtypes from Human Pluripotent Stem Cells. Stem Cell Rep. 2016, 7, 941–954. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Zhang, X.J.; Renier, N.; Wu, Z.; Atkin, T.; Sun, Z.; Ozair, M.Z.; Tchieu, J.; Zimmer, B.; Fattahi, F.; et al. Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat. Biotechnol. 2017, 35, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.; Chan, C. Signal transduction of the physical environment in the neural differentiation of stem cells. Technol. Singap. World Sci. 2016, 4, 1–8. [Google Scholar] [CrossRef]
- Law, S.M.; Zheng, J.J. Premise and peril of Wnt signaling activation through GSK-3β inhibition. iScience 2022, 25, 104159. [Google Scholar] [CrossRef]
- Kirkeby, A.; Parmar, M.; Barker, R.A. Strategies for bringing stem cell-derived dopamine neurons to the clinic: A European approach (STEM-PD). Prog. Brain Res. 2017, 230, 165–190. [Google Scholar]
- Wang, Y.; Song, L.; Zhou, C.J. The canonical Wnt/β-catenin signaling pathway regulates Fgf signaling for early facial development. Dev. Biol. 2011, 349, 250–260. [Google Scholar] [CrossRef]
- Jo, J.; Xiao, Y.; Sun, A.X.; Cukuroglu, E.; Tran, H.D.; Göke, J.; Tan, Z.Y.; Saw, T.Y.; Tan, C.P.; Lokman, H.; et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell 2016, 19, 248–257. [Google Scholar] [CrossRef]
- Cheng, X.Y.; Biswas, S.; Li, J.; Mao, C.J.; Chechneva, O.; Chen, J.; Li, K.; Li, J.; Zhang, J.R.; Liu, C.F.; et al. Human iPSCs derived astrocytes rescue rotenone-induced mitochondrial dysfunction and dopaminergic neurodegeneration in vitro by donating functional mitochondria. Transl. Neurodegener. 2020, 9, 13. [Google Scholar] [CrossRef]
- de Rus Jacquet, A. Preparation and Co-Culture of iPSC-Derived Dopaminergic Neurons and Astrocytes. Curr. Protoc. Cell Biol. 2019, 85, e98. [Google Scholar] [CrossRef] [PubMed]
- de Rus Jacquet, A.; Tancredi, J.L.; Lemire, A.L.; DeSantis, M.C.; Li, W.P.; O’Shea, E.K. The LRRK2 G2019S mutation alters astrocyte-to-neuron communication via extracellular vesicles and induces neuron atrophy in a human iPSC-derived model of Parkinson’s disease. eLife 2021, 10, e73062. [Google Scholar] [CrossRef] [PubMed]
- di Domenico, A.; Carola, G.; Calatayud, C.; Pons-Espinal, M.; Muñoz, J.P.; Richaud-Patin, Y.; Fernandez-Carasa, I.; Gut, M.; Faella, A.; Parameswaran, J.; et al. Patient-Specific iPSC-Derived Astrocytes Contribute to Non-Cell-Autonomous Neurodegeneration in Parkinson’s Disease. Stem Cell Rep. 2019, 12, 213–229. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Luo, C.; Lancaster, M.A.; Castanon, R.; Nery, J.R.; Knoblich, J.A.; Ecker, J.R. Cerebral Organoids Recapitulate Epigenomic Signatures of the Human Fetal Brain. Cell Rep. 2016, 17, 3369–3384. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, J.M.; Saia-Cereda, V.M.; Sartore, R.C.; da Costa, R.M.; Schitine, C.S.; Freitas, H.R.; Murgu, M.; de Melo Reis, R.A.; Rehen, S.K.; Martins-de-Souza, D. Human Cerebral Organoids and Fetal Brain Tissue Share Proteomic Similarities. Front. Cell Dev. Biol. 2019, 7, 303. [Google Scholar] [CrossRef]
- Trujillo, C.A.; Gao, R.; Negraes, P.D.; Gu, J.; Buchanan, J.; Preissl, S.; Wang, A.; Wu, W.; Haddad, G.G.; Chaim, I.A.; et al. Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell 2019, 25, 558–569.e7. [Google Scholar] [CrossRef]
- Birey, F.; Andersen, J.; Makinson, C.D.; Islam, S.; Wei, W.; Huber, N.; Fan, H.C.; Metzler, K.R.C.; Panagiotakos, G.; Thom, N.; et al. Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Eura, N.; Matsui, T.K.; Luginbühl, J.; Matsubayashi, M.; Nanaura, H.; Shiota, T.; Kinugawa, K.; Iguchi, N.; Kiriyama, T.; Zheng, C.; et al. Brainstem Organoids from Human Pluripotent Stem Cells. Front. Neurosci. 2020, 14, 538. [Google Scholar] [CrossRef]
- Muguruma, K.; Nishiyama, A.; Kawakami, H.; Hashimoto, K.; Sasai, Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015, 10, 537–550. [Google Scholar] [CrossRef]
- Pellegrini, L.; Bonfio, C.; Chadwick, J.; Begum, F.; Skehel, M.; Lancaster, M.A. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science 2020, 369, eaaz5626. [Google Scholar] [CrossRef] [PubMed]
- Ozone, C.; Suga, H.; Eiraku, M.; Kadoshima, T.; Yonemura, S.; Takata, N.; Oiso, Y.; Tsuji, T.; Sasai, Y. Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nat. Commun. 2016, 7, 10351. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Tanaka, Y.; Cakir, B.; Patterson, B.; Kim, K.Y.; Sun, P.; Kang, Y.J.; Zhong, M.; Liu, X.; Patra, P.; et al. hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell 2019, 24, 487–497.e7. [Google Scholar] [CrossRef]
- Pomeshchik, Y.; Klementieva, O.; Gil, J.; Martinsson, I.; Hansen, M.G.; de Vries, T.; Sancho-Balsells, A.; Russ, K.; Savchenko, E.; Collin, A.; et al. Human iPSC-Derived Hippocampal Spheroids: An Innovative Tool for Stratifying Alzheimer Disease Patient-Specific Cellular Phenotypes and Developing Therapies. Stem Cell Rep. 2020, 15, 256–273. [Google Scholar] [CrossRef]
- Sakaguchi, H.; Kadoshima, T.; Soen, M.; Narii, N.; Ishida, Y.; Ohgushi, M.; Takahashi, J.; Eiraku, M.; Sasai, Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015, 6, 8896. [Google Scholar] [CrossRef] [PubMed]
- Tieng, V.; Stoppini, L.; Villy, S.; Fathi, M.; Dubois-Dauphin, M.; Krause, K.H. Engineering of midbrain organoids containing long-lived dopaminergic neurons. Stem Cells Dev. 2014, 23, 1535–1547. [Google Scholar] [CrossRef] [PubMed]
- Fiorenzano, A.; Sozzi, E.; Birtele, M.; Kajtez, J.; Giacomoni, J.; Nilsson, F.; Bruzelius, A.; Sharma, Y.; Zhang, Y.; Mattsson, B.; et al. Single-cell transcriptomics captures features of human midbrain development and dopamine neuron diversity in brain organoids. Nat. Commun. 2021, 12, 7302. [Google Scholar] [CrossRef] [PubMed]
- Perrier, A.L.; Tabar, V.; Barberi, T.; Rubio, M.E.; Bruses, J.; Topf, N.; Harrison, N.L.; Studer, L. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2004, 101, 12543–12548. [Google Scholar] [CrossRef]
- Schulz, T.C.; Noggle, S.A.; Palmarini, G.M.; Weiler, D.A.; Lyons, I.G.; Pensa, K.A.; Meedeniya, A.C.; Davidson, B.P.; Lambert, N.A.; Condie, B.G. Differentiation of human embryonic stem cells to dopaminergic neurons in serum-free suspension culture. Stem Cells 2004, 22, 1218–1238. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Yang, D.; Zarnowska, E.D.; Du, Z.; Werbel, B.; Valliere, C.; Pearce, R.A.; Thomson, J.A.; Zhang, S.C. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells 2005, 23, 781–790. [Google Scholar] [CrossRef]
- Park, C.H.; Minn, Y.K.; Lee, J.Y.; Choi, D.H.; Chang, M.Y.; Shim, J.W.; Ko, J.Y.; Koh, H.C.; Kang, M.J.; Kang, J.S.; et al. In vitro and in vivo analyses of human embryonic stem cell-derived dopamine neurons. J. Neurochem. 2005, 92, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Correia, A.S.; Anisimov, S.V.; Roybon, L.; Li, J.Y.; Brundin, P. Fibroblast growth factor-20 increases the yield of midbrain dopaminergic neurons derived from human embryonic stem cells. Front. Neuroanat. 2007, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, K.C.; Pruszak, J.; Yoshizaki, T.; van Arensbergen, J.; Sanchez-Pernaute, R.; Isacson, O. Enhanced yield of neuroepithelial precursors and midbrain-like dopaminergic neurons from human embryonic stem cells using the bone morphogenic protein antagonist noggin. Stem Cells 2007, 25, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.S.; Lee, Y.E.; Kim, J.Y.; Chung, S.; Cho, Y.H.; Kim, D.S.; Kang, S.M.; Lee, H.; Kim, M.H.; Kim, J.H.; et al. Highly efficient and large-scale generation of functional dopamine neurons from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 3392–3397. [Google Scholar] [CrossRef] [PubMed]
- Chin, M.H.; Mason, M.J.; Xie, W.; Volinia, S.; Singer, M.; Peterson, C.; Ambartsumyan, G.; Aimiuwu, O.; Richter, L.; Zhang, J.; et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 2009, 5, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Geeta, R.; Ramnath, R.L.; Rao, H.S.; Chandra, V. One year survival and significant reversal of motor deficits in parkinsonian rats transplanted with hESC derived dopaminergic neurons. Biochem. Biophys. Res. Commun. 2008, 373, 258–264. [Google Scholar] [CrossRef]
- Hayashi, H.; Morizane, A.; Koyanagi, M.; Ono, Y.; Sasai, Y.; Hashimoto, N.; Takahashi, J. Meningeal cells induce dopaminergic neurons from embryonic stem cells. Eur. J. Neurosci. 2008, 27, 261–268. [Google Scholar] [CrossRef]
- Hong, S.; Kang, U.J.; Isacson, O.; Kim, K.S. Neural precursors derived from human embryonic stem cells maintain long-term proliferation without losing the potential to differentiate into all three neural lineages, including dopaminergic neurons. J. Neurochem. 2008, 104, 316–324. [Google Scholar] [CrossRef]
- Swistowski, A.; Peng, J.; Han, Y.; Swistowska, A.M.; Rao, M.S.; Zeng, X. Xeno-free defined conditions for culture of human embryonic stem cells, neural stem cells and dopaminergic neurons derived from them. PLoS ONE 2009, 4, e6233. [Google Scholar] [CrossRef]
- Cai, J.; Yang, M.; Poremsky, E.; Kidd, S.; Schneider, J.S.; Iacovitti, L. Dopaminergic neurons derived from human induced pluripotent stem cells survive and integrate into 6-OHDA-lesioned rats. Stem Cells Dev. 2010, 19, 1017–1023. [Google Scholar] [CrossRef]
- Morizane, A.; Darsalia, V.; Guloglu, M.O.; Hjalt, T.; Carta, M.; Li, J.Y.; Brundin, P. A simple method for large-scale generation of dopamine neurons from human embryonic stem cells. J. Neurosci. Res. 2010, 88, 3467–3478. [Google Scholar] [CrossRef] [PubMed]
- Swistowska, A.M.; da Cruz, A.B.; Han, Y.; Swistowski, A.; Liu, Y.; Shin, S.; Zhan, M.; Rao, M.S.; Zeng, X. Stage-specific role for shh in dopaminergic differentiation of human embryonic stem cells induced by stromal cells. Stem Cells Dev. 2010, 19, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Swistowski, A.; Peng, J.; Liu, Q.; Mali, P.; Rao, M.S.; Cheng, L.; Zeng, X. Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells 2010, 28, 1893–1904. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Morizane, A.; Doi, D.; Onoe, H.; Hayashi, T.; Kawasaki, T.; Saiki, H.; Miyamoto, S.; Takahashi, J. Survival of human induced pluripotent stem cell-derived midbrain dopaminergic neurons in the brain of a primate model of Parkinson’s disease. J. Park. Dis. 2011, 1, 395–412. [Google Scholar] [CrossRef]
- Wu, S.M.; Tan, K.S.; Chen, H.; Beh, T.T.; Yeo, H.C.; Ng, S.K.; Wei, S.; Lee, D.Y.; Choo, A.B.; Chan, K.K. Enhanced production of neuroprogenitors, dopaminergic neurons, and identification of target genes by overexpression of sonic hedgehog in human embryonic stem cells. Stem Cells Dev. 2012, 21, 729–741. [Google Scholar] [CrossRef]
- Daadi, M.M.; Grueter, B.A.; Malenka, R.C.; Redmond, D.E., Jr.; Steinberg, G.K. Dopaminergic neurons from midbrain-specified human embryonic stem cell-derived neural stem cells engrafted in a monkey model of Parkinson’s disease. PLoS ONE 2012, 7, e41120. [Google Scholar] [CrossRef]
- Mak, S.K.; Huang, Y.A.; Iranmanesh, S.; Vangipuram, M.; Sundararajan, R.; Nguyen, L.; Langston, J.W.; Schule, B. Small molecules greatly improve conversion of human-induced pluripotent stem cells to the neuronal lineage. Stem Cells Int. 2012, 2012, 140427. [Google Scholar] [CrossRef]
- Sánchez-Danés, A.; Richaud-Patin, Y.; Carballo-Carbajal, I.; Jiménez-Delgado, S.; Caig, C.; Mora, S.; Di Guglielmo, C.; Ezquerra, M.; Patel, B.; Giralt, A.; et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol. Med. 2012, 4, 380–395. [Google Scholar] [CrossRef]
- Schwartz, C.M.; Tavakoli, T.; Jamias, C.; Park, S.S.; Maudsley, S.; Martin, B.; Phillips, T.M.; Yao, P.J.; Itoh, K.; Ma, W.; et al. Stromal factors SDF1alpha, sFRP1, and VEGFD induce dopaminergic neuron differentiation of human pluripotent stem cells. J. Neurosci. Res. 2012, 90, 1367–1381. [Google Scholar] [CrossRef]
- Xi, J.; Liu, Y.; Liu, H.; Chen, H.; Emborg, M.E.; Zhang, S.C. Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells 2012, 30, 1655–1663. [Google Scholar] [CrossRef]
- Doi, D.; Morizane, A.; Kikuchi, T.; Onoe, H.; Hayashi, T.; Kawasaki, T.; Motono, M.; Sasai, Y.; Saiki, H.; Gomi, M.; et al. Prolonged maturation culture favors a reduction in the tumorigenicity and the dopaminergic function of human ESC-derived neural cells in a primate model of Parkinson’s disease. Stem Cells 2012, 30, 935–945. [Google Scholar] [CrossRef]
- Kim, J.; Sachdev, P.; Sidhu, K. Alginate microcapsule as a 3D platform for the efficient differentiation of human embryonic stem cells to dopamine neurons. Stem Cell Res. 2013, 11, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Pedersen, O.Z.; Peng, J.; Couture, L.A.; Rao, M.S.; Zeng, X. Optimizing dopaminergic differentiation of pluripotent stem cells for the manufacture of dopaminergic neurons for transplantation. Cytotherapy 2013, 15, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Theka, I.; Caiazzo, M.; Dvoretskova, E.; Leo, D.; Ungaro, F.; Curreli, S.; Manago, F.; Dell’Anno, M.T.; Pezzoli, G.; Gainetdinov, R.R.; et al. Rapid generation of functional dopaminergic neurons from human induced pluripotent stem cells through a single-step procedure using cell lineage transcription factors. Stem Cells Transl. Med. 2013, 2, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, M.; Bogetofte, H.; Lawson, T.; Jansson, J.; Smith, G.; Astradsson, A.; Moore, M.; Osborn, T.; Cooper, O.; Spealman, R.; et al. Improved cell therapy protocols for Parkinson’s disease based on differentiation efficiency and safety of hESC-, hiPSC-, and non-human primate iPSC-derived dopaminergic neurons. Stem Cells 2013, 31, 1548–1562. [Google Scholar] [CrossRef] [PubMed]
- Hartfield, E.M.; Yamasaki-Mann, M.; Ribeiro Fernandes, H.J.; Vowles, J.; James, W.S.; Cowley, S.A.; Wade-Martins, R. Physiological characterisation of human iPS-derived dopaminergic neurons. PLoS ONE 2014, 9, e87388. [Google Scholar] [CrossRef] [PubMed]
- Sagal, J.; Zhan, X.; Xu, J.; Tilghman, J.; Karuppagounder, S.S.; Chen, L.; Dawson, V.L.; Dawson, T.M.; Laterra, J.; Ying, M. Proneural transcription factor Atoh1 drives highly efficient differentiation of human pluripotent stem cells into dopaminergic neurons. Stem Cells Transl. Med. 2014, 3, 888–898. [Google Scholar] [CrossRef] [PubMed]
- Stanslowsky, N.; Haase, A.; Martin, U.; Naujock, M.; Leffler, A.; Dengler, R.; Wegner, F. Functional differentiation of midbrain neurons from human cord blood-derived induced pluripotent stem cells. Stem Cell Res. Ther. 2014, 5, 35. [Google Scholar] [CrossRef]
- Yang, F.; Liu, Y.; Tu, J.; Wan, J.; Zhang, J.; Wu, B.; Chen, S.; Zhou, J.; Mu, Y.; Wang, L. Activated astrocytes enhance the dopaminergic differentiation of stem cells and promote brain repair through bFGF. Nat. Commun. 2014, 5, 5627. [Google Scholar] [CrossRef]
- Komatsu, M.; Konagaya, S.; Egawa, E.Y.; Iwata, H. Maturation of human iPS cell-derived dopamine neuron precursors in alginate-Ca(2+) hydrogel. Biochim. Biophys. Acta 2015, 1850, 1669–1675. [Google Scholar] [CrossRef]
- Lim, M.S.; Shin, M.S.; Lee, S.Y.; Minn, Y.K.; Hoh, J.K.; Cho, Y.H.; Kim, D.W.; Lee, S.H.; Kim, C.H.; Park, C.H. Noggin Over-Expressing Mouse Embryonic Fibroblasts and MS5 Stromal Cells Enhance Directed Differentiation of Dopaminergic Neurons from Human Embryonic Stem Cells. PLoS ONE 2015, 10, e0138460. [Google Scholar] [CrossRef] [PubMed]
- Kirkeby, A.; Grealish, S.; Wolf, D.A.; Nelander, J.; Wood, J.; Lundblad, M.; Lindvall, O.; Parmar, M. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 2012, 1, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, P.; Glatza, M.; Hemmer, K.; Tsytsyura, Y.; Thiel, C.S.; Höing, S.; Moritz, S.; Parga, J.A.; Wagner, L.; Bruder, J.M.; et al. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS ONE 2013, 8, e59252. [Google Scholar] [CrossRef]
- Kikuchi, T.; Morizane, A.; Doi, D.; Magotani, H.; Onoe, H.; Hayashi, T.; Mizuma, H.; Takara, S.; Takahashi, R.; Inoue, H.; et al. Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature 2017, 548, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Nolbrant, S.; Heuer, A.; Parmar, M.; Kirkeby, A. Generation of high-purity human ventral midbrain dopaminergic progenitors for in vitro maturation and intracerebral transplantation. Nat. Protoc. 2017, 12, 1962–1979. [Google Scholar] [CrossRef]
- Fedele, S.; Collo, G.; Behr, K.; Bischofberger, J.; Muller, S.; Kunath, T.; Christensen, K.; Gundner, A.L.; Graf, M.; Jagasia, R.; et al. Expansion of human midbrain floor plate progenitors from induced pluripotent stem cells increases dopaminergic neuron differentiation potential. Sci. Rep. 2017, 7, 6036. [Google Scholar] [CrossRef]
- Deflorio, C.; Blanchard, S.; Carisi, M.C.; Bohl, D.; Maskos, U. Human polymorphisms in nicotinic receptors: A functional analysis in iPS-derived dopaminergic neurons. FASEB J. 2017, 31, 828–839. [Google Scholar] [CrossRef]
- Adil, M.M.; Rodrigues, G.M.; Kulkarni, R.U.; Rao, A.T.; Chernavsky, N.E.; Miller, E.W.; Schaffer, D.V. Efficient generation of hPSC-derived midbrain dopaminergic neurons in a fully defined, scalable, 3D biomaterial platform. Sci. Rep. 2017, 7, 40573. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Li, Q.; Lei, Y. Three-dimensional tissues using human pluripotent stem cell spheroids as biofabrication building blocks. Biofabrication 2017, 9, 025007. [Google Scholar] [CrossRef] [PubMed]
- Adil, M.M.; Schaffer, D.V. hPSC-derived Midbrain Dopaminergic Neurons Generated in a Scalable 3-D Biomaterial. Curr. Protoc. Stem Cell Biol. 2018, 44, 2D.21.1–2D.21.17. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, V.M.; Salti, A.; Tilleman, H.; Zega, K.; Jukic, M.M.; Zou, H.; Friedel, R.H.; Prakash, N.; Blaess, S.; Edenhofer, F.; et al. BMP/SMAD Pathway Promotes Neurogenesis of Midbrain Dopaminergic Neurons In Vivo and in Human Induced Pluripotent and Neural Stem Cells. J. Neurosci. 2018, 38, 1662–1676. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.K.B.; Lim, W.W.M.; Chai, C.; Kukumberg, M.; Lim, K.L.; Goh, E.L.K.; Yim, E.K.F. Sequential Application of Discrete Topographical Patterns Enhances Derivation of Functional Mesencephalic Dopaminergic Neurons from Human Induced Pluripotent Stem Cells. Sci. Rep. 2018, 8, 9567. [Google Scholar] [CrossRef] [PubMed]
- Paik, E.J.; O’Neil, A.L.; Ng, S.Y.; Sun, C.; Rubin, L.L. Using intracellular markers to identify a novel set of surface markers for live cell purification from a heterogeneous hIPSC culture. Sci. Rep. 2018, 8, 804. [Google Scholar] [CrossRef] [PubMed]
- Sommer, A.; Marxreiter, F.; Krach, F.; Fadler, T.; Grosch, J.; Maroni, M.; Graef, D.; Eberhardt, E.; Riemenschneider, M.J.; Yeo, G.W.; et al. Th17 Lymphocytes Induce Neuronal Cell Death in a Human iPSC-Based Model of Parkinson’s Disease. Cell Stem Cell 2018, 23, 123–131.e6. [Google Scholar] [CrossRef] [PubMed]
- Arioka, Y.; Shishido, E.; Kubo, H.; Kushima, I.; Yoshimi, A.; Kimura, H.; Ishizuka, K.; Aleksic, B.; Maeda, T.; Ishikawa, M.; et al. Single-cell trajectory analysis of human homogenous neurons carrying a rare RELN variant. Transl. Psychiatry 2018, 8, 129. [Google Scholar] [CrossRef]
- Tofoli, F.A.; Semeano, A.T.S.; Oliveira-Giacomelli, A.; Goncalves, M.C.B.; Ferrari, M.F.R.; Veiga Pereira, L.; Ulrich, H. Midbrain Dopaminergic Neurons Differentiated from Human-Induced Pluripotent Stem Cells. Methods Mol. Biol. 2019, 1919, 97–118. [Google Scholar] [PubMed]
- Xue, Y.; Zhan, X.; Sun, S.; Karuppagounder, S.S.; Xia, S.; Dawson, V.L.; Dawson, T.M.; Laterra, J.; Zhang, J.; Ying, M. Synthetic mRNAs Drive Highly Efficient iPS Cell Differentiation to Dopaminergic Neurons. Stem Cells Transl. Med. 2019, 8, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Leitner, D.; Ramamoorthy, M.; Dejosez, M.; Zwaka, T.P. Immature mDA neurons ameliorate motor deficits in a 6-OHDA Parkinson’s disease mouse model and are functional after cryopreservation. Stem Cell Res. 2019, 41, 101617. [Google Scholar] [CrossRef]
- Uberbacher, C.; Obergasteiger, J.; Volta, M.; Venezia, S.; Muller, S.; Pesce, I.; Pizzi, S.; Lamonaca, G.; Picard, A.; Cattelan, G.; et al. Application of CRISPR/Cas9 editing and digital droplet PCR in human iPSCs to generate novel knock-in reporter lines to visualize dopaminergic neurons. Stem Cell Res. 2019, 41, 101656. [Google Scholar] [CrossRef]
- Schweitzer, J.S.; Song, B.; Herrington, T.M.; Park, T.Y.; Lee, N.; Ko, S.; Jeon, J.; Cha, Y.; Kim, K.; Li, Q.; et al. Personalized iPSC-Derived Dopamine Progenitor Cells for Parkinson’s Disease. N. Engl. J. Med. 2020, 382, 1926–1932. [Google Scholar] [CrossRef]
- Stathakos, P.; Jimenez-Moreno, N.; Crompton, L.A.; Nistor, P.A.; Badger, J.L.; Barbuti, P.A.; Kerrigan, T.L.; Randall, A.D.; Caldwell, M.A.; Lane, J.D. A monolayer hiPSC culture system for autophagy/mitophagy studies in human dopaminergic neurons. Autophagy 2021, 17, 855–871. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Cha, Y.; Ko, S.; Jeon, J.; Lee, N.; Seo, H.; Park, K.J.; Lee, I.H.; Lopes, C.; Feitosa, M.; et al. Human autologous iPSC-derived dopaminergic progenitors restore motor function in Parkinson’s disease models. J. Clin. Investig. 2020, 130, 904–920. [Google Scholar] [CrossRef] [PubMed]
- Drummond, N.J.; Singh Dolt, K.; Canham, M.A.; Kilbride, P.; Morris, G.J.; Kunath, T. Cryopreservation of Human Midbrain Dopaminergic Neural Progenitor Cells Poised for Neuronal Differentiation. Front. Cell Dev. Biol. 2020, 8, 578907. [Google Scholar] [CrossRef]
- Dhingra, A.; Tager, J.; Bressan, E.; Rodriguez-Nieto, S.; Bedi, M.S.; Broer, S.; Sadikoglou, E.; Fernandes, N.; Castillo-Lizardo, M.; Rizzu, P.; et al. Automated Production of Human Induced Pluripotent Stem Cell-Derived Cortical and Dopaminergic Neurons with Integrated Live-Cell Monitoring. J. Vis. Exp. 2020, 162, e61525. [Google Scholar]
- Yamaguchi, A.; Ishikawa, K.I.; Inoshita, T.; Shiba-Fukushima, K.; Saiki, S.; Hatano, T.; Mori, A.; Oji, Y.; Okuzumi, A.; Li, Y.; et al. Identifying Therapeutic Agents for Amelioration of Mitochondrial Clearance Disorder in Neurons of Familial Parkinson Disease. Stem Cell Rep. 2020, 14, 1060–1075. [Google Scholar] [CrossRef]
- Jefri, M.; Bell, S.; Peng, H.; Hettige, N.; Maussion, G.; Soubannier, V.; Wu, H.; Silveira, H.; Theroux, J.F.; Moquin, L.; et al. Stimulation of L-type calcium channels increases tyrosine hydroxylase and dopamine in ventral midbrain cells induced from somatic cells. Stem Cells Transl. Med. 2020, 9, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Piao, J.; Zabierowski, S.; Dubose, B.N.; Hill, E.J.; Navare, M.; Claros, N.; Rosen, S.; Ramnarine, K.; Horn, C.; Fredrickson, C.; et al. Preclinical Efficacy and Safety of a Human Embryonic Stem Cell-Derived Midbrain Dopamine Progenitor Product, MSK-DA01. Cell Stem Cell 2021, 28, 217–229.e7. [Google Scholar] [CrossRef]
- Kim, T.W.; Piao, J.; Koo, S.Y.; Kriks, S.; Chung, S.Y.; Betel, D.; Socci, N.D.; Choi, S.J.; Zabierowski, S.; Dubose, B.N.; et al. Biphasic Activation of WNT Signaling Facilitates the Derivation of Midbrain Dopamine Neurons from hESCs for Translational Use. Cell Stem Cell 2021, 28, 343–355.e5. [Google Scholar] [CrossRef]
- Walter, J.; Bolognin, S.; Poovathingal, S.K.; Magni, S.; Gérard, D.; Antony, P.M.A.; Nickels, S.L.; Salamanca, L.; Berger, E.; Smits, L.M.; et al. The Parkinson’s-disease-associated mutation LRRK2-G2019S alters dopaminergic differentiation dynamics via NR2F1. Cell Rep. 2021, 37, 109864. [Google Scholar] [CrossRef]
- Ishikawa, K.I.; Nonaka, R.; Akamatsu, W. Differentiation of Midbrain Dopaminergic Neurons from Human iPS Cells. Methods Mol. Biol. 2021, 2322, 73–80. [Google Scholar]
- Tong, Z.; Kwak, E.; Aguiar, A.; Peng, B.; Pouton, C.W.; Voelcker, N.H.; Haynes, J.M. Compartmentalized microfluidic chambers enable long-term maintenance and communication between human pluripotent stem cell-derived forebrain and midbrain neurons. Lab Chip 2021, 21, 4016–4030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Q.; Zhang, S.C. Differentiation of neural precursors and dopaminergic neurons from human embryonic stem cells. Methods Mol. Biol. 2010, 584, 355–366. [Google Scholar] [PubMed]
- Sato, T.; Imaizumi, K.; Watanabe, H.; Ishikawa, M.; Okano, H. Generation of region-specific and high-purity neurons from human feeder-free iPSCs. Neurosci. Lett. 2021, 746, 135676. [Google Scholar] [CrossRef] [PubMed]
- Rossignoli, G.; Kramer, K.; Lugara, E.; Alrashidi, H.; Pope, S.; De La Fuente Barrigon, C.; Barwick, K.; Bisello, G.; Ng, J.; Counsell, J.; et al. Aromatic l-amino acid decarboxylase deficiency: A patient-derived neuronal model for precision therapies. Brain 2021, 144, 2443–2456. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Owens, M.J.S.; Toledo, E.M.; Arenas, E.; Bradley, M.; Ffrench-Constant, C. Combinatorial ECM Arrays Identify Cooperative Roles for Matricellular Proteins in Enhancing the Generation of TH+ Neurons from Human Pluripotent Cells. Front. Cell Dev. Biol. 2021, 9, 755406. [Google Scholar] [CrossRef]
- Xu, P.; He, H.; Gao, Q.; Zhou, Y.; Wu, Z.; Zhang, X.; Sun, L.; Hu, G.; Guan, Q.; You, Z.; et al. Human midbrain dopaminergic neuronal differentiation markers predict cell therapy outcomes in a Parkinson’s disease model. J. Clin. Investig. 2022, 132, 156768. [Google Scholar] [CrossRef]
- Tay, S.H.; Winanto; Khong, Z.J.; Koh, Y.H.; Ng, S.Y. Generation of Cortical, Dopaminergic, Motor, and Sensory Neurons from Human Pluripotent Stem Cells. Methods Mol. Biol. 2022, 2549, 359–377. [Google Scholar] [PubMed]
- Li, H.; Jiang, H.; Li, H.; Li, L.; Yan, Z.; Feng, J. Generation of human A9 dopaminergic pacemakers from induced pluripotent stem cells. Mol. Psychiatry 2022, 27, 4407–4418. [Google Scholar] [CrossRef]
- Virdi, G.S.; Choi, M.L.; Evans, J.R.; Yao, Z.; Athauda, D.; Strohbuecker, S.; Nirujogi, R.S.; Wernick, A.I.; Pelegrina-Hidalgo, N.; Leighton, C.; et al. Protein aggregation and calcium dysregulation are hallmarks of familial Parkinson’s disease in midbrain dopaminergic neurons. NPJ Park. Dis. 2022, 8, 162. [Google Scholar] [CrossRef]
- Hiller, B.M.; Marmion, D.J.; Thompson, C.A.; Elliott, N.A.; Federoff, H.; Brundin, P.; Mattis, V.B.; McMahon, C.W.; Kordower, J.H. Optimizing maturity and dose of iPSC-derived dopamine progenitor cell therapy for Parkinson’s disease. NPJ Regen. Med. 2022, 7, 24. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Ishikawa, K.I.; Akamatsu, W. Methods to Induce Small-Scale Differentiation of iPS Cells into Dopaminergic Neurons and to Detect Disease Phenotypes. Methods Mol. Biol. 2022, 2549, 271–279. [Google Scholar] [PubMed]
- Alekseenko, Z.; Dias, J.M.; Adler, A.F.; Kozhevnikova, M.; van Lunteren, J.A.; Nolbrant, S.; Jeggari, A.; Vasylovska, S.; Yoshitake, T.; Kehr, J.; et al. Robust derivation of transplantable dopamine neurons from human pluripotent stem cells by timed retinoic acid delivery. Nat. Commun. 2022, 13, 3046. [Google Scholar] [CrossRef] [PubMed]
- Monzel, A.S.; Smits, L.M.; Hemmer, K.; Hachi, S.; Moreno, E.L.; van Wuellen, T.; Jarazo, J.; Walter, J.; Brüggemann, I.; Boussaad, I.; et al. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Rep. 2017, 8, 1144–1154. [Google Scholar] [CrossRef] [PubMed]
- Berger, E.; Magliaro, C.; Paczia, N.; Monzel, A.S.; Antony, P.; Linster, C.L.; Bolognin, S.; Ahluwalia, A.; Schwamborn, J.C. Millifluidic culture improves human midbrain organoid vitality and differentiation. Lab Chip 2018, 18, 3172–3183. [Google Scholar] [CrossRef]
- Smits, L.M.; Reinhardt, L.; Reinhardt, P.; Glatza, M.; Monzel, A.S.; Stanslowsky, N.; Rosato-Siri, M.D.; Zanon, A.; Antony, P.M.; Bellmann, J.; et al. Modeling Parkinson’s disease in midbrain-like organoids. NPJ Parkinsons Dis. 2019, 5, 5. [Google Scholar] [CrossRef]
- Chlebanowska, P.; Tejchman, A.; Sułkowski, M.; Skrzypek, K.; Majka, M. Use of 3D Organoids as a Model to Study Idiopathic Form of Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 694. [Google Scholar] [CrossRef]
- Kano, M.; Takanashi, M.; Oyama, G.; Yoritaka, A.; Hatano, T.; Shiba-Fukushima, K.; Nagai, M.; Nishiyama, K.; Hasegawa, K.; Inoshita, T.; et al. Reduced astrocytic reactivity in human brains and midbrain organoids with PRKN mutations. NPJ Parkinsons Dis. 2020, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Kwak, T.H.; Kang, J.H.; Hali, S.; Kim, J.; Kim, K.P.; Park, C.; Lee, J.H.; Ryu, H.K.; Na, J.E.; Jo, J.; et al. Generation of homogeneous midbrain organoids with in vivo-like cellular composition facilitates neurotoxin-based Parkinson’s disease modeling. Stem Cells 2020, 38, 727–740. [Google Scholar] [CrossRef]
- Monzel, A.S.; Hemmer, K.; Kaoma, T.; Smits, L.M.; Bolognin, S.; Lucarelli, P.; Rosety, I.; Zagare, A.; Antony, P.; Nickels, S.L.; et al. Machine learning-assisted neurotoxicity prediction in human midbrain organoids. Park. Relat. Disord. 2020, 75, 105–109. [Google Scholar] [CrossRef]
- Ha, J.; Kang, J.S.; Lee, M.; Baek, A.; Kim, S.; Chung, S.K.; Lee, M.O.; Kim, J. Simplified Brain Organoids for Rapid and Robust Modeling of Brain Disease. Front. Cell Dev. Biol. 2020, 8, 594090. [Google Scholar] [CrossRef]
- Nickels, S.L.; Modamio, J.; Mendes-Pinheiro, B.; Monzel, A.S.; Betsou, F.; Schwamborn, J.C. Reproducible generation of human midbrain organoids for in vitro modeling of Parkinson’s disease. Stem Cell Res. 2020, 46, 101870. [Google Scholar] [CrossRef] [PubMed]
- Renner, H.; Grabos, M.; Becker, K.J.; Kagermeier, T.E.; Wu, J.; Otto, M.; Peischard, S.; Zeuschner, D.; TsyTsyura, Y.; Disse, P.; et al. A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. eLife 2020, 9, e52904. [Google Scholar] [CrossRef] [PubMed]
- Tejchman, A.; Znój, A.; Chlebanowska, P.; Frączek-Szczypta, A.; Majka, M. Carbon Fibers as a New Type of Scaffold for Midbrain Organoid Development. Int. J. Mol. Sci. 2020, 21, 5959. [Google Scholar] [CrossRef] [PubMed]
- Boussaad, I.; Cruciani, G.; Bolognin, S.; Antony, P.; Dording, C.M.; Kwon, Y.J.; Heutink, P.; Fava, E.; Schwamborn, J.C.; Krüger, R. Integrated, automated maintenance, expansion and differentiation of 2D and 3D patient-derived cellular models for high throughput drug screening. Sci. Rep. 2021, 11, 1439. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, N.V.; Sirois, J.; Ramamurthy, J.; Mathur, M.; Lépine, P.; Deneault, E.; Maussion, G.; Nicouleau, M.; Chen, C.X.; Abdian, N.; et al. Midbrain organoids with an SNCA gene triplication model key features of synucleinopathy. Brain Commun. 2021, 3, fcab223. [Google Scholar] [CrossRef]
- Kim, S.W.; Woo, H.J.; Kim, E.H.; Kim, H.S.; Suh, H.N.; Kim, S.H.; Song, J.J.; Wulansari, N.; Kang, M.; Choi, S.Y.; et al. Neural stem cells derived from human midbrain organoids as a stable source for treating Parkinson’s disease: Midbrain organoid-NSCs (Og-NSC) as a stable source for PD treatment. Prog. Neurobiol. 2021, 204, 102086. [Google Scholar] [CrossRef]
- Renner, H.; Becker, K.J.; Kagermeier, T.E.; Grabos, M.; Eliat, F.; Günther, P.; Schöler, H.R.; Bruder, J.M. Cell-Type-Specific High Throughput Toxicity Testing in Human Midbrain Organoids. Front. Mol. Neurosci. 2021, 14, 715054. [Google Scholar] [CrossRef]
- Jo, J.; Yang, L.; Tran, H.D.; Yu, W.; Sun, A.X.; Chang, Y.Y.; Jung, B.C.; Lee, S.J.; Saw, T.Y.; Xiao, B.; et al. Lewy Body-like Inclusions in Human Midbrain Organoids Carrying Glucocerebrosidase and α-Synuclein Mutations. Ann. Neurol. 2021, 90, 490–505. [Google Scholar] [CrossRef]
- Sarrafha, L.; Parfitt, G.M.; Reyes, R.; Goldman, C.; Coccia, E.; Kareva, T.; Ahfeldt, T. High-throughput generation of midbrain dopaminergic neuron organoids from reporter human pluripotent stem cells. STAR Protoc. 2021, 2, 100463. [Google Scholar] [CrossRef]
- Wulansari, N.; Darsono, W.H.W.; Woo, H.J.; Chang, M.Y.; Kim, J.; Bae, E.J.; Sun, W.; Lee, J.H.; Cho, I.J.; Shin, H.; et al. Neurodevelopmental defects and neurodegenerative phenotypes in human brain organoids carrying Parkinson’s disease-linked DNAJC6 mutations. Sci. Adv. 2021, 7, eabb1540. [Google Scholar] [CrossRef]
- Zanetti, C.; Spitz, S.; Berger, E.; Bolognin, S.; Smits, L.M.; Crepaz, P.; Rothbauer, M.; Rosser, J.M.; Marchetti-Deschmann, M.; Schwamborn, J.C.; et al. Monitoring the neurotransmitter release of human midbrain organoids using a redox cycling microsensor as a novel tool for personalized Parkinson’s disease modelling and drug screening. Analyst 2021, 146, 2358–2367. [Google Scholar] [CrossRef]
- Dong, J.; Duchesne, A.; Bayne, A.N.; Mohamed, N.V.; Yi, W.; Mathur, M.; Chen, C.X.Q.; You, Z.; Abdian, N.; Taylor, L.; et al. An approach to measuring protein turnover in human induced pluripotent stem cell organoids by mass spectrometry. Methods 2022, 203, 17–27. [Google Scholar] [CrossRef]
- Jarazo, J.; Barmpa, K.; Modamio, J.; Saraiva, C.; Sabaté-Soler, S.; Rosety, I.; Griesbeck, A.; Skwirblies, F.; Zaffaroni, G.; Smits, L.M.; et al. Parkinson’s Disease Phenotypes in Patient Neuronal Cultures and Brain Organoids Improved by 2-Hydroxypropyl-β-Cyclodextrin Treatment. Mov. Disord. 2022, 37, 80–94. [Google Scholar] [CrossRef]
- Lee, Y.; Kang, J.S.; Ham, O.J.; Son, M.Y.; Lee, M.O. Gut metabolite trimethylamine N-oxide induces aging-associated phenotype of midbrain organoids for the induced pluripotent stem cell-based modeling of late-onset disease. Front. Aging Neurosci. 2022, 14, 925227. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, N.V.; Lépine, P.; Lacalle-Aurioles, M.; Sirois, J.; Mathur, M.; Reintsch, W.; Beitel, L.K.; Fon, E.A.; Durcan, T.M. Microfabricated disk technology: Rapid scale up in midbrain organoid generation. Methods 2022, 203, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Pamies, D.; Wiersma, D.; Katt, M.E.; Zhao, L.; Burtscher, J.; Harris, G.; Smirnova, L.; Searson, P.C.; Hartung, T.; Hogberg, H.T. Human IPSC 3D brain model as a tool to study chemical-induced dopaminergic neuronal toxicity. Neurobiol. Dis. 2022, 169, 105719. [Google Scholar] [CrossRef] [PubMed]
- Sabate-Soler, S.; Nickels, S.L.; Saraiva, C.; Berger, E.; Dubonyte, U.; Barmpa, K.; Lan, Y.J.; Kouno, T.; Jarazo, J.; Robertson, G.; et al. Microglia integration into human midbrain organoids leads to increased neuronal maturation and functionality. Glia 2022, 70, 1267–1288. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Li, B.; Fan, H.; Liu, P.; Zhao, W.; Chen, T.; Chen, P.; Yang, L. Sevoflurane promotes premature differentiation of dopaminergic neurons in hiPSC-derived midbrain organoids. Front. Cell Dev. Biol. 2022, 10, 941984. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, E.; Nilsson, F.; Kajtez, J.; Parmar, M.; Fiorenzano, A. Generation of Human Ventral Midbrain Organoids Derived from Pluripotent Stem Cells. Curr. Protoc. 2022, 2, e555. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Tao, M.; Hong, Y.; Wu, S.; Chu, C.; Zheng, Z.; Han, X.; Zhu, Q.; Xu, M.; Ewing, A.G.; et al. Dysfunction of vesicular storage in young-onset Parkinson’s patient-derived dopaminergic neurons and organoids revealed by single cell electrochemical cytometry. Chem. Sci. 2022, 13, 6217–6223. [Google Scholar] [CrossRef]
- Morizane, A.; Doi, D.; Kikuchi, T.; Nishimura, K.; Takahashi, J. Small-molecule inhibitors of bone morphogenic protein and activin/nodal signals promote highly efficient neural induction from human pluripotent stem cells. J. Neurosci. Res. 2011, 89, 117–126. [Google Scholar] [CrossRef]
- Tojo, M.; Hamashima, Y.; Hanyu, A.; Kajimoto, T.; Saitoh, M.; Miyazono, K.; Node, M.; Imamura, T. The ALK-5 inhibitor A-83-01 inhibits Smad signaling and epithelial-to-mesenchymal transition by transforming growth factor-beta. Cancer Sci. 2005, 96, 791–800. [Google Scholar] [CrossRef]
- Zhang, J.; Götz, S.; Vogt Weisenhorn, D.M.; Simeone, A.; Wurst, W.; Prakash, N. A WNT1-regulated developmental gene cascade prevents dopaminergic neurodegeneration in adult En1(+/−) mice. Neurobiol. Dis. 2015, 82, 32–45. [Google Scholar] [CrossRef]
- Shao, S.; Wang, G.L.; Raymond, C.; Deng, X.H.; Zhu, X.L.; Wang, D.; Hong, L.P. Activation of Sonic hedgehog signal by Purmorphamine, in a mouse model of Parkinson’s disease, protects dopaminergic neurons and attenuates inflammatory response by mediating PI3K/AKt signaling pathway. Mol. Med. Rep. 2017, 16, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.K.; Taipale, J.; Young, K.E.; Maiti, T.; Beachy, P.A. Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA 2002, 99, 14071–14076. [Google Scholar] [CrossRef] [PubMed]
- Hébert, J.M. FGFs: Neurodevelopment’s Jack-of-all-Trades-How Do They Do it? Front. Neurosci. 2011, 5, 133. [Google Scholar] [CrossRef]
- Gonzalez-Quevedo, R.; Lee, Y.; Poss, K.D.; Wilkinson, D.G. Neuronal Regulation of the Spatial Patterning of Neurogenesis. Dev. Cell 2010, 18, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Ng, Y.H.; Chanda, S.; Janas, J.A.; Yang, N.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Efficient generation of dopaminergic induced neuronal cells with midbrain characteristics. Stem Cell Rep. 2021, 16, 1763–1776. [Google Scholar] [CrossRef]
- Poulsen, K.T.; Armanini, M.P.; Klein, R.D.; Hynes, M.A.; Phillips, H.S.; Rosenthal, A. TGFβ2 and TGFβ3 are potent survival factors for midbrain dopaminergic neurons. Neuron 1994, 13, 1245–1252. [Google Scholar] [CrossRef]
- Rharass, T.; Lantow, M.; Gbankoto, A.; Weiss, D.G.; Panáková, D.; Lucas, S. Ascorbic acid alters cell fate commitment of human neural progenitors in a WNT/β-catenin/ROS signaling dependent manner. J. Biomed. Sci. 2017, 24, 78. [Google Scholar] [CrossRef]
- Stayte, S.; Rentsch, P.; Li, K.M.; Vissel, B. Activin A protects midbrain neurons in the 6-hydroxydopamine mouse model of Parkinson’s disease. PLoS ONE 2015, 10, e0124325. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, W.; Zhang, Y.; Wei, W.; Ambasudhan, R.; Xia, P.; Talantova, M.; Lin, T.; Kim, J.; Wang, X.; et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc. Natl. Acad. Sci. USA 2011, 108, 8299–8304. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Kobayashi, Y.; Inoshita, T.; Meng, H.; Arano, T.; Uemura, K.; Asano, T.; Yoshimi, K.; Zhang, C.L.; Matsumoto, G.; et al. The Parkinson’s Disease-Associated Protein Kinase LRRK2 Modulates Notch Signaling through the Endosomal Pathway. PLoS Genet. 2015, 11, e1005503. [Google Scholar] [CrossRef]
- Kurokawa, D.; Ohmura, T.; Sakurai, Y.; Inoue, K.; Suda, Y.; Aizawa, S. Otx2 expression in anterior neuroectoderm and forebrain/midbrain is directed by more than six enhancers. Dev. Biol. 2014, 387, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Ables, J.L.; Dickel, L.K.; Eisch, A.J.; Johnson, J.E. Ascl1 (Mash1) Defines Cells with Long-Term Neurogenic Potential in Subgranular and Subventricular Zones in Adult Mouse Brain. PLoS ONE 2011, 6, e18472. [Google Scholar] [CrossRef]
- Ellis, P.; Fagan, B.M.; Magness, S.T.; Hutton, S.; Taranova, O.; Hayashi, S.; McMahon, A.; Rao, M.; Pevny, L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev. Neurosci. 2004, 26, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Soltani, M.H.; Pichardo, R.; Song, Z.; Sangha, N.; Camacho, F.; Satyamoorthy, K.; Sangueza, O.P.; Setaluri, V. Microtubule-associated protein 2, a marker of neuronal differentiation, induces mitotic defects, inhibits growth of melanoma cells, and predicts metastatic potential of cutaneous melanoma. Am. J. Pathol. 2005, 166, 1841–1850. [Google Scholar] [CrossRef] [PubMed]
- Crawford, T.Q.; Roelink, H. The Notch response inhibitor DAPT enhances neuronal differentiation in embryonic stem cell-derived embryoid bodies independently of sonic hedgehog signaling. Dev. Dyn. 2007, 236, 886–892. [Google Scholar] [CrossRef]
- Renner, H.; Grabos, M.; Schöler, H.R.; Bruder, J.M. Generation and Maintenance of Homogeneous Human Midbrain Organoids. Bio. Protoc. 2021, 11, e4049. [Google Scholar] [CrossRef]
- Halevy, T.; Urbach, A. Comparing ESC and iPSC-Based Models for Human Genetic Disorders. J. Clin. Med. 2014, 3, 1146–1162. [Google Scholar] [CrossRef]
- Okita, K.; Ichisaka, T.; Yamanaka, S. Generation of germline-competent induced pluripotent stem cells. Nature 2007, 448, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Gore, A.; Li, Z.; Fung, H.L.; Young, J.E.; Agarwal, S.; Antosiewicz-Bourget, J.; Canto, I.; Giorgetti, A.; Israel, M.A.; Kiskinis, E.; et al. Somatic coding mutations in human induced pluripotent stem cells. Nature 2011, 471, 63–67. [Google Scholar] [CrossRef]
- Yoshihara, M.; Hayashizaki, Y.; Murakawa, Y. Genomic Instability of iPSCs: Challenges Towards Their Clinical Applications. Stem Cell Rev. Rep. 2017, 13, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Uzquiano, A.; Kedaigle, A.J.; Pigoni, M.; Paulsen, B.; Adiconis, X.; Kim, K.; Faits, T.; Nagaraja, S.; Antón-Bolaños, N.; Gerhardinger, C.; et al. Proper acquisition of cell class identity in organoids allows definition of fate specification programs of the human cerebral cortex. Cell 2022, 185, 3770–3788.e27. [Google Scholar] [CrossRef]
- Foo, J.N.; Liany, H.; Tan, L.C.; Au, W.L.; Prakash, K.M.; Liu, J.; Tan, E.K. DNAJ mutations are rare in Chinese Parkinson’s disease patients and controls. Neurobiol. Aging 2014, 35, e1–e2. [Google Scholar] [CrossRef]
- Edvardson, S.; Cinnamon, Y.; Ta-Shma, A.; Shaag, A.; Yim, Y.I.; Zenvirt, S.; Jalas, C.; Lesage, S.; Brice, A.; Taraboulos, A.; et al. A deleterious mutation in DNAJC6 encoding the neuronal-specific clathrin-uncoating co-chaperone auxilin, is associated with juvenile parkinsonism. PLoS ONE 2012, 7, e36458. [Google Scholar] [CrossRef]
- Marton, R.M.; Pașca, S.P. Organoid and Assembloid Technologies for Investigating Cellular Crosstalk in Human Brain Development and Disease. Trends Cell Biol. 2020, 30, 133–143. [Google Scholar] [CrossRef]
- Chen, X.; Saiyin, H.; Liu, Y.; Wang, Y.; Li, X.; Ji, R.; Ma, L. Human striatal organoids derived from pluripotent stem cells recapitulate striatal development and compartments. PLoS Biol. 2022, 20, e3001868. [Google Scholar] [CrossRef]
- Rietdijk, C.D.; Perez-Pardo, P.; Garssen, J.; van Wezel, R.J.; Kraneveld, A.D. Exploring Braak’s Hypothesis of Parkinson’s Disease. Front. Neurol. 2017, 8, 37. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Berkers, G.; Kruisselbrink, E.; Vonk, A.; De Jonge, H.R.; Janssens, H.M.; Bronsveld, I.; van de Graaf, E.A.; Nieuwenhuis, E.E.; Houwen, R.H. Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 2016, 8, ra84–ra344. [Google Scholar] [CrossRef]
- Leung, C.M.; de Haan, P.; Ronaldson-Bouchard, K.; Kim, G.-A.; Ko, J.; Rho, H.S.; Chen, Z.; Habibovic, P.; Jeon, N.L.; Takayama, S.; et al. A guide to the organ-on-a-chip. Nat. Rev. Methods Primers 2022, 2, 33. [Google Scholar] [CrossRef]
- Kim, M.-H.; Kim, D.; Sung, J.H. A Gut-Brain Axis-on-a-Chip for studying transport across epithelial and endothelial barriers. J. Ind. Eng. Chem. 2021, 101, 126–134. [Google Scholar] [CrossRef]
- Raimondi, M.T.; Albani, D.; Giordano, C. An Organ-On-A-Chip Engineered Platform to Study the Microbiota–Gut–Brain Axis in Neurodegeneration. Trends Mol. Med. 2019, 25, 737–740. [Google Scholar] [CrossRef]
- Trapecar, M.; Wogram, E.; Svoboda, D.; Communal, C.; Omer, A.; Lungjangwa, T.; Sphabmixay, P.; Velazquez, J.; Schneider, K.; Wright, C.W.; et al. Human physiomimetic model integrating microphysiological systems of the gut, liver, and brain for studies of neurodegenerative diseases. Sci. Adv. 2021, 7, eabd1707. [Google Scholar] [CrossRef] [PubMed]
- Nashimoto, Y.; Hayashi, T.; Kunita, I.; Nakamasu, A.; Torisawa, Y.-S.; Nakayama, M.; Takigawa-Imamura, H.; Kotera, H.; Nishiyama, K.; Miura, T.; et al. Integrating perfusable vascular networks with a three-dimensional tissue in a microfluidic device. Integr. Biol. 2017, 9, 506–518. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, E.; Kajtez, J.; Bruzelius, A.; Wesseler, M.F.; Nilsson, F.; Birtele, M.; Larsen, N.B.; Ottosson, D.R.; Storm, P.; Parmar, M.; et al. Silk scaffolding drives self-assembly of functional and mature human brain organoids. Front. Cell Dev. Biol. 2022, 10, 1023279. [Google Scholar] [CrossRef] [PubMed]
- Bolognin, S.; Fossépré, M.; Qing, X.; Jarazo, J.; Ščančar, J.; Moreno, E.L.; Nickels, S.L.; Wasner, K.; Ouzren, N.; Walter, J.; et al. 3D Cultures of Parkinson’s Disease-Specific Dopaminergic Neurons for High Content Phenotyping and Drug Testing. Adv. Sci. 2019, 6, 1800927. [Google Scholar] [CrossRef]
- Kirkeby, A.; Nolbrant, S.; Tiklova, K.; Heuer, A.; Kee, N.; Cardoso, T.; Ottosson, D.R.; Lelos, M.J.; Rifes, P.; Dunnett, S.B.; et al. Predictive Markers Guide Differentiation to Improve Graft Outcome in Clinical Translation of hESC-Based Therapy for Parkinson’s Disease. Cell Stem Cell 2017, 20, 135–148. [Google Scholar] [CrossRef]
- Doi, D.; Samata, B.; Katsukawa, M.; Kikuchi, T.; Morizane, A.; Ono, Y.; Sekiguchi, K.; Nakagawa, M.; Parmar, M.; Takahashi, J. Isolation of human induced pluripotent stem cell-derived dopaminergic progenitors by cell sorting for successful transplantation. Stem Cell Rep. 2014, 2, 337–350. [Google Scholar] [CrossRef]
- Bye, C.R.; Jönsson, M.E.; Björklund, A.; Parish, C.L.; Thompson, L.H. Transcriptome analysis reveals transmembrane targets on transplantable midbrain dopamine progenitors. Proc. Natl. Acad. Sci. USA 2015, 112, E1946–E1955. [Google Scholar] [CrossRef] [PubMed]
- Grealish, S.; Jönsson, M.E.; Li, M.; Kirik, D.; Björklund, A.; Thompson, L.H. The A9 dopamine neuron component in grafts of ventral mesencephalon is an important determinant for recovery of motor function in a rat model of Parkinson’s disease. Brain 2010, 133, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Grealish, S.; Diguet, E.; Kirkeby, A.; Mattsson, B.; Heuer, A.; Bramoulle, Y.; Van Camp, N.; Perrier, A.L.; Hantraye, P.; Björklund, A.; et al. Human ESC-derived dopamine neurons show similar preclinical efficacy and potency to fetal neurons when grafted in a rat model of Parkinson’s disease. Cell Stem Cell 2014, 15, 653–665. [Google Scholar] [CrossRef]
- Mansour, A.A.; Gonçalves, J.T.; Bloyd, C.W.; Li, H.; Fernandes, S.; Quang, D.; Johnston, S.; Parylak, S.L.; Jin, X.; Gage, F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018, 36, 432–441. [Google Scholar] [CrossRef]
- Daviaud, N.; Friedel, R.H.; Zou, H. Vascularization and engraftment of transplanted human cerebral organoids in mouse cortex. eNeuro 2018, 5, ENEURO.0219-18.2018. [Google Scholar] [CrossRef]
- Chen, H.I.; Song, H.; Ming, G.L. Applications of human brain organoids to clinical problems. Dev. Dyn. 2019, 248, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, T.; Sakaguchi, H.; Morizane, A.; Kikuchi, T.; Miyamoto, S.; Takahashi, J. Axonal extensions along corticospinal tracts from transplanted human cerebral organoids. Stem Cell Rep. 2020, 15, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Grebenyuk, S.; Ranga, A. Engineering Organoid Vascularization. Front. Bioeng. Biotechnol. 2019, 7, 39. [Google Scholar] [CrossRef]
- Doi, D.; Magotani, H.; Kikuchi, T.; Ikeda, M.; Hiramatsu, S.; Yoshida, K.; Amano, N.; Nomura, M.; Umekage, M.; Morizane, A.; et al. Pre-clinical study of induced pluripotent stem cell-derived dopaminergic progenitor cells for Parkinson’s disease. Nat. Commun. 2020, 11, 3369. [Google Scholar] [CrossRef]
- Takahashi, J. iPS cell-based therapy for Parkinson’s disease: A Kyoto trial. Regen. Ther. 2020, 13, 18–22. [Google Scholar] [CrossRef]
- Garitaonandia, I.; Gonzalez, R.; Christiansen-Weber, T.; Abramihina, T.; Poustovoitov, M.; Noskov, A.; Sherman, G.; Semechkin, A.; Snyder, E.; Kern, R. Neural Stem Cell Tumorigenicity and Biodistribution Assessment for Phase I Clinical Trial in Parkinson’s Disease. Sci. Rep. 2016, 6, 34478. [Google Scholar] [CrossRef] [PubMed]
- Garitaonandia, I.; Gonzalez, R.; Sherman, G.; Semechkin, A.; Evans, A.; Kern, R. Novel Approach to Stem Cell Therapy in Parkinson’s Disease. Stem Cells Dev. 2018, 27, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Center for iPS Cell Research and Application, K.U. For Patients and Their Families (Parkinson’s Disease Research). Available online: https://www.cira.kyoto-u.ac.jp/e//faq/faq_patient.html (accessed on 20 November 2022).
- Barker, R.A.; Studer, L.; Cattaneo, E.; Takahashi, J. G-Force PD: A global initiative in coordinating stem cell-based dopamine treatments for Parkinson’s disease. NPJ Park. Dis. 2015, 1, 15017. [Google Scholar] [CrossRef] [PubMed]
- Therapeutics, B.; Center, M.S.K.C. Phase 1 Safety and Tolerability Study of MSK-DA01 Cell Therapy for Advanced Parkinson’s Disease. Available online: https://www.clinicaltrials.gov/ct2/show/NCT04802733 (accessed on 20 November 2022).
- Safety and Efficacy Study of Human ESC-derived Neural Precursor Cells in the Treatment of Parkinson’s Disease. 2016. Available online: https://clinicaltrials.gov/ct2/show/NCT03119636 (accessed on 20 November 2022).
- Corporation International Stem Cell. International Stem Cell Corporation Announces Successful Completion of Its Phase 1 Clinical Trial in Parkinson’s Disease. 2021. Available online: https://investors.internationalstemcell.com/International-Stem-Cell-Corporation-Announces-Successful-Completion-of-Its-Phase-1-Clinical-Trial-in-Parkinson--s-Disease-6-30-2021 (accessed on 20 November 2022).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeap, Y.J.; Teddy, T.J.W.; Lee, M.J.; Goh, M.; Lim, K.L. From 2D to 3D: Development of Monolayer Dopaminergic Neuronal and Midbrain Organoid Cultures for Parkinson’s Disease Modeling and Regenerative Therapy. Int. J. Mol. Sci. 2023, 24, 2523. https://doi.org/10.3390/ijms24032523
Yeap YJ, Teddy TJW, Lee MJ, Goh M, Lim KL. From 2D to 3D: Development of Monolayer Dopaminergic Neuronal and Midbrain Organoid Cultures for Parkinson’s Disease Modeling and Regenerative Therapy. International Journal of Molecular Sciences. 2023; 24(3):2523. https://doi.org/10.3390/ijms24032523
Chicago/Turabian StyleYeap, Yee Jie, Tng J. W. Teddy, Mok Jung Lee, Micaela Goh, and Kah Leong Lim. 2023. "From 2D to 3D: Development of Monolayer Dopaminergic Neuronal and Midbrain Organoid Cultures for Parkinson’s Disease Modeling and Regenerative Therapy" International Journal of Molecular Sciences 24, no. 3: 2523. https://doi.org/10.3390/ijms24032523
APA StyleYeap, Y. J., Teddy, T. J. W., Lee, M. J., Goh, M., & Lim, K. L. (2023). From 2D to 3D: Development of Monolayer Dopaminergic Neuronal and Midbrain Organoid Cultures for Parkinson’s Disease Modeling and Regenerative Therapy. International Journal of Molecular Sciences, 24(3), 2523. https://doi.org/10.3390/ijms24032523





