Identification of Novel Senescent Markers in Small Extracellular Vesicles
Abstract
1. Introduction
2. Results
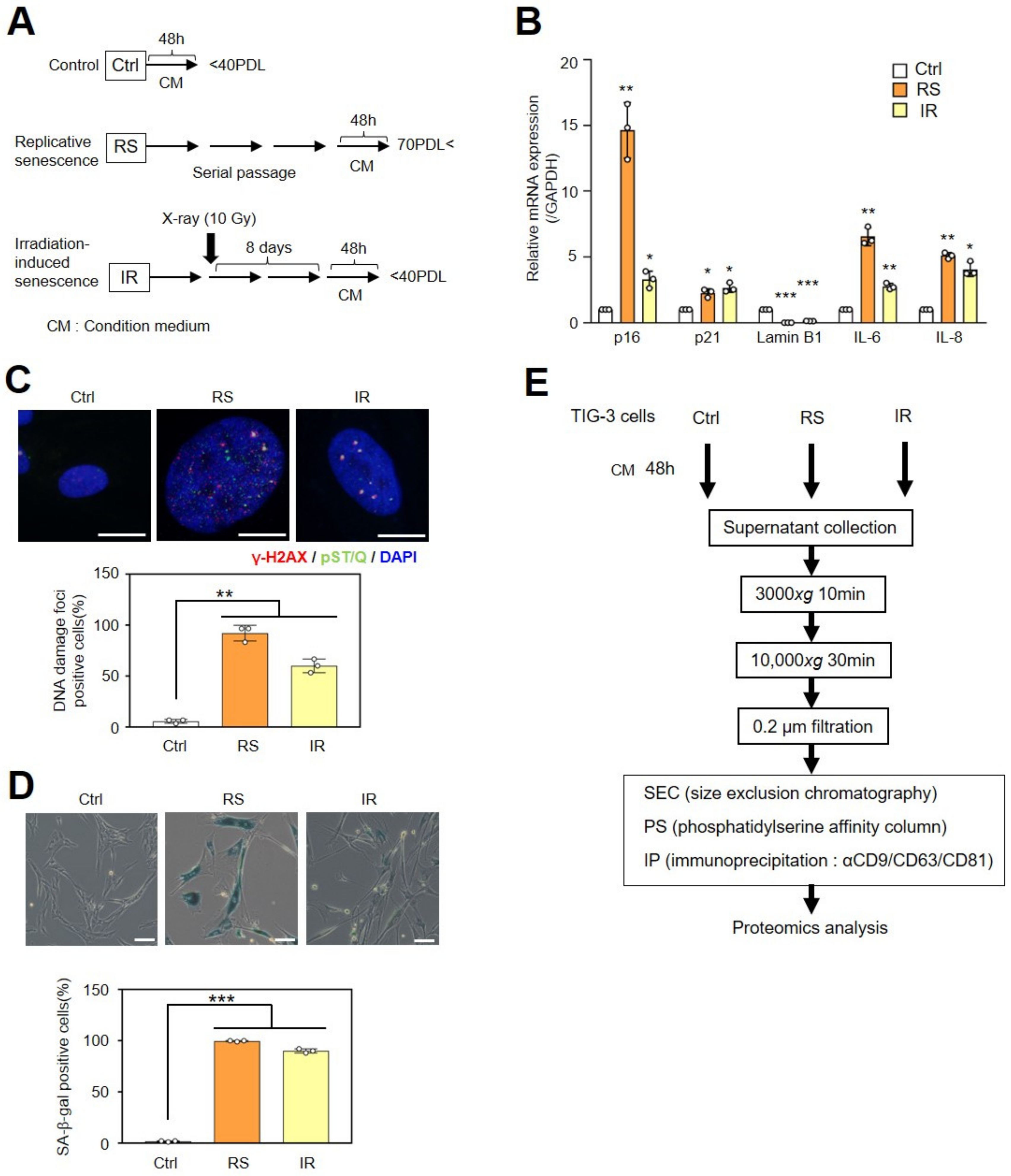
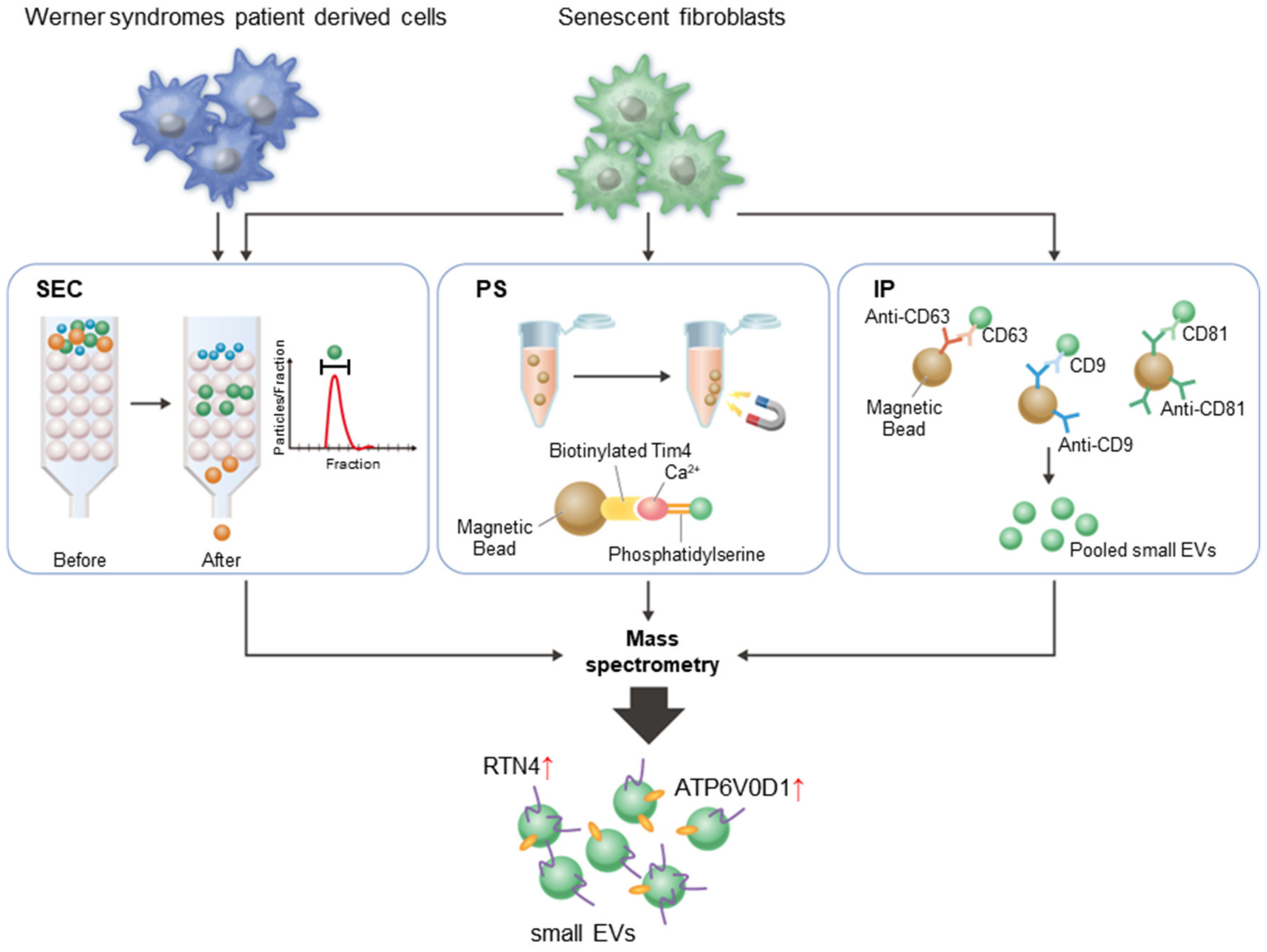
2.1. Small EVs Collected by Three Different Methods
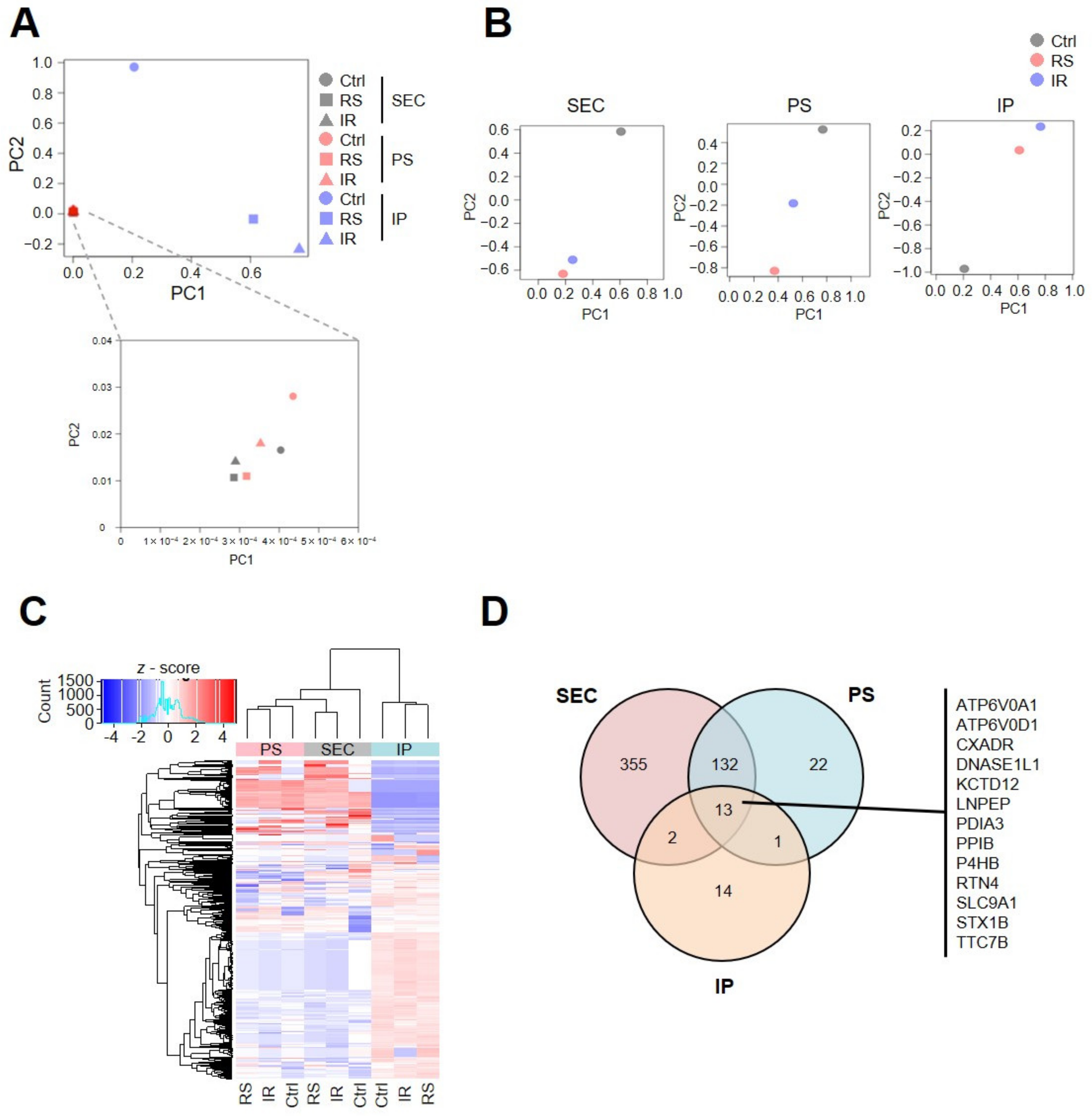
2.2. Senescent Proteomic Profiles Are Different Depending on the Collection Procedure
2.3. Characterization of Small EVs from Patient Cells with Werner Syndrome
2.4. Two Proteins Are Enriched in Small EVs from Senescent Cells, Patient Cells with Progeria, and Serum from Aged Mice
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Small EV Isolation from Cells
4.3. Isolation of Small EVs from Mouse Serum
4.4. Fluorescence Microscopic Analysis
4.5. β-Galactosidase (β-Gal) Staining
4.6. Mass Spectrometry
4.7. Quantitative Real-Time PCR
4.8. Western Blot Analysis
4.9. Clinical Samples
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef]
- Yamakoshi, K.; Takahashi, A.; Hirota, F.; Nakayama, R.; Ishimaru, N.; Kubo, Y.; Mann, D.J.; Ohmura, M.; Hirao, A.; Saya, H.; et al. Real-time in vivo imaging of p16Ink4a reveals cross talk with p53. J. Cell Biol. 2009, 186, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Loo, T.M.; Miyata, K.; Tanaka, Y.; Takahashi, A. Cellular senescence and senescence-associated secretory phenotype via the cGAS-STING signaling pathway in cancer. Cancer Sci. 2020, 111, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Faget, D.V.; Ren, Q.; Stewart, S.A. Unmasking senescence: Context-dependent effects of SASP in cancer. Nat. Rev. Cancer 2019, 19, 439–453. [Google Scholar] [CrossRef]
- Takasugi, M.; Okada, R.; Takahashi, A.; Virya Chen, D.; Watanabe, S.; Hara, E. Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through EphA2. Nat. Commun. 2017, 8, 15729. [Google Scholar] [CrossRef] [PubMed]
- Borghesan, M.; Fafián-Labora, J.; Eleftheriadou, O.; Carpintero-Fernández, P.; Paez-Ribes, M.; Vizcay-Barrena, G.; Swisa, A.; Kolodkin-Gal, D.; Ximénez-Embún, P.; Lowe, R.; et al. Small Extracellular Vesicles Are Key Regulators of Non-cell Autonomous Intercellular Communication in Senescence via the Interferon Protein IFITM3. Cell Rep. 2019, 27, 3956–3971.e6. [Google Scholar] [CrossRef]
- Lananna, B.V.; Imai, S.I. Friends and foes: Extracellular vesicles in aging and rejuvenation. FASEB Bioadv. 2021, 3, 787–801. [Google Scholar] [CrossRef]
- Miyata, K.; Imai, Y.; Hori, S.; Nishio, M.; Loo, T.M.; Okada, R.; Yang, L.; Nakadai, T.; Maruyama, R.; Fujii, R.; et al. Pericentromeric noncoding RNA changes DNA binding of CTCF and inflammatory gene expression in senescence and cancer. Proc. Natl. Acad. Sci. USA 2021, 118, e2025647118. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, 6478. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Takahashi, A. Senescence-associated extracellular vesicle release plays a role in senescence-associated secretory phenotype (SASP) in age-associated diseases. J. Biochem. 2021, 169, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C.; et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287. [Google Scholar] [CrossRef]
- Jeon, O.H.; Wilson, D.R.; Clement, C.C.; Rathod, S.; Cherry, C.; Powell, B.; Lee, Z.; Khalil, A.M.; Green, J.J.; Campisi, J.; et al. Senescence cell-associated extracellular vesicles serve as osteoarthritis disease and therapeutic markers. JCI Insight 2019, 4, 7. [Google Scholar] [CrossRef]
- Hitomi, K.; Okada, R.; Loo, T.M.; Miyata, K.; Nakamura, A.J.; Takahashi, A. DNA Damage Regulates Senescence-Associated Extracellular Vesicle Release via the Ceramide Pathway to Prevent Excessive Inflammatory Responses. Int. J. Mol. Sci. 2020, 21, 3720. [Google Scholar] [CrossRef] [PubMed]
- Misawa, T.; Tanaka, Y.; Okada, R.; Takahashi, A. Biology of extracellular vesicles secreted from senescent cells as senescence-associated secretory phenotype factors. Geriatr. Gerontol. Int. 2020, 20, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Chen, H.; Wang, Y.; Zhang, L.; Wang, X. Roles of extracellular vesicles in the aging microenvironment and age-related diseases. J. Extracell. Vesicles 2021, 10, e12154. [Google Scholar] [CrossRef]
- Childs, B.G.; Gluscevic, M.; Baker, D.J.; Laberge, R.M.; Marquess, D.; Dananberg, J.; van Deursen, J.M. Senescent cells: An emerging target for diseases of ageing. Nat. Rev. Drug Discov. 2017, 16, 718–735. [Google Scholar] [CrossRef]
- Yoshida, M.; Satoh, A.; Lin, J.B.; Mills, K.F.; Sasaki, Y.; Rensing, N.; Wong, M.; Apte, R.S.; Imai, S.I. Extracellular Vesicle-Contained eNAMPT Delays Aging and Extends Lifespan in Mice. Cell Metab. 2019, 30, 329–342.e5. [Google Scholar] [CrossRef]
- Wakita, M.; Takahashi, A.; Sano, O.; Loo, T.M.; Imai, Y.; Narukawa, M.; Iwata, H.; Matsudaira, T.; Kawamoto, S.; Ohtani, N.; et al. A BET family protein degrader provokes senolysis by targeting NHEJ and autophagy in senescent cells. Nat. Commun. 2020, 11, 1935. [Google Scholar] [CrossRef]
- Guan, L.; Crasta, K.C.; Maier, A.B. Assessment of cell cycle regulators in human peripheral blood cells as markers of cellular senescence. Ageing Res. Rev. 2022, 78, 101634. [Google Scholar] [CrossRef] [PubMed]
- Buratta, S.; Urbanelli, L.; Sagini, K.; Giovagnoli, S.; Caponi, S.; Fioretto, D.; Mitro, N.; Caruso, D.; Emiliani, C. Extracellular vesicles released by fibroblasts undergoing H-Ras induced senescence show changes in lipid profile. PLoS ONE 2017, 12, e0188840. [Google Scholar] [CrossRef]
- Alibhai, F.J.; Lim, F.; Yeganeh, A.; DiStefano, P.V.; Binesh-Marvasti, T.; Belfiore, A.; Wlodarek, L.; Gustafson, D.; Millar, S.; Li, S.H.; et al. Cellular senescence contributes to age-dependent changes in circulating extracellular vesicle cargo and function. Aging Cell 2020, 19, e13103. [Google Scholar] [CrossRef] [PubMed]
- Basisty, N.; Kale, A.; Jeon, O.H.; Kuehnemann, C.; Payne, T.; Rao, C.; Holtz, A.; Shah, S.; Sharma, V.; Ferrucci, L.; et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020, 18, e3000599. [Google Scholar] [CrossRef] [PubMed]
- Fafián-Labora, J.A.; O’Loghlen, A. NF-κB/IKK activation by small extracellular vesicles within the SASP. Aging Cell 2021, 20, e13426. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Sugawara, S.; Okada, R.; Loo, T.M.; Tanaka, H.; Miyata, K.; Chiba, M.; Kawasaki, H.; Katoh, K.; Kaji, S.; Maezawa, Y.; et al. RNaseH2A downregulation drives inflammatory gene expression via genomic DNA fragmentation in senescent and cancer cells. Commun. Biol. 2022, 5, 1420. [Google Scholar] [CrossRef]
- Yu, X.; Harris, S.L.; Levine, A.J. The regulation of exosome secretion: A novel function of the p53 protein. Cancer Res. 2006, 66, 4795–4801. [Google Scholar] [CrossRef]
- Lespagnol, A.; Duflaut, D.; Beekman, C.; Blanc, L.; Fiucci, G.; Marine, J.C.; Vidal, M.; Amson, R.; Telerman, A. Exosome secretion, including the DNA damage-induced p53-dependent secretory pathway, is severely compromised in TSAP6/Steap3-null mice. Cell Death Differ. 2008, 15, 1723–1733. [Google Scholar] [CrossRef]
- Terlecki-Zaniewicz, L.; Lämmermann, I.; Latreille, J.; Bobbili, M.R.; Pils, V.; Schosserer, M.; Weinmüllner, R.; Dellago, H.; Skalicky, S.; Pum, D.; et al. Small extracellular vesicles and their miRNA cargo are anti-apoptotic members of the senescence-associated secretory phenotype. Aging 2018, 10, 1103–1132. [Google Scholar] [CrossRef]
- Kirschner, K.; Rattanavirotkul, N.; Quince, M.F.; Chandra, T. Functional heterogeneity in senescence. Biochem. Soc. Trans. 2020, 48, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, T.; Koiwa, M.; Yonemura, A.; Miyake, K.; Kariya, R.; Kubota, S.; Yokomizo-Nakano, T.; Yasuda-Yoshihara, N.; Uchihara, T.; Itoyama, R.; et al. Inflammation-driven senescence-associated secretory phenotype in cancer-associated fibroblasts enhances peritoneal dissemination. Cell Rep. 2021, 34, 108779. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Loo, T.M.; Okada, R.; Kamachi, F.; Watanabe, Y.; Wakita, M.; Watanabe, S.; Kawamoto, S.; Miyata, K.; Barber, G.N.; et al. Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat. Commun. 2018, 9, 1249. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, N.; Miyata, K.; Loo, T.M.; Chiba, M.; Hanyu, A.; Nishio, M.; Kawasaki, H.; Zheng, H.; Toyokuni, S.; Kon, S.; et al. Hepatocyte growth factor derived from senescent cells attenuates cell competition-induced apical elimination of oncogenic cells. Nat. Commun. 2022, 13, 4157. [Google Scholar] [CrossRef]
- Jakhar, R.; Crasta, K. Exosomes as Emerging Pro-Tumorigenic Mediators of the Senescence-Associated Secretory Phenotype. Int. J. Mol. Sci. 2019, 20, 2547. [Google Scholar] [CrossRef]
- Borghesan, M.; Hoogaars, W.M.H.; Varela-Eirin, M.; Talma, N.; Demaria, M. A Senescence-Centric View of Aging: Implications for Longevity and Disease. Trends Cell Biol. 2020, 30, 777–791. [Google Scholar] [CrossRef]
- Kirkland, J.L.; Tchkonia, T. Senolytic drugs: From discovery to translation. J. Intern. Med. 2020, 288, 518–536. [Google Scholar] [CrossRef]
- Fleury, H.; Malaquin, N.; Tu, V.; Gilbert, S.; Martinez, A.; Olivier, M.A.; Sauriol, A.; Communal, L.; Leclerc-Desaulniers, K.; Carmona, E.; et al. Exploiting interconnected synthetic lethal interactions between PARP inhibition and cancer cell reversible senescence. Nat. Commun. 2019, 10, 2556. [Google Scholar] [CrossRef]
- Fleischer, J.G.; Schulte, R.; Tsai, H.H.; Tyagi, S.; Ibarra, A.; Shokhirev, M.N.; Huang, L.; Hetzer, M.W.; Navlakha, S. Predicting age from the transcriptome of human dermal fibroblasts. Genome Biol. 2018, 19, 221. [Google Scholar] [CrossRef]
- Dodd, D.A.; Niederoest, B.; Bloechlinger, S.; Dupuis, L.; Loeffler, J.P.; Schwab, M.E. Nogo-A, -B, and -C are found on the cell surface and interact together in many different cell types. J. Biol. Chem. 2005, 280, 12494–12502. [Google Scholar] [CrossRef]
- Zhao, H.; Su, W.; Zhu, C.; Zeng, T.; Yang, S.; Wu, W.; Wang, D. Cell fate regulation by reticulon-4 in human prostate cancers. J. Cell. Physiol. 2019, 234, 10372–10385. [Google Scholar] [CrossRef] [PubMed]
- Ullah, H.M.A.; Elfadl, A.K.; Park, S.; Kim, Y.D.; Chung, M.J.; Son, J.Y.; Yun, H.H.; Park, J.M.; Yim, J.H.; Jung, S.J.; et al. Nogo-A Is Critical for Pro-Inflammatory Gene Regulation in Myocytes and Macrophages. Cells 2021, 10, 282. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.S.; Jun, S.; Kim, M.J.; Lee, S.H.; Suh, H.N.; Lien, E.M.; Jung, H.Y.; Lee, S.; Zhang, J.; Yang, J.I.; et al. TMEM9 promotes intestinal tumorigenesis through vacuolar-ATPase-activated Wnt/β-catenin signalling. Nat. Cell Biol. 2018, 20, 1421–1433. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Misawa, T.; Hitomi, K.; Miyata, K.; Tanaka, Y.; Fujii, R.; Chiba, M.; Loo, T.M.; Hanyu, A.; Kawasaki, H.; Kato, H.; et al. Identification of Novel Senescent Markers in Small Extracellular Vesicles. Int. J. Mol. Sci. 2023, 24, 2421. https://doi.org/10.3390/ijms24032421
Misawa T, Hitomi K, Miyata K, Tanaka Y, Fujii R, Chiba M, Loo TM, Hanyu A, Kawasaki H, Kato H, et al. Identification of Novel Senescent Markers in Small Extracellular Vesicles. International Journal of Molecular Sciences. 2023; 24(3):2421. https://doi.org/10.3390/ijms24032421
Chicago/Turabian StyleMisawa, Tomoka, Kazuhiro Hitomi, Kenichi Miyata, Yoko Tanaka, Risa Fujii, Masatomo Chiba, Tze Mun Loo, Aki Hanyu, Hiroko Kawasaki, Hisaya Kato, and et al. 2023. "Identification of Novel Senescent Markers in Small Extracellular Vesicles" International Journal of Molecular Sciences 24, no. 3: 2421. https://doi.org/10.3390/ijms24032421
APA StyleMisawa, T., Hitomi, K., Miyata, K., Tanaka, Y., Fujii, R., Chiba, M., Loo, T. M., Hanyu, A., Kawasaki, H., Kato, H., Maezawa, Y., Yokote, K., Nakamura, A. J., Ueda, K., Yaegashi, N., & Takahashi, A. (2023). Identification of Novel Senescent Markers in Small Extracellular Vesicles. International Journal of Molecular Sciences, 24(3), 2421. https://doi.org/10.3390/ijms24032421





