Defining the Role of the Gut Microbiome in the Pathogenesis and Treatment of Lymphoid Malignancies
Abstract
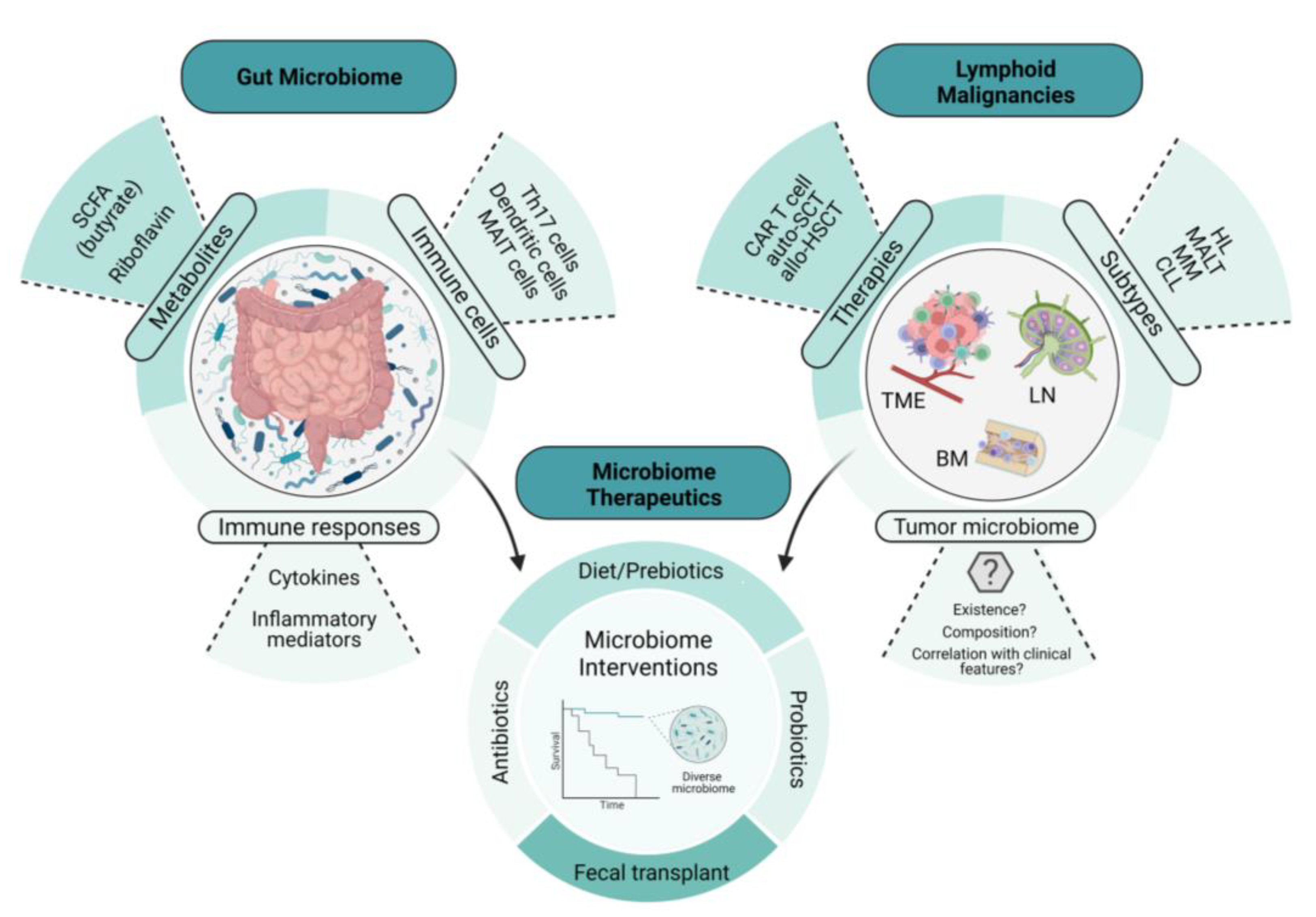
1. Introduction
2. Microbiome Environments and Lymphomagenesis
3. Immune-Mediated Mechanisms of Lymphoma Development and Therapy
3.1. Dendritic Cell Activation
3.2. T Cell Activation
3.3. Effects of Gut-Associated Metabolites
4. Clinical Implications of the Gut Microbiome in Lymphoma Diagnosis and Therapy
4.1. Lymphoma/Myeloma Diagnosis
4.2. Chemotherapy/Immunotherapy
4.3. Autologous Stem Cell Transplantation
4.4. Allogeneic Stem Cell Transplantation
4.5. CAR T Cell Therapy
4.6. Gut-Microbiome-Directed Interventions
5. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef]
- Severyn, C.J.; Brewster, R.; Andermann, T.M. Microbiota modification in hematology: Still at the bench or ready for the bedside? Blood Adv. 2019, 3, 3461–3472. [Google Scholar] [CrossRef]
- Frick, J.-S.; Autenrieth, I.B. The gut microflora and its variety of roles in health and disease. Between Pathog. Commens. 2012, 358, 273–289. [Google Scholar]
- Savage, D.C. Microbial Ecology of the Gastrointestinal Tract. Annu. Rev. Microbiol. 1977, 31, 107–133. [Google Scholar] [CrossRef]
- Purcell, R.V.; Pearson, J.; Aitchison, A.; Dixon, L.; Frizelle, F.A.; Keenan, J.I. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS ONE 2017, 12, e0171602. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.-J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.-R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Varadi, L.; Luo, J.L.; Hibbs, D.E.; Perry, J.D.; Anderson, R.J.; Orenga, S.; Groundwater, P.W. Methods for the detection and identification of pathogenic bacteria: Past, present, and future. Chem. Soc. Rev. 2017, 46, 4818–4832. [Google Scholar] [CrossRef]
- Tang, Q.; Jin, G.; Wang, G.; Liu, T.; Liu, X.; Wang, B.; Cao, H. Current Sampling Methods for Gut Microbiota: A Call for More Precise Devices. Front. Cell. Infect. Microbiol. 2020, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Allaband, C.; McDonald, D.; Vázquez-Baeza, Y.; Minich, J.J.; Tripathi, A.; Brenner, D.A.; Loomba, R.; Smarr, L.; Sandborn, W.J.; Schnabl, B.; et al. Microbiome 101: Studying, Analyzing, and Interpreting Gut Microbiome Data for Clinicians. Clin. Gastroenterol. Hepatol. 2019, 17, 218–230. [Google Scholar] [CrossRef]
- Ringel, Y.; Maharshak, N.; Ringel-Kulka, T.; Wolber, E.A.; Sartor, R.B.; Carroll, I.M. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes 2015, 6, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.H.; Peterson, C.M.; Raggio, A.; Keenan, M.J.; Martin, R.J.; Ravussin, E.; Marco, M.L. Impact of Different Fecal Processing Methods on Assessments of Bacterial Diversity in the Human Intestine. Front. Microbiol. 2016, 7, 1643. [Google Scholar] [CrossRef]
- Yamamoto, M.L.; Schiestl, R.H. Intestinal Microbiome and Lymphoma Development. Cancer J. 2014, 20, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Tegla, C.A.; Herrera, A.M.; Seffens, A.M.; Fanok, M.H.; Dean, G.; Kawaoka, J.; Laird, M.E.; Fulmer, Y.; Willerslev-Olsen, A.; Hymes, K.B.; et al. Skin Associated Staphylococcus Aureus Contributes to Disease Progression in CTCL. Blood 2019, 134, 659. [Google Scholar] [CrossRef]
- Lindahl, L.M.; Willerslev-Olsen, A.; Gjerdrum, L.M.R.; Nielsen, P.R.; Blumel, E.; Rittig, A.H.; Celis, P.; Herpers, B.; Becker, J.C.; Stausbol-Gron, B.; et al. Antibiotics inhibit tumor and disease activity in cutaneous T-cell lymphoma. Blood 2019, 134, 1072–1083. [Google Scholar] [CrossRef]
- Wotherspoon, A.C.; Ortiz-Hidalgo, C.; Falzon, M.R.; Isaacson, P.G. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet 1991, 338, 1175–1176. [Google Scholar] [CrossRef]
- Farinha, P.; Gascoyne, R.D. Helicobacter pylori and MALT lymphoma. Gastroenterology 2005, 128, 1579–1605. [Google Scholar] [CrossRef]
- Kuo, S.H.; Wu, M.S.; Yeh, K.H.; Lin, C.W.; Hsu, P.N.; Chen, L.T.; Cheng, A.L. Novel Insights of Lymphomagenesis of Helicobacter pylori-Dependent Gastric Mucosa-Associated Lymphoid Tissue Lymphoma. Cancers 2019, 11, 547. [Google Scholar] [CrossRef]
- Wotherspoon, A.C.; Doglioni, C.; Diss, T.C.; Pan, L.; Moschini, A.; de Boni, M.; Isaacson, P.G. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 1993, 342, 575–577. [Google Scholar] [CrossRef]
- Hussell, T.; Isaacson, P.G.; Crabtree, J.E.; Spencer, J. The response of cells from low-grade B-cell gastric lymphomas of mucosa-associated lymphoid tissue to Helicobacter pylori. Lancet 1993, 342, 571–574. [Google Scholar] [CrossRef]
- Greiner, A.; Knorr, C.; Qin, Y.; Sebald, W.; Schimpl, A.; Banchereau, J.; Muller-Hermelink, H.K. Low-grade B cell lymphomas of mucosa-associated lymphoid tissue (MALT-type) require CD40-mediated signaling and Th2-type cytokines for in vitro growth and differentiation. Am. J. Pathol. 1997, 150, 1583–1593. [Google Scholar]
- Craig, V.J.; Cogliatti, S.B.; Arnold, I.; Gerke, C.; Balandat, J.E.; Wundisch, T.; Muller, A. B-cell receptor signaling and CD40 ligand-independent T cell help cooperate in Helicobacter-induced MALT lymphomagenesis. Leukemia 2010, 24, 1186–1196. [Google Scholar] [CrossRef]
- Garcia, M.; Bellosillo, B.; Sanchez-Gonzalez, B.; Garcia-Payarols, F.; Seoane, A.; Ferrer, A.M.; Gimeno, E.; Barranco, L.E.; Torner, A.; Sole, F.; et al. Study of regulatory T-cells in patients with gastric malt lymphoma: Influence on treatment response and outcome. PLoS ONE 2012, 7, e51681. [Google Scholar] [CrossRef]
- de Jong, D.; Vyth-Dreese, F.; Dellemijn, T.; Verra, N.; Ruskone-Fourmestraux, A.; Lavergne-Slove, A.; Hart, G.; Boot, H. Histological and immunological parameters to predict treatment outcome of Helicobacter pylori eradication in low-grade gastric MALT lymphoma. J. Pathol. 2001, 193, 318–324. [Google Scholar] [CrossRef]
- Yang, Z.J.; Wang, B.Y.; Wang, T.T.; Wang, F.F.; Guo, Y.X.; Hua, R.X.; Shang, H.W.; Lu, X.; Xu, J.D. Functions of Dendritic Cells and Its Association with Intestinal Diseases. Cells 2021, 10, 583. [Google Scholar] [CrossRef]
- Owen, J.L.; Mohamadzadeh, M. Microbial activation of gut dendritic cells and the control of mucosal immunity. J. Interferon Cytokine Res. 2013, 33, 619–631. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Artis, D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 2008, 8, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Cebra, J.J. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 1999, 69, 1046s–1051s. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, D.; Wei, B.; Presley, L.L.; Brewer, S.; McPherson, M.; Lewinski, M.A.; Borneman, J.; Braun, J. Systemic control of plasmacytoid dendritic cells by CD8+ T cells and commensal microbiota. J. Immunol. 2008, 180, 5843–5852. [Google Scholar] [CrossRef]
- Cozen, W.; Hamilton, A.S.; Zhao, P.; Salam, M.T.; Deapen, D.M.; Nathwani, B.N.; Weiss, L.M.; Mack, T.M. A protective role for early oral exposures in the etiology of young adult Hodgkin lymphoma. Blood 2009, 114, 4014–4020. [Google Scholar] [CrossRef]
- Andrlová, H.; Miltiadous, O.; Kousa, A.I.; Dai, A.; DeWolf, S.; Violante, S.; Park, H.Y.; Janaki-Raman, S.; Gardner, R.; El Daker, S.; et al. MAIT and Vδ2 unconventional T cells are supported by a diverse intestinal microbiome and correlate with favorable patient outcome after allogeneic HCT. Sci. Transl. Med. 2022, 14, eabj2829. [Google Scholar] [CrossRef]
- Godfrey, D.I.; Uldrich, A.P.; McCluskey, J.; Rossjohn, J.; Moody, D.B. The burgeoning family of unconventional T cells. Nat. Immunol. 2015, 16, 1114–1123. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Hanafi, L.A.; Sheih, A.; Golob, J.L.; Srinivasan, S.; Boeckh, M.J.; Pergam, S.A.; Mahmood, S.; Baker, K.K.; Gooley, T.A.; et al. Graft-Derived Reconstitution of Mucosal-Associated Invariant T Cells after Allogeneic Hematopoietic Cell Transplantation. Biol. Blood Marrow Transplant. 2018, 24, 242–251. [Google Scholar] [CrossRef]
- Gao, M.-G.; Hong, Y.; Zhao, X.-Y.; Pan, X.-A.; Sun, Y.-Q.; Kong, J.; Wang, Z.-D.; Wang, F.-R.; Wang, J.-Z.; Yan, C.-H. The Potential Roles of Mucosa-Associated Invariant T Cells in the Pathogenesis of Gut Graft-Versus-Host Disease After Hematopoietic Stem Cell Transplantation. Front. Immunol. 2021, 12, 720354. [Google Scholar] [CrossRef]
- Kawaguchi, K.; Umeda, K.; Hiejima, E.; Iwai, A.; Mikami, M.; Nodomi, S.; Saida, S.; Kato, I.; Hiramatsu, H.; Yasumi, T.; et al. Influence of post-transplant mucosal-associated invariant T cell recovery on the development of acute graft-versus-host disease in allogeneic bone marrow transplantation. Int. J. Hematol. 2018, 108, 66–75. [Google Scholar] [CrossRef]
- Minculescu, L.; Marquart, H.V.; Ryder, L.P.; Andersen, N.S.; Schjoedt, I.; Friis, L.S.; Kornblit, B.T.; Petersen, S.L.; Haastrup, E.; Fischer-Nielsen, A.; et al. Improved Overall Survival, Relapse-Free-Survival, and Less Graft-vs.-Host-Disease in Patients With High Immune Reconstitution of TCR Gamma Delta Cells 2 Months After Allogeneic Stem Cell Transplantation. Front. Immunol. 2019, 10, 1997. [Google Scholar] [CrossRef] [PubMed]
- Calcinotto, A.; Brevi, A.; Chesi, M.; Ferrarese, R.; Garcia Perez, L.; Grioni, M.; Kumar, S.; Garbitt, V.M.; Sharik, M.E.; Henderson, K.J.; et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat. Commun. 2018, 9, 4832. [Google Scholar] [CrossRef] [PubMed]
- Noonan, K.; Marchionni, L.; Anderson, J.; Pardoll, D.; Roodman, G.D.; Borrello, I. A novel role of IL-17–producing lymphocytes in mediating lytic bone disease in multiple myeloma. Blood J. Am. Soc. Hematol. 2010, 116, 3554–3563. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Abid, M.B.; Shah, N.N.; Maatman, T.C.; Hari, P.N. Gut microbiome and CAR-T therapy. Exp. Hematol. Oncol. 2019, 8, 31. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Levy, M.; Thaiss, C.A.; Elinav, E. Metabolites: Messengers between the microbiota and the immune system. Genes Dev. 2016, 30, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Blacher, E.; Elinav, E. Microbiome, metabolites and host immunity. Curr. Opin. Microbiol. 2017, 35, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Huang, S.; Wang, Y.; Cai, S.; Yu, H.; Liu, H.; Zeng, X.; Zhang, G.; Qiao, S. Bridging intestinal immunity and gut microbiota by metabolites. Cell. Mol. Life Sci. 2019, 76, 3917–3937. [Google Scholar] [CrossRef]
- Smith, K.; McCoy, K.D.; Macpherson, A.J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007, 19, 59–69. [Google Scholar] [CrossRef]
- Chung, H.; Pamp, S.J.; Hill, J.A.; Surana, N.K.; Edelman, S.M.; Troy, E.B.; Reading, N.C.; Villablanca, E.J.; Wang, S.; Mora, J.R.; et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 2012, 149, 1578–1593. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid. Res. 2006, 47, 241–259. [Google Scholar] [CrossRef]
- Zheng, X.; Xie, G.; Zhao, A.; Zhao, L.; Yao, C.; Chiu, N.H.; Zhou, Z.; Bao, Y.; Jia, W.; Nicholson, J.K.; et al. The footprints of gut microbial-mammalian co-metabolism. J. Proteome Res. 2011, 10, 5512–5522. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef]
- Wei, W.; Sun, W.; Yu, S.; Yang, Y.; Ai, L. Butyrate production from high-fiber diet protects against lymphoma tumor. Leuk. Lymphoma 2016, 57, 2401–2408. [Google Scholar] [CrossRef]
- Lu, H.; Xu, X.; Fu, D.; Gu, Y.; Fan, R.; Yi, H.; He, X.; Wang, C.; Ouyang, B.; Zhao, P.; et al. Butyrate-producing Eubacterium rectale suppresses lymphomagenesis by alleviating the TNF-induced TLR4/MyD88/NF-kappaB axis. Cell Host Microbe 2022, 30, 1139–1150.e1137. [Google Scholar] [CrossRef]
- Jian, X.; Zhu, Y.; Ouyang, J.; Wang, Y.; Lei, Q.; Xia, J.; Guan, Y.; Zhang, J.; Guo, J.; He, Y.; et al. Alterations of gut microbiome accelerate multiple myeloma progression by increasing the relative abundances of nitrogen-recycling bacteria. Microbiome 2020, 8, 74. [Google Scholar] [CrossRef]
- Faitová, T.; Svanberg, R.; da Cunha-Bang, C.; Ilett, E.E.; Jørgensen, M.; Noguera-Julian, M.; Paredes, R.; MacPherson, C.R.; Niemann, C.U. The gut microbiome in patients with chronic lymphocytic leukemia. Hematologica 2022, 107, 2238–2243. [Google Scholar] [CrossRef]
- Zhang, B.; Gu, J.; Liu, J.; Huang, B.; Li, J. Fecal Microbiota Taxonomic Shifts in Chinese Multiple Myeloma Patients Analyzed by Quantitative Polimerase Chain Reaction (QPCR) and 16S rRNA High-Throughput Sequencing. Med. Sci. Monit. 2019, 25, 8269–8280. [Google Scholar] [CrossRef]
- Shi, Z.; Hu, G.; Li, M.W.; Zhang, L.; Li, X.; Li, L.; Wang, X.; Fu, X.; Sun, Z.; Zhang, X.; et al. Gut microbiota as non-invasive diagnostic and prognostic biomarkers for natural killer/T-cell lymphoma. Gut 2022, gutjnl-2022-328256. [Google Scholar] [CrossRef]
- Diefenbach, C.S.; Peters, B.A.; Li, H.; Raphael, B.; Moskovits, T.; Hymes, K.; Schluter, J.; Chen, J.; Bennani, N.N.; Witzig, T.E.; et al. Microbial dysbiosis is associated with aggressive histology and adverse clinical outcome in B-cell non-Hodgkin lymphoma. Blood Adv. 2021, 5, 1194–1198. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Pflug, N.; Kluth, S.; Vehreschild, J.J.; Bahlo, J.; Tacke, D.; Biehl, L.; Eichhorst, B.; Fischer, K.; Cramer, P.; Fink, A.-M.; et al. Efficacy of antineoplastic treatment is associated with the use of antibiotics that modulate intestinal microbiota. Oncoimmunology 2016, 5, e1150399. [Google Scholar] [CrossRef]
- Montassier, E.; Al-Ghalith, G.; Ward, T.; Corvec, S.; Gastinne, T.; Potel, G.; Moreau, P.; de la Cochetiere, M.F.; Batard, E.; Knights, D. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med. 2016, 8, 49. [Google Scholar] [CrossRef]
- Daillere, R.; Vetizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Hwang, S.R.; Higgins, A.; Castillo Almeida, N.E.; LaPlant, B.; Maurer, M.J.; Ansell, S.M.; Witzig, T.E.; Thanarajasingam, G.; Bennani, N.N. Effect of antibiotic use on outcomes in patients with Hodgkin lymphoma treated with immune checkpoint inhibitors. Leuk. Lymphoma 2021, 62, 247–251. [Google Scholar] [CrossRef]
- Lalani, A.A.; Xie, W.; Braun, D.A.; Kaymakcalan, M.; Bossé, D.; Steinharter, J.A.; Martini, D.J.; Simantov, R.; Lin, X.; Wei, X.X.; et al. Effect of Antibiotic Use on Outcomes with Systemic Therapies in Metastatic Renal Cell Carcinoma. Eur. Urol. Oncol. 2020, 3, 372–381. [Google Scholar] [CrossRef]
- Pinato, D.J.; Howlett, S.; Ottaviani, D.; Urus, H.; Patel, A.; Mineo, T.; Brock, C.; Power, D.; Hatcher, O.; Falconer, A.; et al. Association of Prior Antibiotic Treatment With Survival and Response to Immune Checkpoint Inhibitor Therapy in Patients With Cancer. JAMA Oncol. 2019, 5, 1774–1778. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Peled, J.U.; Devlin, S.M.; Staffas, A.; Lumish, M.; Khanin, R.; Littmann, E.R.; Ling, L.; Kosuri, S.; Maloy, M.; Slingerland, J.B.; et al. Intestinal Microbiota and Relapse After Hematopoietic-Cell Transplantation. J. Clin. Oncol. 2017, 35, 1650–1659. [Google Scholar] [CrossRef]
- Montassier, E.; Gastinne, T.; Vangay, P.; Al-Ghalith, G.A.; Bruley des Varannes, S.; Massart, S.; Moreau, P.; Potel, G.; de La Cochetière, M.F.; Batard, E.; et al. Chemotherapy-driven dysbiosis in the intestinal microbiome. Aliment. Pharmacol. Ther. 2015, 42, 515–528. [Google Scholar] [CrossRef]
- El Jurdi, N.; Filali-Mouhim, A.; Salem, I.; Retuerto, M.; Dambrosio, N.M.; Baer, L.; Lazarus, H.M.; Caimi, P.; Cooper, B.; Tomlinson, B.; et al. Gastrointestinal Microbiome and Mycobiome Changes during Autologous Transplantation for Multiple Myeloma: Results of a Prospective Pilot Study. Biol. Blood Marrow Transplant. 2019, 25, 1511–1519. [Google Scholar] [CrossRef]
- D’Angelo, C.; Sudakaran, S.; Asimakopoulos, F.; Hematti, P.; El-Gamal, D.; Safdar, N.; Callander, N. Perturbation of the gut microbiome and association with outcomes following autologous stem cell transplantation in patients with multiple myeloma. Leuk. Lymphoma 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Lindner, S.; Gomes, A.L.C.; Devlin, S.M.; Shah, G.L.; Sung, A.D.; Sauter, C.S.; Landau, H.J.; Dahi, P.B.; Perales, M.-A.; et al. Fecal microbiota diversity disruption and clinical outcomes after auto-HCT: A multicenter observational study. Blood 2021, 137, 1527–1537. [Google Scholar] [CrossRef]
- Taur, Y.; Jenq, R.R.; Perales, M.-A.; Littmann, E.R.; Morjaria, S.; Ling, L.; No, D.; Gobourne, A.; Viale, A.; Dahi, P.B.; et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014, 124, 1174–1182. [Google Scholar] [CrossRef]
- Peled, J.U.; Gomes, A.L.C.; Devlin, S.M.; Littmann, E.R.; Taur, Y.; Sung, A.D.; Weber, D.; Hashimoto, D.; Slingerland, A.E.; Slingerland, J.B.; et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N. Engl. J. Med. 2020, 382, 822–834. [Google Scholar] [CrossRef] [PubMed]
- Harris, B.; Morjaria, S.M.; Littmann, E.R.; Geyer, A.I.; Stover, D.E.; Barker, J.N.; Giralt, S.A.; Taur, Y.; Pamer, E.G. Gut Microbiota Predict Pulmonary Infiltrates after Allogeneic Hematopoietic Cell Transplantation. Am. J. Respir. Crit. Care Med. 2016, 194, 450–463. [Google Scholar] [CrossRef]
- Chong, E.A.; Ruella, M.; Schuster, S.J. Five-Year Outcomes for Refractory B-Cell Lymphomas with CAR T-Cell Therapy. N. Engl. J. Med. 2021, 384, 673–674. [Google Scholar] [CrossRef]
- Santomasso, B.; Bachier, C.; Westin, J.; Rezvani, K.; Shpall, E.J. The Other Side of CAR T-Cell Therapy: Cytokine Release Syndrome, Neurologic Toxicity, and Financial Burden. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.; Dai, A.; Ghilardi, G.; Amelsberg, K.V.; Devlin, S.M.; Pajarillo, R.; Slingerland, J.B.; Beghi, S.; Herrera, P.S.; Giardina, P.; et al. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat. Med. 2022, 28, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Tóth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef] [PubMed]
- van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; de Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.; Tijssen, J.G.; et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [Google Scholar] [CrossRef]
- Park, C.H.; Hong, C.; Lee, A.r.; Sung, J.; Hwang, T.H. Multi-omics reveals microbiome, host gene expression, and immune landscape in gastric carcinogenesis. iScience 2022, 25, 103956. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Lythgoe, M.P.; Mullish, B.H.; Frampton, A.E.; Krell, J. Polymorphic microbes: A new emerging hallmark of cancer. Trends Microbiol. 2022, 30, 1131–1134. [Google Scholar] [CrossRef]
- Brooks, A.W.; Priya, S.; Blekhman, R.; Bordenstein, S.R. Gut microbiota diversity across ethnicities in the United States. PLoS Biol. 2018, 16, e2006842. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, F.-H.; Wu, P.-Q.; Xing, H.-Y.; Ma, T. The Role of The Tumor Microbiome in Tumor Development and Its Treatment. Front. Immunol. 2022, 13, 935846. [Google Scholar] [CrossRef]
| Brief Study Title | Disease/Population | Intervention | Status |
|---|---|---|---|
| Choosing the best antibiotic to protect the gut microbiota during stem-cell transplant | Allo-HSCT for any hematologic malignancies | Piperacillin-tazobactam versus cefepime for the treatment of neutropenic fever | Recruiting (NCT03078010) |
| Prebiotics during ASCT for lymphoma/myeloma (PRIMAL) | Allo-SCT for lymphoma /myeloma | Resistant potato starch | Recruiting (NCT05135351) |
| Plant-based diet for MGUS and smoldering myeloma | Monoclonal gammopathy of undetermined significance or smoldering myeloma | Plant-based diet | Recruiting (NCT04920084) |
| Effects of prebiotics on the gut microbiome in patients undergoing HSCT (HCTDiet) | Allo-HSCT for lymphoma | Prebiotic foods | Recruiting (NCT04629430) |
| Dietary manipulation of the microbiome-metabolomic axis for GVHD mitigation | Allo-HSCT for lymphoma | Resistant potato starch | Recruiting (NCT02763033) |
| Intermittent fasting and CLL/SLL | Chronic lymphocytic leukemia or small lymphocytic leukemia | Intermittent fasting | Active, Not recruiting (NCT04629430) |
| Fecal microbial transplant after allogeneic stem cell Transplantation | Allo-HSCT for any hematologic malignancies | FMT | Not yet recruiting (NCT04935684) |
| A novel vaccine as monotherapy or combination therapy in indolent NHL | Follicular and marginal zone lymphoma | Tumor-antigen or microbiome peptide vaccine | Recruiting (NCT04669171) |
| Safety and efficacy of curcumin in children with ALL (CurCumPedALL) | Acute lymphoblastic leukemia | Curcumin | Recruiting (NCT05045443) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Upadhyay Banskota, S.; Skupa, S.A.; El-Gamal, D.; D’Angelo, C.R. Defining the Role of the Gut Microbiome in the Pathogenesis and Treatment of Lymphoid Malignancies. Int. J. Mol. Sci. 2023, 24, 2309. https://doi.org/10.3390/ijms24032309
Upadhyay Banskota S, Skupa SA, El-Gamal D, D’Angelo CR. Defining the Role of the Gut Microbiome in the Pathogenesis and Treatment of Lymphoid Malignancies. International Journal of Molecular Sciences. 2023; 24(3):2309. https://doi.org/10.3390/ijms24032309
Chicago/Turabian StyleUpadhyay Banskota, Shristi, Sydney A. Skupa, Dalia El-Gamal, and Christopher R. D’Angelo. 2023. "Defining the Role of the Gut Microbiome in the Pathogenesis and Treatment of Lymphoid Malignancies" International Journal of Molecular Sciences 24, no. 3: 2309. https://doi.org/10.3390/ijms24032309
APA StyleUpadhyay Banskota, S., Skupa, S. A., El-Gamal, D., & D’Angelo, C. R. (2023). Defining the Role of the Gut Microbiome in the Pathogenesis and Treatment of Lymphoid Malignancies. International Journal of Molecular Sciences, 24(3), 2309. https://doi.org/10.3390/ijms24032309






