Abstract
The oral cavity is the second most colonized site of Helicobacter pylori after the stomach. This study aimed to compare the genetic relatedness between gastric and oral H. pylori in Japanese patients with early gastric cancer through multilocus sequence typing (MLST) analysis using eight housekeeping genes. Gastric biopsy specimens and oral samples were collected from 21 patients with a fecal antigen test positive for H. pylori. The number of H. pylori allelic profiles ranged from zero to eight since the yield of DNA was small even when the nested PCR was performed. MLST analysis revealed that only one patient had a matching oral and gastric H. pylori genotype, suggesting that different genotypes of H. pylori inhabit the oral cavity and gastric mucosa. The phylogenetic analysis showed that oral H. pylori in six patients was similar to gastric H. pylori, implying that the two strains are related but not of the same origin, and those strains may be infected on separate occasions. It is necessary to establish a culture method for oral H. pylori to elucidate whether the oral cavity acts as the source of gastric infection, as our analysis was based on a limited number of allele sequences.
1. Introduction
Helicobacter pylori is a major causative pathogen of gastritis, peptic ulcers, and gastric carcinoma [1,2,3,4]. Transmission of H. pylori is thought to occur mainly via the oral cavity during childhood [5,6]. H. pylori persists throughout the life of the patient without specific therapy, and most people infected with H. pylori usually remain asymptomatic. However, 30% of individuals may develop mild to severe upper gastrointestinal disease [6]. The prevalence of H. pylori infections among Japanese children and adolescents has been markedly decreasing with socioeconomic development. A meta-regression analysis of H. pylori infections in Japan from 1908 to 2003 revealed that the predicted prevalence in persons born in 1990 and 2000 was only 15.6% and 6.6%, respectively [7]. Thus, it is thought that the incidence of gastric cancer will continue to decrease over time [8].
As the oral cavity is the inlet port, researchers considered that it may also be the secondary habitat of H. pylori following colonization of the stomach. H. pylori grows spirally in the stomach, which is a microaerobic environment. In contrast, the bacteria are considered viable but non-culturable in the human oral cavity, as they do not appear to grow under aerobic or anaerobic conditions [9]. Therefore, nested polymerase chain reaction (PCR) is typically used to confirm their presence in oral specimens [10,11,12,13]. The specific DNA of H. pylori has been detected in a broad range of oral specimens, including saliva [13,14,15,16,17], supra- or sub-gingival biofilm [13,15,16,17,18], dentin caries [18], dental pulp [17,19], infected root canal [18], and the coating of the tongue [13,15]. We have reported that one-fifth of Japanese young adults have oral H. pylori DNA [13].
It has not been fully investigated whether H. pylori exists alive in the oral cavity and serves as a supply source for the stomach. The speculation that the oral cavity could also be a potential reservoir for H. pylori is based on epidemiological studies demonstrating a correlation between the presence of oral and gastric H. pylori [18,20,21]. However, so far, no evidence of a genotypic correlation between strains in the gastric mucosa and the oral cavity exists. Moreover, although there were some studies demonstrating that H. pylori in the oral cavity affected the outcome of eradication therapy [11] and that adjunctive periodontal therapy could enhance the efficiency of H. pylori treatment [22,23], there is still no evidence that the recurrence after eradication therapy of gastric H. pylori is caused by oral H. pylori. In their systematic review and meta-analysis, López-Valverde et al. reported that there is no clear evidence that H. pylori present in oral bacterial biofilm cause gastric infections and vice versa [24].
Since there are few studies comparing genotype differences in multiple genes, we aimed to investigate the genetic relatedness between gastric and oral H. pylori in patients with H. pylori infection in the stomach. To overcome the difficulty of the analysis, we applied a multilocus sequence typing (MLST) technique. MLST, which consists of a combination of partial nucleotide sequences of several housekeeping genes, is valid to address the intraspecific phylogenetic structure between different strains of the bacteria [25,26,27].
2. Results
2.1. Characteristics of Patients
The characteristics of the participants are summarized in Table 1. We recruited 21 patients (seven women and 14 men; mean age: 72.8 years, range: 59 to 86 years). Of the 21 participants, 20 had adenocarcinoma in the stomach, and one had squamous cell carcinoma and adenocarcinoma in the esophagus and stomach. The No. of remaining teeth and DMFT ranged from 5 to 30 and from 5 to 28, respectively. The average number of oral cleanings per day was 1.8, and 47.6% (10/21) of the patients used oral cleaning aids such as dental floss and interdental brush. Further, 19.0% (4/21) of the participants were smokers.

Table 1.
Demographic and clinical characteristics of the patients.
2.2. Identification of H. pylori in Stomach Tissue
The presence of H. pylori in gastric tissue was confirmed by both histopathology and PCR amplification of the 16s rRNA gene from the gastric biopsy. Hematoxylin and eosin (H-E) and Giemsa staining of the tissues showed spiral rods that penetrated between the gastric mucosa and mucus-producing cells (Figure 1 and Figure S1). This is consistent with what has been reported as a characteristic of H. pylori [28,29]. H. pylori DNA was detected in all participants by single-step PCR from an isolate on the agar plate (Figure S1).
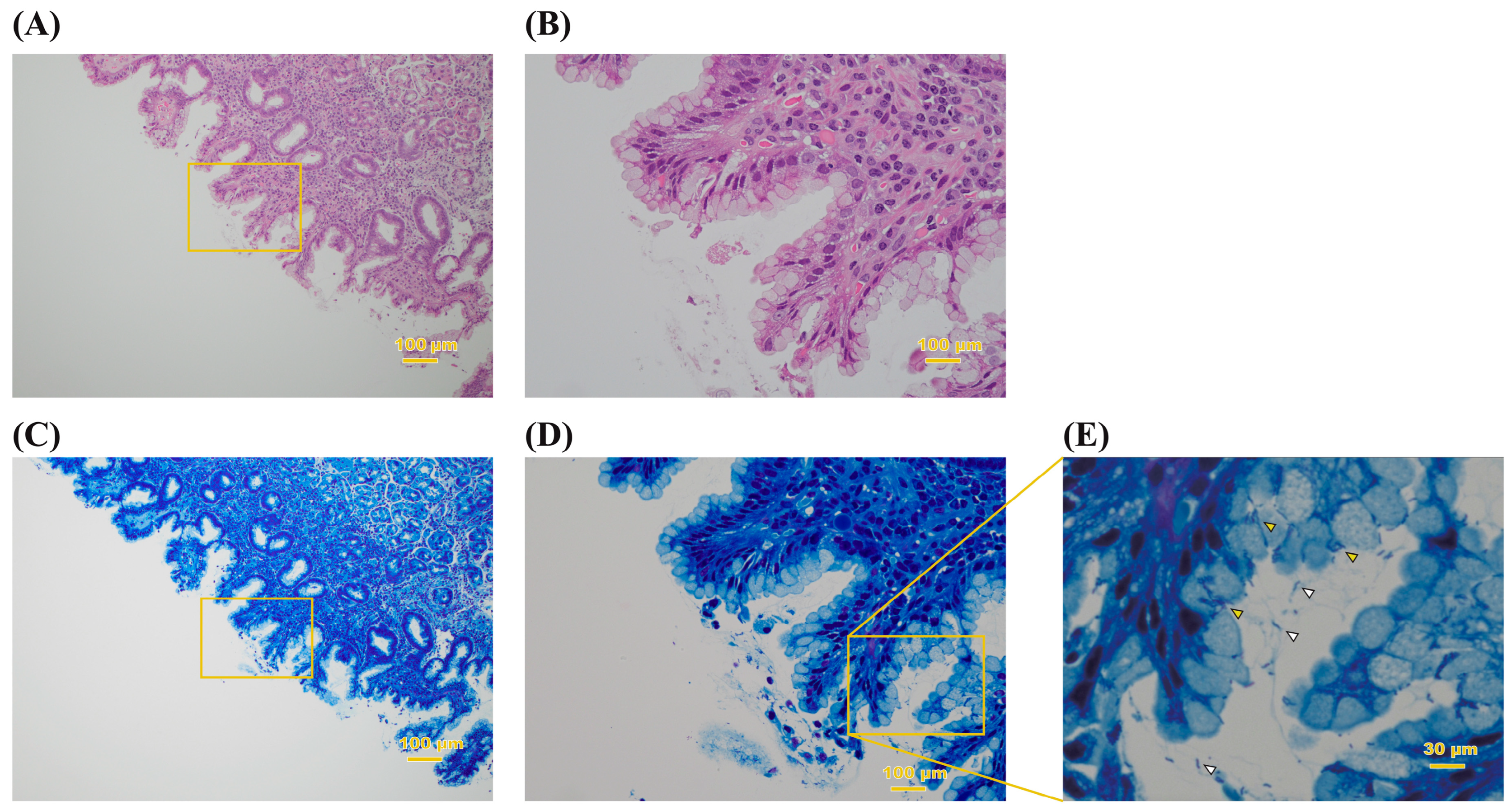
Figure 1.
Histopathological observation of biopsy specimen. The gastric mucosa stained with HE (A,B) and Giemsa (C–E) of patient no. 15. The original magnification is ×10. (B,D) Higher magnification of the area indicated by the squares in (A,C), respectively. (E) High-magnification image of the box in (D). Spiral-like and seagull-like bacilli can be confirmed in the mucus (white arrowhead) and the interior of mucous cells (yellow arrowhead) of the stomach.
2.3. Nested PCR and Allele Analysis
H. pylori DNA was detected in at least one site of the oral cavity in all patients using nested PCR (Table 2). The upper incisors had the highest abundance of the organism (71.4%, p < 0.05). Further, H. pylori DNA was detected in the biopsy specimens of all patients.

Table 2.
Prevalence of H. pylori DNA in gastric tissue and by location in the oral cavity using nested PCR.
Table 3 compares the H. pylori allelic profiles obtained from oral and gastric samples based on eight housekeeping genes. The number of H. pylori allelic profiles ranged from zero to eight since the yield of DNA was small even when the nested PCR was performed. The alleles of eight loci from both collection sites were determined from only one patient (patient no. 3), and two out of eight alleles matched between oral and gastric samples. Moreover, for one sample set (patient no. 4), no amplification was observed for any of the eight housekeeping genes, and therefore, no allele analysis was performed for the sample set. For the rest, allele analysis was performed on the obtained sequences to determine the allele number, of which some were novel and, therefore, unclassified.

Table 3.
Allele analysis of H. pylori DNA sequences obtained from oral and gastric samples targeting eight housekeeping genes.
First, we evaluated the validity of the sequence obtained from the sample by creating a phylogenic tree using the allele type sequence here. Figure S2 shows the phylogenetic analysis of the oral and gastric H. pylori based on the allelic profiles of the seven housekeeping genes. In addition, a total of 159 isolates from Asia, Oceania, Europe, North America, South America, and Africa were included in the phylogentic analysis. In this analysis, Asia was further categorized as East Asia, Southeast Asia, and South Asia. The H. pylori allelic profiles from the oral and gastric samples in this study mainly clustered with the Asian isolates. Partial sequences of the seven genes per patient are presented in the Supplementary Materials.
2.4. MLST
The genetic relatedness between oral and gastric H. pylori was investigated using MLST. DNA sequences of up to seven of the housekeeping genes were used for MLST to obtain sequence types. Although the vacuolating cytotoxin A (vacA) gene was able to generate an allele number, it could not be used for MLST analysis on the PubMLST database (http://pubmlst.org/helicobacter/. Accessed on 17 December 2022) [30,31]. Table 4 summarizes the MLST profiles of the sample sets. Of the genotypes obtained from the oral cavity and stomach, only one sample set (patient no. 13) had a matching sequence type. The genotypes of the other 20 sample sets were various, and the gastric and oral H. pylori were not identified to be similar. In particular, MLST analysis of patient no. 3 was performed using seven alleles, suggesting that different genotypes of H. pylori inhabit the oral cavity and gastric mucosa.

Table 4.
MLST profiles of oral and gastric H. pylori.
2.5. Phylogenetic Analysis
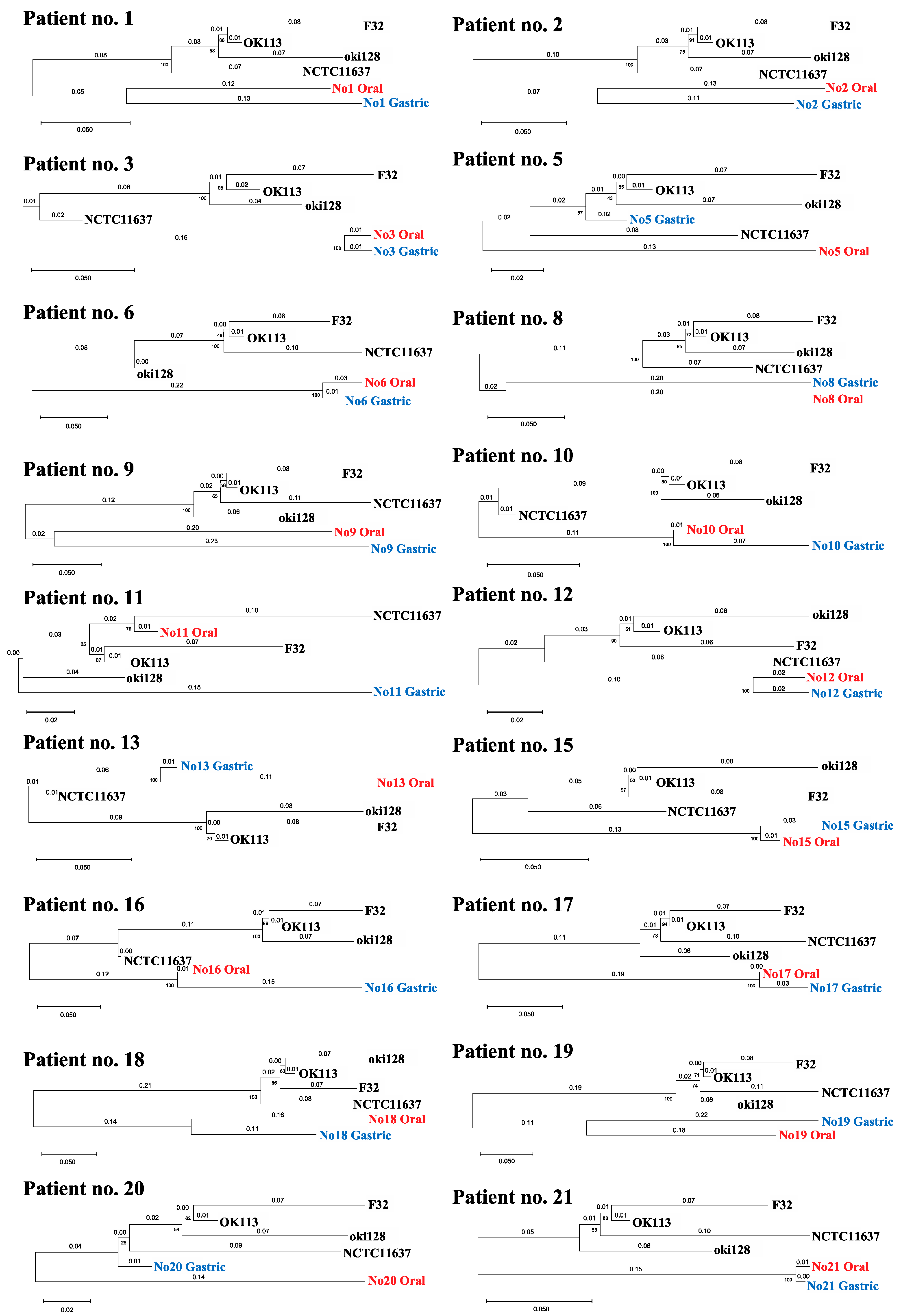
Phylogenetic trees were created using the samples that obtained a combination of two or more alleles (Figure 2). There was a large sequence of diversity between strains of oral and gastric origin in most samples. On the other hand, the oral-derived H. pylori strains in patients no. 3, 6, 12, 15, 17 and 21 had markedly similar sequences to their respective counterpart gastric-derived strains, revealing that evolution distances were 0.03 or less, with bootstrap values of 100. These data imply the possibility that oral and gastric H. pylori are related in a small number of patients. Partial sequences per patient are demonstrated in the Supplementary Materials (File S1).
Figure 2.
Phylogenetic tree showing the correlation of oral and gastric H. pylori based on sequence arrangements. Phylogenetic tree was created using the samples that obtained a combination of two or more alleles, including patient no. 13. The gene sequences of H. pylori OK113, oki128, and F32 clinical isolates from East Asia were downloaded from the BLAST website. The numbers indicate distances between the strains tested (substitutions/site).
3. Discussion
In the present study, we compared the genome sequences of gastric and oral H. pylori from 21 patients with early gastrointestinal cancer using MLST. MLST analysis is an unambiguous procedure that characterizes isolates of bacterial species using the sequences of internal fragments of housekeeping genes. MLST analysis can also help predict the history of human migrations [25]. To our knowledge, this is the first report of the molecular epidemiological analysis of the homology between gastric and oral H. pylori.
We were able to obtain H. pylori DNA from both the stomach and the oral cavity of all patients who tested positive through a fecal antigen test. These findings supported previous epidemiological studies showing that H. pylori colonization in the oral cavity is associated with its presence in gastric tissue [18,32].
Oral H. pylori DNA was frequently detected in the supragingival biofilm on the upper incisor teeth (Table 2). The characteristic distribution may be due to local oxygen concentration and environmental acidity. Since anterior teeth are frequently exposed to oxygen, the local oxygen concentration may create a favorable environment for the bacteria. Supragingival biofilm may also provide an optimum environment for H. pylori due to its acidic-maintaining property.
In almost all the phylogenetic trees obtained using the seven housekeeping genes, H. pylori sequences in this study classified close to the Asian strain that is considered to be high pathogenic (Figure S2). Some H. pylori strains involved in carcinogenesis have cytotoxin-associated genes such as cagA and vacA [33,34]. An isolate from East Asia often carries these genes with unique sequences, which is associated with enhanced virulence [35]. Yamaoka et al. reported that the carrying rate of the highly pathogenic vacA gene was significantly higher in East Asian strains (94.7%) than that in non-Asian strains (0.1%) [36]. Conversely, bacteria with weak expression of these genes are classified as attenuated or non-toxic strains [37].
The findings obtained from the MLST analysis were extremely limited. The number of H. pylori allelic profiles of eight loci obtained from oral samples ranged from zero to eight. One of the reasons for the failure to acquire the allelic profile is that the yield of DNA was small even when the nested PCR was performed. At present, no reliable isolation culture method for oral H. pylori has been established. Therefore, researchers are using the nested PCR method to detect the very rare oral H. pylori. However, the sensitivity of the nested PCR varies according to the primer set. For instance, the nested PCR of the combinations of EHC/ET-5 is incapable of detecting H. pylori unless oral H. pylori exist at concentrations exceeding one in a million [13]. Sulo et al. reported that the abundance and threshold of H. pylori present in the oral cavity are low, and the possibility of DNA breakage is high, so the sensitivity of primers with long bp is considerably reduced [38]. Most of the primers targeting the house-keeping DNA used in this study have more than 500 bp in the final amplification product, which is longer than the 228 bp of EHC/ET-5. To overcome this limitation, scholars need to establish an isolation culture method that enables genetic heterogeneity analysis, such as through whole-genome sequencing [39], Core Genome MLST [40], and pulse-field gel electrophoresis [41].
MLST results showed that one out of the 21 patients (patient no. 13) harbored the same genotype, indicating that the strains in the stomach and oral cavity may be of the same origin (Table 4). However, the specificity was low because only two housekeeping genes were used for the analysis. Moreover, although MLST was possible with one gene, the number of candidate MLST numbers increased accordingly, and the specificity and reliability were low due to the fact that six or more alleles are required to generate reliable results [42]. MLST analysis of patient no. 3, which was performed using seven alleles, showed the different candidates between gastric and oral H. pylori (Table 4). This result indicated with high confidence that the oral and gastric H. pylori were of different origin.
We created a phylogenetic tree using the samples that obtained a combination of two or more alleles, including vacA (Figure 2). The phylogenetic analysis of patient no. 3 was performed using the sequences of eight genes. The result showed that the oral H. pylori strains were closely related to the corresponding gastric H. pylori. In addition, the analyses of patient no. 6, 12, 15, 17 and 21 were performed using the sequences of several genes (no. 6; mutY and trpC, no. 12; ppa and yphC, no. 15; efp, mutY and vacA, no. 17; Urel, mutY and yphC, no. 21; Urel and yphC), and those two strains were closely related. Thus, gastric and oral H. pylori in some patients are genetically similar but not of the same origin, and those strains may be infected on separate occasions. Although the analyses of other patients were also performed using only a few sequences, the results suggested that the oral and gastric H. pylori were likely of completely different origins. Another genotype of H. Pylori may have infected the oral cavity after the initial infection of the stomach. Since the patients with an H. pylori infection in the stomach continue to live in an environment where H. pylori are abundant, they may have more opportunities for H. pylori re-infection.
Our findings are consistent with a study by Wongphutorn et al. The genotypes of H. pylori from saliva and stool samples were compared using a partial vacA gene sequence. For seven out of 12 individuals, saliva and stool sequences fell into different clusters on a phylogenetic tree, indicating intra-host genetic variation of H. pylori. Although this study used only one gene for their comparison, nearly half of the pairs had different genotypes [43].
However, the identity of gastric and oral H. pylori cannot be completely denied because of the following two possibilities. One possibility is a genetical mutation due to the environmental difference. The oral cavity is a challenging environment for microbial survival, since it undergoes high daily fluctuations in nutrient supply, temperature, pH, shear and mechanical forces from mastication and hygiene practices, and chemical exposure such as hygiene and pharmaceutical products [44]. Although the two related strains of six patients were originally of the same origin, the environmental difference may cause a mutation in the housekeeping genes, resulting in different allelic profiles. Although housekeeping genes are highly conserved [45,46], studies have reported that some housekeeping genes can mutate in response to changes in a specific environment [47,48]. Linz et al. reported that the mutation rate during the acute phase of H. pylori infection is more than 10 times faster than during the chronic infection phase [49]. Based on this, it is possible that H. pylori which originally invaded the oral cavity may have acquired some gene mutations during infection of the stomach, hence the discrepancy in the strain types.
Another possibility is mixed infections of H. pylori in the oral cavity and the stomach. Although the two H. pylori analyzed in this study were genetically mismatched, the same strain may be present in both the oral cavity and the stomach. There are some studies reporting mixed infections of multiple types of H. pylori in the oral cavity and stomach [50,51,52]. Palau et al. reported that, based on housekeeping genes, different strains of H. pylori were detected from the same site in the stomach of the same patient. This may also occur in the oral cavity [52]. Thus, a future study comparing H. pylori DNA extracted from samples taken at different locations is necessary.
In contrast, there were some studies reporting the homology of H. pylori in stomach and oral cavity [35,53,54]. Wang et al. compared cagA and vacA genotypes of H. pylori strains from both saliva and stomach in 31 patients with gastritis and peptic ulcer by PCR. The gastric sample was collected via biopsy from the antrum. The results showed 95% agreement between stomach H. pylori isolates and their corresponding saliva DNA in at least one cytotoxin genotype. The authors concluded that the same H. pylori strain may exist in the saliva and stomach in the same patient. However, the concordance rate of all four cytotoxin genotypes was only 27%, indicating considerable diversity between two comparison targets. Although DNA sequencing from three patients showed 66.9% to 78% homology of H. pylori from both sources, data for the other 28 subjects were not provided [55]. Since most studies that analyzed the homology of H. pylori in the stomach and oral cavity compared the genotypes of a limited number of pathogenic genes, it is not possible to draw conclusions about the presence or absence of homology in these strains.
Analyzing whether oral and gastric H. pylori are genetically identical is important for future strategies of H. pylori control. If the genetic relatedness between oral and gastric H. pylori was confirmed, the trigger of infection is either the mouth and the stomach at the same time, or the oral cavity functions as a reservoir for H. pylori. If H. pylori exist alive in the oral cavity and serve as a supply source for the stomach, eradication therapy for H. pylori will need to be performed simultaneously in the oral cavity and the stomach. It has been reported that the eradication efficiency in the stomach was 85.8%, while in the oral cavity, it was only 5.7%, revealing 55.6 of the pooled odds ratio (OR) [56].
If a different genotype of H. pylori exists in the oral cavity independently of H. pylori present in the stomach, the presence of oral H. pylori is not a risk factor for gastric infection. Oral H. pylori may exist as a part of the normal microflora in the adult oral cavity [13].
Several clinical studies suggested that periodontal therapy combined with H. pylori eradication treatment increased the eradication rate of gastric H. pylori compared with eradication treatment alone (OR 2.15; 95% Cl 1.47 to 3.14). In addition, the non-recurrence rate of gastric H. pylori infection increased in participants treated with periodontal therapy compared with those who received eradication therapy alone (OR 3.60; 95% Cl 2.11 to 6.15) [22]. Although these studies support the claim that the oral cavity is an important reservoir for gastric H. pylori infection, it has not been proven that H. pylori exist alive in the oral cavity, nor that a species genetically identical to oral H. pylori was detected in the stomach after the eradication. Within the limitations of this study, the methodology for analyzing the genetic relationship between oral and gastric H. pylori using MLST may be useful for discussing the necessity of eradication of oral H. pylori.
4. Materials and Methods
4.1. Participants
The study was conducted at Niigata University Medical and Dental Hospital (Niigata, Japan) between November 2019 and October 2020. We recruited patients, with a confirmed H. pylori infection using a fecal antigen test, who planned to admit for endoscopic surgery on the upper gastrointestinal cancers. The presence of at least one tooth in the oral cavity was part of the inclusion criteria, and participants using dentures were not excluded. The study protocol was approved by the Niigata University Ethics Committee (approval number 2019-0220), and the study was carried out in accordance with the approved guidelines. All participants signed an informed consent form before participating in the study.
4.2. Oral Sample Collection
An oral examination, followed by a collection of saliva and biofilm on teeth and tongue, was performed prior to endoscopic surgery. Smoking and oral hygiene status, such as frequency of oral cleaning and use of oral cleaning aids, were recorded. The number of decayed, missing, and filled teeth (DMFT) was also recorded. Unstimulated saliva (2 mL) was collected by spitting into a tube. Supragingival dental biofilm samples were collected by scraping from the upper incisors, lower incisors, upper right molars, and lower left molars using a sterile curette. If the participant had no teeth in the designated location, the sample was taken from the opposite site or denture. Each sample was transferred into a tube containing phosphate-buffered saline (PBS; pH 7). The superficial layers of the tongue were collected using five gentle strokes from the papillae circumvallatae to the anterior part of the tongue dorsum with a tongue brush (Tongue Cleaner Plus, Ci Medical, Ishikawa, Japan). Bacterial cells were retrieved by vigorous stirring in 20 mL PBS. The samples were centrifuged for 10 min at 10,000 rpm, washed twice with PBS, and stored at −80 °C until further use.
4.3. Gastrointestinal Endoscopy and Histologic Examination
Biopsy specimens were taken from three locations in the stomach using an endoscope: one from the vestibular region and the other two from the body of the stomach. A part of the samples was fixed with 10% formaldehyde and histologically examined by both Giemsa and H-E staining. The pieces were mixed and crushed with a Power masher II (Nippi, Incorporated, Tokyo, Japan) immediately after collection, and half of the amount was inoculated onto Helicobacter Selective Agar medium (Nissui Pharmaceutical co., Ltd., Tokyo, Japan) and incubated at 37 °C under slightly aerobic conditions (5% O2, 10% CO2, and 85% N2), for 72 h. The other half was stored at −80 °C until further use.
4.4. DNA Extraction and Nested PCR
DNA was extracted from the oral samples and biopsy specimens in the stomach as well as from a control strain, H. pylori NCTC11637, using the NucleoSpin® Microbial DNA Kit (TaKaRa Bio, Shiga, Japan) according to the manufacturer’s instructions. The quantity and purity of the DNA were assessed by spectrophotometry at 260/280 nm. The DNA was stored at −80 °C until processing.
An aliquot of the DNA extract was amplified using nested PCR as previously described [13], and the presence of H. pylori DNA in the sample was confirmed by 1.5% agarose gel electrophoresis. Briefly, the first round of PCR amplification was performed with Takara Ex Taq® Hot-Start Version (RR006A; TaKaRa Bio, Shiga, Japan) using the EHC-U (5′-CCCTCACGCCATCAGTCCCAAAAA-3′) and EHC-L (5′-AAGAAGTCAAAAACGCCCCAAAAC-3′) primers targeting an 860-bp fragment of H. pylori genomic DNA. The amplification comprised 40 cycles of denaturation at 98 °C for 10 s, annealing at 57 °C for 30 s, and extension at 72 °C for 1 min on a MiniAmpTM Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA). The expected product size was 417 bp, covering the area from 80,076 bp to 80,492 bp in the H. pylori genome [10].
For the nested PCR assay, the amplification product (1 µL) obtained by single-step PCR was re-amplified over 20 cycles under the same conditions as in the first round. The internal primer pair ET-5U (5′-GGCAAATCATAAGTCCGCAGAA-3′) and ET-5L (5′- TGAGACTTTCCTAGAAGCGGTGTT-3′) was used at a concentration of 50 pmol/µL. The expected size was 228 bp. A sample, in which the presence of H. pylori DNA was confirmed, was used for the subsequent analysis.
4.5. Sequencing and MLST Analysis
MLST was performed as described on PubMLST (http://pubmlst.org/helicobacter/, accessed on 8 January 2023) [30]. Primer sets targeting seven housekeeping genes, atpA, ureI, efp, mutY, ppa, trpC, and yphC were used. The sequences were designed with reference to the MLST website or an article by Osaki et al. (Table 5). Gene fragments containing these genes were amplified from H. pylori-positive specimens by the nested PCR as described above.

Table 5.
Primers for amplification of housekeeping genes using nested PCR.
Briefly, the first round of PCR amplification was performed using Takara Ex Taq® Hot-Start Version. The amplification comprised 40 cycles of denaturation at 98 °C for 10 s, annealing at 57 °C of each primer for 30 s, and extension at 72 °C for 1 min on a MiniAmp™ Thermal Cycler. For the nested PCR assay, the amplification product (1 µL) obtained by single-step PCR was re-amplified over 40 cycles under the same conditions as in the first round. H. pylori NCTC11637 DNA served as the positive control, and water was used as the negative control. Each PCR product was confirmed by 1.5% agarose gel electrophoresis.
The band visualized under LED light was cut out from the gel using a gel band cutter (FastGene ™ Agarose Gel Band Cutter, Nippon Genetics co., Ltd., Tokyo, Japan). Where multiple bands were present, bands of the same size as the positive control band were collected. Amplicons were purified from the gel slices using the Freeze’N squeeze DNA Gel Extraction Spin Clumns (Bio-Rad, Hercules, CA, USA) and NucleoSpin®︎ Gel and PCR Clean-up (TaKaRa Bio, Shiga, Japan). The amplified DNA was then sequenced and analyzed using the Applied Biosystems 3730xl DNA analyzer at Macrogen Japan corp. (Tokyo, Japan).
The sequences were uploaded onto PubMLST (http://pubmlst.org/helicobacter/, accessed on 8 January 2023) to determine the closest allele type for each gene. Using the allelic profile of each gene, the sequence type of the sample was determined.
4.6. Phylogenic Tree Analysis
To analyze the phylogeny of H. pylori obtained from the oral cavity and stomach, housekeeping gene sequences of 159 H. pylori isolates from 29 countries, in eight regions of the world, were downloaded from the BLAST website (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 8 January 2023) (Table S1).
Multiple alignments of the MLST genes was then performed using MEGA (V11). The aligned sequences were used to develop a phylogenetic tree. The amplicons obtained from the mouth and stomach were then sequenced and aligned with isolates from Europe, Africa, Asia, America and Oceania using multiple alignments with the MUSCLE program. Asia was classified into East Asia, Southeast Asia, and South Asia. Thereafter, a phylogenetic tree was constructed using the construct/Test Maximum Likelihood tree with the alignment result in MEGA. Unbiased bootstrap values were used to support the tree using MEGA’s Bootstrap Replications. A cluster that was bootstrapped 1000 times and had a bootstrap value of 95 or higher was considered supported.
4.7. Statistical Analysis
Data analysis was carried out using SPSS® 11.0 (SPSS, Chicago, IL, USA). The chi-squared test and Fisher’s exact probability test were used when applicable, and the results were considered statistically significant when the p-value was <0.05. Prevalence was expressed as a proportion and the OR was used to measure the strength of the association between the variables.
5. Conclusions
In conclusion, within the limitations that require careful interpretation, different genotypes of H. pylori exist in the oral cavity independently of H. pylori present in the stomach in most cases. There are some cases in which nearly related H. pylori strains with close evolutionary distance are present in the stomach and the oral cavity. Oral H. pylori may exist as a part of the normal microflora in the adult oral cavity.
A method using MLST for analyzing the genetic relationship between oral and gastric H. pylori may be useful for scientific elucidation of infection routes and discussing the necessity of eradication of oral H. pylori. It is necessary to establish a culture method for oral H. pylori to elucidate whether the oral cavity acts as the source of gastric infection, as our analysis was based on a limited number of combinations of allele sequences.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24032211/s1.
Author Contributions
Conception and design of the study, R.N. and Y.N.; acquisition of data and analysis, R.N. and H.S.; interpretation of data, S.T. (Shoji Takenaka) and Y.N.; Drafting the article, R.N. and S.T. (Shoji Takenaka); revising the article critically for important intellectual content, R.N., H.S., S.T. (Shoji Takenaka), J.Y., S.T. (Shuji Terai), H.M. and Y.N.; funding acquisition, Y.N.; project administration, Y.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by JSPS KAKENHI (grant number 19K22704, 21K21081).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Niigata University Ethics Committee (approval number 2019-0220, 2 October 2019). Data presented in this study are available upon reasonable request from the corresponding author due to restrictions, such as privacy or ethical concerns. The data are not publicly available due to assured participant confidentiality.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below DDBJ. Accession numbers are shown in the article/Table S2.
Acknowledgments
The authors are grateful to Tatsuya Ohsumi for his technical support regarding the nested PCR procedure.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Marshall, B.J.; Warren, J.R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1984, 1, 1311–1315. [Google Scholar] [CrossRef] [PubMed]
- Dooley, C.P.; Cohen, H.; Fitzgibbons, P.L.; Bauer, M.; Appleman, M.D.; Perez-Perez, G.I.; Blaser, M.J. Prevalence of Helicobacter pylori infection and histologic gastritis in asymptomatic persons. N. Engl. J. Med. 1989, 321, 1562–1566. [Google Scholar] [CrossRef] [PubMed]
- Parsonnet, J.; Friedman, G.D.; Vandersteen, D.P.; Chang, Y.; Vogelman, J.H.; Orentreich, N.; Sibley, R.K. Helicobacter pylori Infection and the Risk of Gastric Carcinoma. N. Engl. J. Med. 1991, 325, 1127–1131. [Google Scholar] [CrossRef]
- Nomura, A.; Stemmermann, G.N.; Chyou, P.H.; Kato, I.; Perez-Perez, G.I.; Blaser, M.J. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N. Engl. J. Med. 1991, 325, 1132–1136. [Google Scholar] [CrossRef]
- Pounder, R.E.; Ng, D. The prevalence of Helicobacter pylori infection in different countries. Aliment. Pharmacol. Ther. 1995, 9 (Suppl. 2), 33–39. [Google Scholar] [PubMed]
- Kayali, S.; Manfredi, M.; Gaiani, F.; Bianchi, L.; Bizzarri, B.; Leandro, G.; Mario, F.D.; Angelis, G.L. Helicobacter pylori, transmission routes and recurrence of infection; State of the art. Acta Biomed. 2018, 89, 72–76. [Google Scholar] [CrossRef]
- Wang, C.; Nishiyama, T.; Kikuchi, S.; Inoue, M.; Sawada, N.; Tsugane, S.; Lin, Y. Changing trends in the prevalence of H. pylori infection in Japan (1908–2003): A systematic review and meta-regression analysis of 170,752 individuals. Sci. Rep. 2017, 7, 15491. [Google Scholar] [CrossRef]
- Miyamoto, R.; Okuda, M.; Lin, Y.; Murotani, K.; Okumura, A.; Kikuchi, S. Rapidly decreasing prevalence of Helicobacter pylori among Japanese children and adolescents. J. Infect. Chemother. 2019, 25, 526–530. [Google Scholar] [CrossRef]
- Dowsett, S.A.; Kowolik, M.J. Oral Helicobacter pylori: Can we stomach it? Crit. Rev. Oral Biol. Med. 2003, 14, 226–233. [Google Scholar] [CrossRef]
- Song, Q.; Lange, T.; Spahr, A.; Adler, G.; Bode, G. Characteristic distribution pattern of Helicobacter pylori in dental plaque and saliva detected with nested PCR. J. Med. Microbiol. 2000, 49, 349–353. [Google Scholar] [CrossRef]
- Miyabayashi, H.; Furihata, K.; Shimizu, T.; Ueno, I.; Akamatsu, T. Infuluence of oral Helicobacter pylori on the success of eradication therapy against gastric Helicobacter pylori. Helicobacter 2000, 5, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Román-Román, A.; Giono-Cerezo, S.; Camorlinga-Ponce, M.; Martínez-Carrillo, D.N.; Loaiza-Loeza, S.; Fernández-Tilapa, G. VacA genotypes of Helicobacter pylori in the oral cavity and stomach of patients with chronic gastritis and gastric ulcer. Enferm. Infecc. Microbiol. Clin. 2013, 31, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Nagata, R.; Ohsumi, T.; Takenaka, S.; Noiri, Y. Current prevalence of oral Helicobacter pylori among Japanese adults determined using a nested polymerase chain reaction assay. Pathogens 2020, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Wu, J.Y.; Abudayyeh, S.; Hoffman, J.; Brahem, H.; Al-Khatib, K.; Yamaoka, Y.; Graham, D. Unreliability of results of PCR detection of Helicobacter pylori in clinical or environmental samples. J. Clin. Microbiol. 2009, 47, 738–742. [Google Scholar] [CrossRef] [PubMed]
- Adler, I.; Muiño, A.; Aguas, S.; Harada, L.; Diaz, M.; Lence, A.; Labbrozzi, M.; Muiño, J.M.; Elsner, B.; Avagnina, A.; et al. Helicobacter pylori and oral pathology: Relationship with the gastric infection. World J. Gastroenterol. 2014, 20, 9922–9935. [Google Scholar] [CrossRef]
- Anand, P.S.; Kamath, K.P.; Anil, S. Role of dental plaque, saliva and periodontal disease in Helicobacter pylori infection. World J. Gastroenterol. 2014, 20, 5639–5653. [Google Scholar] [CrossRef]
- Kadota, T.; Hamada, M.; Nomura, R.; Ogaya, Y.; Okawa, R.; Uzawa, N.; Nakano, K. Distribution of Helicobacter pylori and periodontopathic bacterial species in the oral cavity. Biomedicines 2020, 8, 161. [Google Scholar] [CrossRef]
- Yee, J.K.C. Are the view of Helicobacter pylori colonized in the oral cavity an illusion? Exp. Mol. Med. 2017, 49, e397. [Google Scholar] [CrossRef]
- Iwai, K.; Watanabe, I.; Yamamoto, T.; Kuriyama, N.; Matsui, D.; Nomura, R.; Ogaya, Y.; Oseko, F.; Adachi, K.; Takizawa, S.; et al. Association between Helicobacter pylori infection and dental pulp reservoirs in Japanese adults. BMC Oral Health 2019, 19, 267. [Google Scholar] [CrossRef]
- Lauritano, D.; Cura, F.; Candotto, V.; Gaudio, R.M.; Mucchi, D.; Carinci, F. Periodontal pockets as a reservoir of Helicobacter pylori causing relapse of gastric ulcer: A review of the literature. J. Biol. Regul. Homeost. Agents 2015, 29 (Suppl. 1), 123–126. [Google Scholar]
- Payão, S.L.M.; Rasmussen, L.T. Helicobacter pylori and its reservoirs: A correlation with the gastric infection. World J. Gastrointest. Pharmacol. Ther. 2016, 7, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Yan, X.; Zhou, Y.; Li, W.X. Periodontal therapy as adjunctive treatment for gastric Helicobacter pylori infection. Cochrane Database Syst. Rev. 2016, 2, CD009477. [Google Scholar] [CrossRef] [PubMed]
- Tongtawee, T.; Wattanawongdon, W.; Simawaranon, T. Effects of periodontal therapy on eradication and recurrence of Helicobacter pylori infection after successful treatment. J. Int. Med. Res. 2019, 47, 875–883. [Google Scholar] [CrossRef] [PubMed]
- López-Valverde, N.; Macedo de Sousa, B.M.; López-Valverde, A.; Suárez, A.; Rodríguez, C.; Aragoneses, J.M. Possible association of periodontal diseases with Helicobacter pylori gastric infection: A systematic review and meta-analysis. Front. Med. 2022, 9, 822194. [Google Scholar] [CrossRef] [PubMed]
- Osaki, T.; Okuda, M.; Ueda, J.; Konno, M.; Yonezawa, H.; Hojo, F.; Yagyu, K.; Lin, Y.; Fukuda, Y.; Kikuchi, M.; et al. Multilocus sequence typing of DNA from faecal specimens for the analysis of intra-familial transmission of Helicobacter pylori. J. Med. Microbiol. 2013, 62, 761–765. [Google Scholar] [CrossRef]
- Hashi, K.; Imai, C.; Yahara, K.; Tahmina, K.; Hayashi, T.; Azuma, T.; Miyabe-Nishiwaki, T.; Sato, H.; Matsuoka, M.; Niimi, A.; et al. Evaluating the origin and virulence of a Helicobacter pylori cagA-positive strain isolated from a non-human primate. Sci. Rep. 2018, 8, 15981. [Google Scholar] [CrossRef]
- Floridia-Yapur, N.; Rusman, F.; Diosque, P.; Tomasini, N. Genome data vs MLST for exploring intraspecific evolutionary history in bacteria: Much is not always better. Infect. Genet. Evol. 2021, 93, 104990. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, N. Diagnosis of Helicobacter pylori by invasive test: Histology. Ann. Transl. Med. 2015, 3, 10. [Google Scholar] [CrossRef]
- Khan, H.; Rauf, F.; Muhammad, N.; Javaid, M.; Alam, S.; Nasir, S. Comparison of special stains (Giemsa stain and Modified toluidine blue stain) with immunohistochemistry as gold standard for the detection of H. pylori in gastric biopsies. Arab J. Gastroenterol. 2022, 23, 75–81. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Yokota, S.; Konno, M.; Fujiwara, S.; Toita, N.; Takahashi, M.; Yamamoto, S.; Ogasawara, N.; Shiraishi, T. Intrafamilial, preferentially mother-to-child and intraspousal, Helicobacter pylori infection in japan determined by mutilocus sequence typing and random amplified polymorphic DNA fingerprinting. Helicobacter 2015, 20, 334–342. [Google Scholar] [CrossRef]
- Morales-Espinosa, R.; Fernandez-Presas, A.; Gonzalez-Valencia, G.; Flores-Hernandez, S.; Delgado-Sapien, G.; Mendez-Sanchez, J.L.; Sanshez-Quezada, E.; Muñoz-Pérez, L.; Leon-Aguilar, R.; Hernandez-Guerrero, J.; et al. Helicobacter pylori in the oral cavity is associated with gastroesophageal disease. Oral Microbiol. Immunol. 2009, 24, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Yahiro, K.; Niidome, T.; Kimura, M.; Hatakeyama, T.; Aoyagi, H.; Kurazono, H.; Imagawa, K.; Wada, A.; Moss, J.; Hirayama, T. Activation of Helicobacter pylori VacA toxin by alkaline or acid conditions increases its binding to a 250-kDa receptor protein-tyrosine phosphatase beta. J. Biol. Chem. 1999, 274, 36693–36699. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Osato, M.S.; Sepulveda, A.R.; Gutierrez, O.; Figura, N.; Kim, J.G.; Kodama, T.; Kashima, K.; Graham, D.Y. Molecular epidemiology of Helicobacter pylori: Separation of H. pylori from East Asian and non-Asian countries. Epidemiol. Infect. 2000, 124, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; EI-Zimaity, H.M.; Gutierrez, O.; Figura, N.; Kim, J.G.; Kodama, T.; Kashima, K.; Graham, D.Y. Relationship between the cagA3′ repeat region of Helicobacter pylori, gastric histology, and susceptibility to low pH. Gastroenterology 1999, 117, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Orito, E.; Mizokami, M.; Gutierrez, O.; Saitou, N.; Kodama, T.; Osato, M.S.; Kim, J.G.; Ramirez, F.C.; Mahachai, V. Helicobacter pylori in North and South America before Columbus. FEBS letters. 2002, 517, 180–184. [Google Scholar] [CrossRef]
- Silva, D.G.; Tinoco, E.M.B.; Rocha, G.A.; Rocha, A.M.C.; Guerra, J.B.; Saraiva, I.E.B.; Queiroz, D.M.M. Helicobacter pylori transiently in the mouth may participate in the transmission of infection. Mem. Inst. Oswaldo Cruz. 2010, 105, 657–660. [Google Scholar] [CrossRef]
- Sulo, P.; Šipková, B. DNA diagnostics for reliable and universal identification of Helicobacter pylori. World J. Gastroenterol. 2021, 27, 7100–7112. [Google Scholar] [CrossRef]
- Palau, M.; Piqué, N.; Ramírez-Lázaro, M.J.; Lario, S.; Calvet, X.; Miñana-Galbis, D. Whole-genome sequencing and comparative genomics of three Helicobacter pylori strains isolated from the stomach of a patient with adenocaricinoma. Pathogens 2021, 10, 331. [Google Scholar] [CrossRef]
- Yassine, I.; Lefèvre, S.; Hansen, E.E.; Ruckly, C.; Carle, I.; Lejay-Collin, M.; Fabre, L.; Rafei, R.; Clermont, D.; Gandara, M.P.; et al. Population structure analysis and laboratory monitoring of Shigella by core-genome multilocus sequence typing. Nat. Commun. 2022, 13, 551. [Google Scholar] [CrossRef]
- Takami, S.; Hayashi, T.; Akashi, H.; Shimoyama, T.; Tamura, T. Genetic heterogeneity of Helicobacter pylori by pulse-field gel electrophoresis and re-evaluation of DNA homology. Eur. J. Gastroenterol. Hepatol. 1994, 6 (Suppl. 1), S53–S56. [Google Scholar]
- Maiden, M.C.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russell, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.; Zurth, K.; Caugant, D.A.; et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 1998, 95, 3140–3145. [Google Scholar] [CrossRef] [PubMed]
- Wongphutorn, P.; Chomvarin, C.; Sripa, B.; Namwat, W.; Faksri, K. Detection and genotyping of Helicobacter pylori in saliva versus stool samples from asymptomatic individuals in Northeastern Thailand reveals intra-host tissue-specific H. pylori subtypes. BMC Microbiol. 2018, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Radaic, A.; Kapila, Y.L. The oralome and its dysbiosis; New insights into oral microbiome-host interactions. Comput. Struct. Biotechnol. J. 2021, 19, 1335–1360. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Harayama, S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 1995, 61, 1104–1109. [Google Scholar] [CrossRef]
- Clarridge, J.E., III. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef]
- Mendoza-Elizalde, S.; Arteaga-Resendiz, N.K.; Valencia-Mayoral, P.; Luna, R.C.; Moreno-Espinosa, S.; Arenas-Huertero, F.; Zúñiga, G.; Velázquez-Guadarrama, N. Diversification of the vacAs1m1 and vacAs2m2 strains of Helicobacter pylori in Meriones unguiculatus. Front. Microbiol. 2016, 7, 1758. [Google Scholar] [CrossRef]
- Mendoza-Elizalde, S.; Cortés-Márquez, A.C.; Zuñiga, G.; Cerritos, R.; Valencia-Mayoral, P.; Sánchez, A.C.; Olivares-Clavijo, H.; Velázquez-Guadarrama, N. Inference from the analysis of genetic structure of Helicobacter pylori strains isolates from two paediatric patients with recurrent infection. BMC Microbiol. 2019, 19, 184. [Google Scholar] [CrossRef]
- Linz, B.; Windsor, H.M.; McGraw, J.J.; Hansen, L.M.; Gajewski, J.P.; Tomsho, L.P.; Hake, C.M.; Solnick, J.V.; Schuster, S.C.; Marshall, B.J. A mutation burst during the acute phase of Helicobacter pylori infection in humans and rhesus macaques. Nat. Commun. 2014, 5, 4165. [Google Scholar] [CrossRef]
- Raymond, J.; Thiberg, J.M.; Chevalier, C.; Kalach, N.; Bergeret, M.; Labigne, A.; Dauga, C. Genetic and transmission analysis of Helicobacter pylori strains within a family. Emerg. Infect. Dis. 2004, 10, 1816–1821. [Google Scholar] [CrossRef]
- Sheu, S.M.; Sheu, B.S.; Lu, C.C.; Yang, H.B.; Wu, J.J. Mixed infections of Helicobacter pylori: Tissue tropism and histological significance. Clin. Microbiol. Infect. 2009, 15, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Palau, M.; Kulmann, M.; Ramírez-Lázaro, M.J.; Lario, S.; Quilez, M.E.; Campo, R.; Piqué, N.; Calvet, X.; Miñana-Galbis, D. Usefulness of housekeeping genes for the diagnosis of Helicobacter pylori infection, strain discrimination and detection of multiple infection. Helicobacter 2016, 21, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Momtaz, H.; Souod, N.; Dabiri, H.; Sarshar, M. Study of Helicobacter pylori genotype status in saliva, dental plaques, stool and gastric biopsy samples. World J. Gastroenterol. 2012, 18, 2105–2111. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Li, W.; Shu, X.; Peng, K.; Zhang, Y.; Jiang, M. Genetic variation of Helicobacter pylori in the oral cavity and stomach detected using thymine adenine cloning in children with chronic gastritis. Pediatr. Infect. Dis. J. 2014, 33, e1–e6. [Google Scholar] [CrossRef]
- Wang, J.; Chi, D.S.; Laffan, J.J.; Li, C.; Ferguson Jr, D.A.; Litchfield, P.; Thomas, E. Comparison of cytotoxin genotypes of Helicobacter pylori in stomach and saliva. Dig. Dis. Sci. 2002, 47, 1850–1856. [Google Scholar] [CrossRef]
- Zou, Q.-H.; Li, R.-Q. Helicobacter pylori in the oral cavity and gastric mucosa: A meta-analysis. J. Oral. Pathol. Med. 2011, 40, 317–324. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).