T-Cell Immunity in COVID-19-Recovered Individuals and Individuals Vaccinated with the Combined Vector Vaccine Gam-COVID-Vac
Abstract
1. Introduction
2. Results
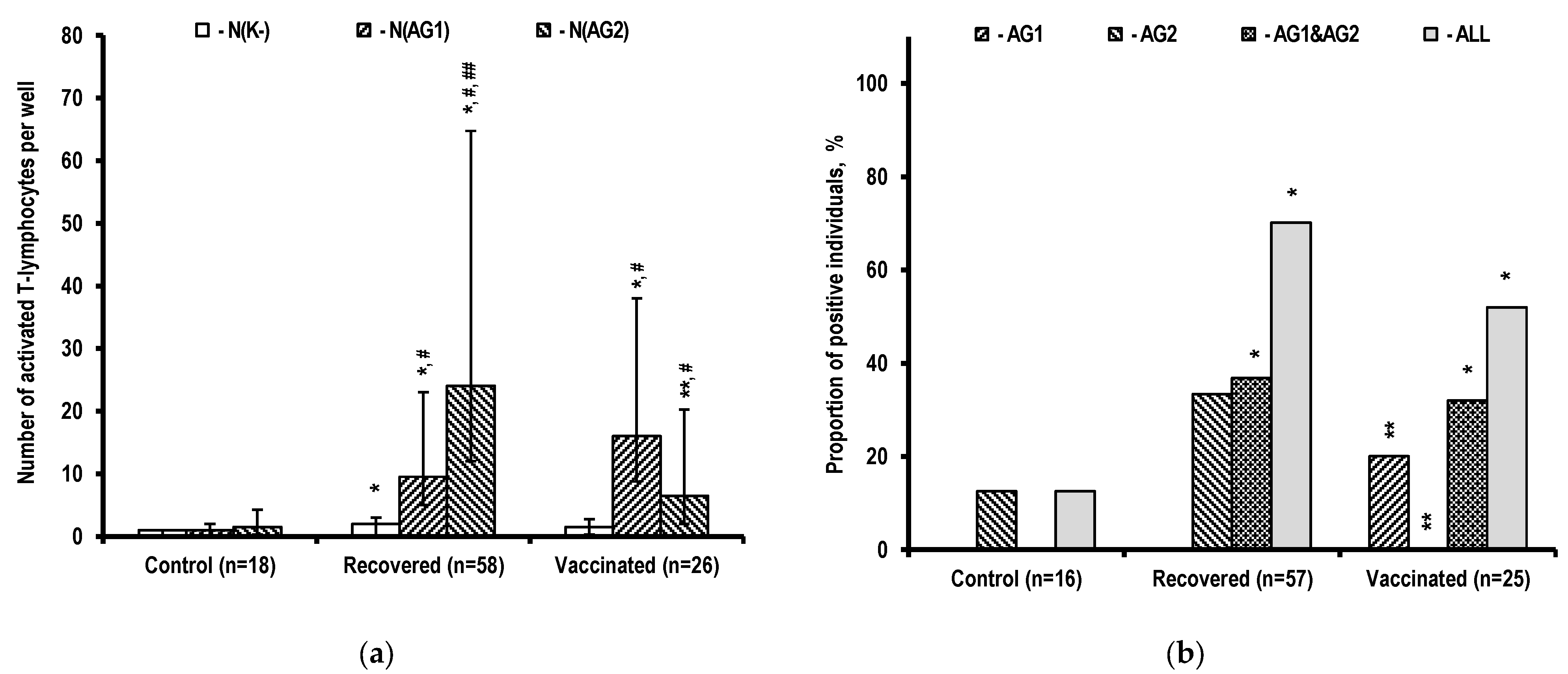
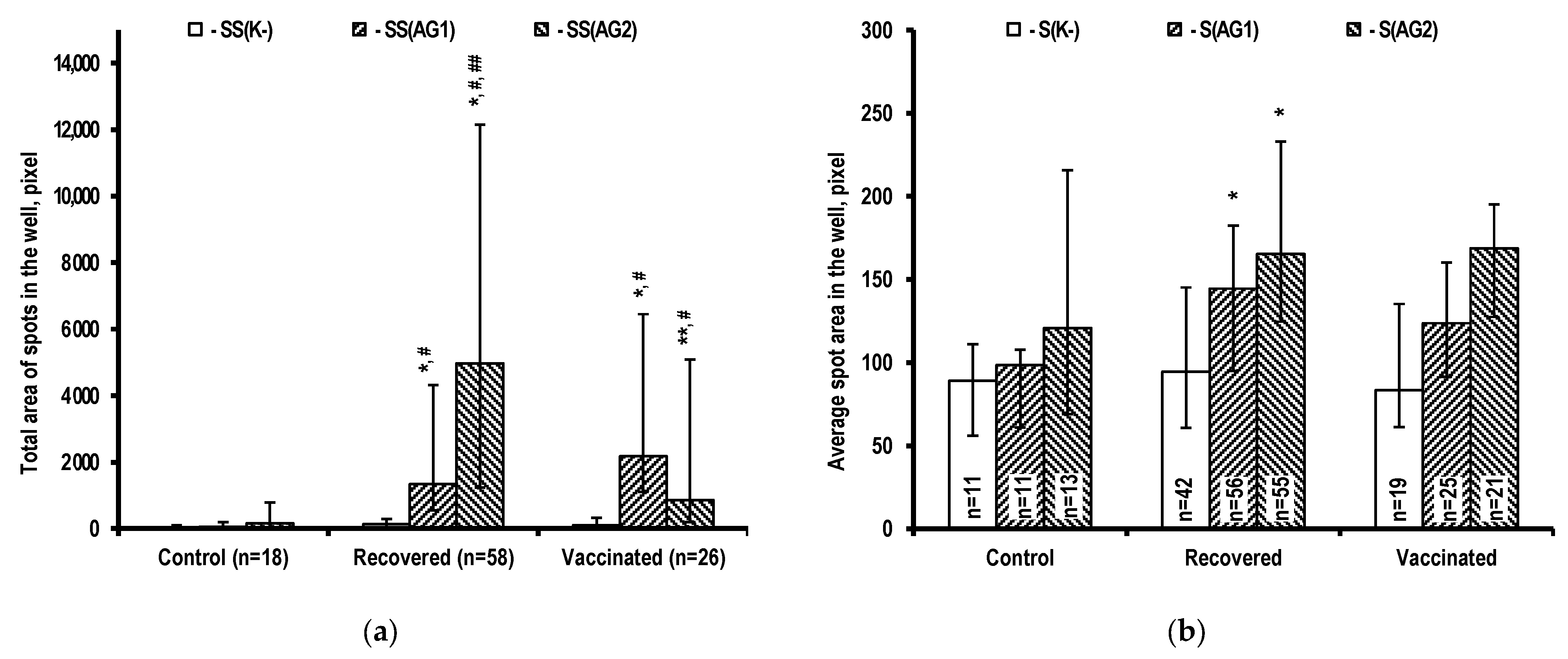
2.1. Assessment of T-Cell Immunity
2.2. Determination of Antibodies Specific to SARS-CoV-2 Antigens
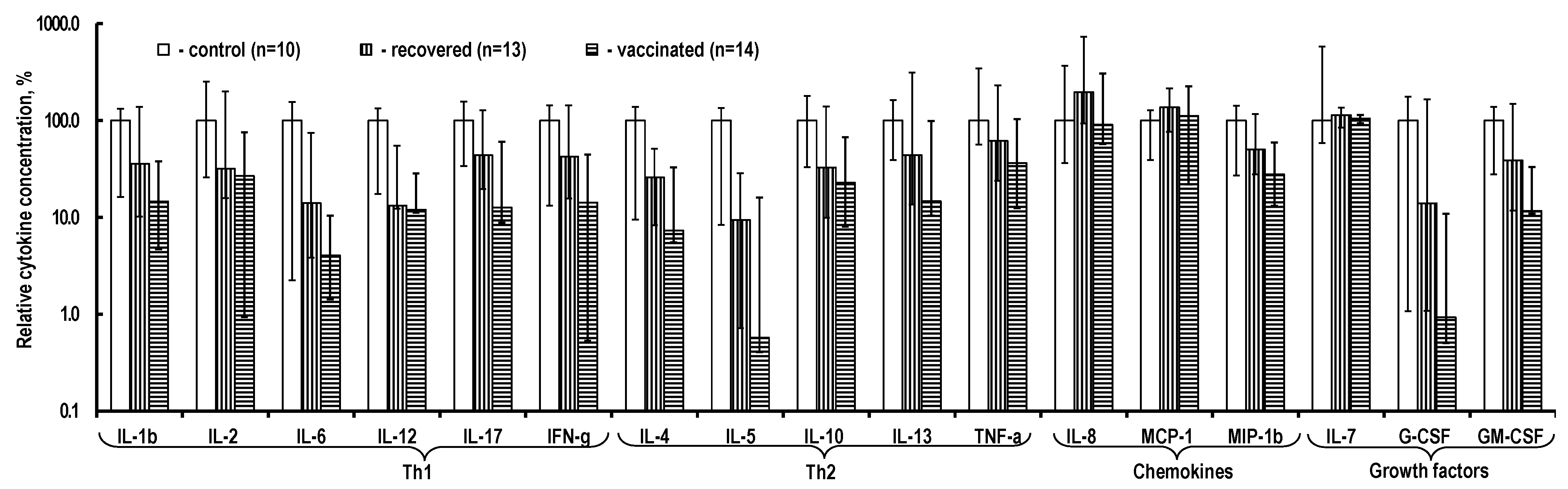
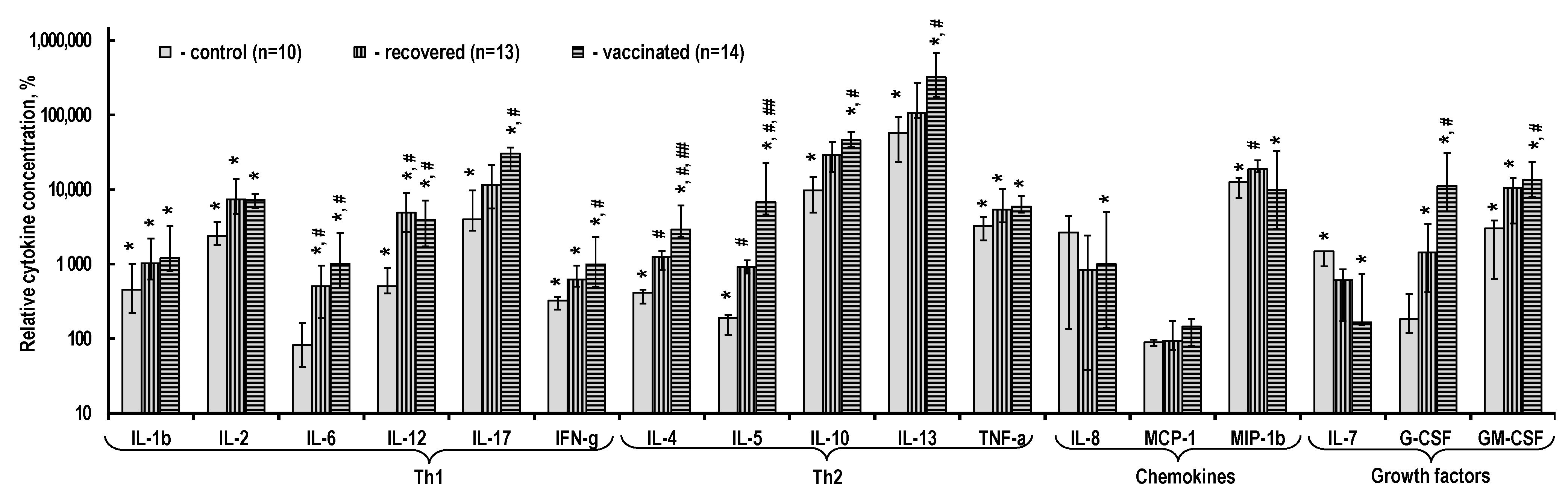
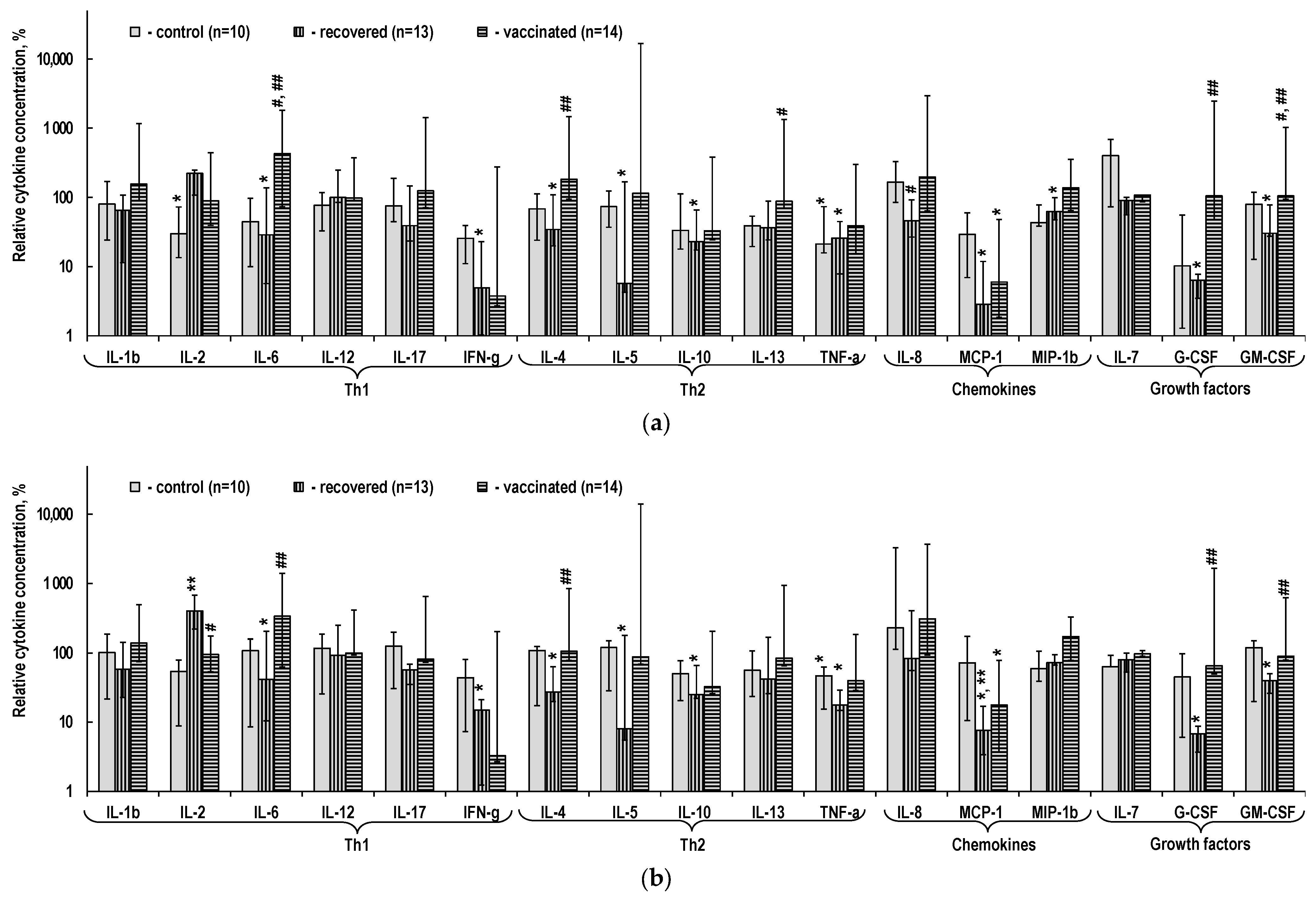
2.3. Cytokine Content in the PBMC Culture Medium
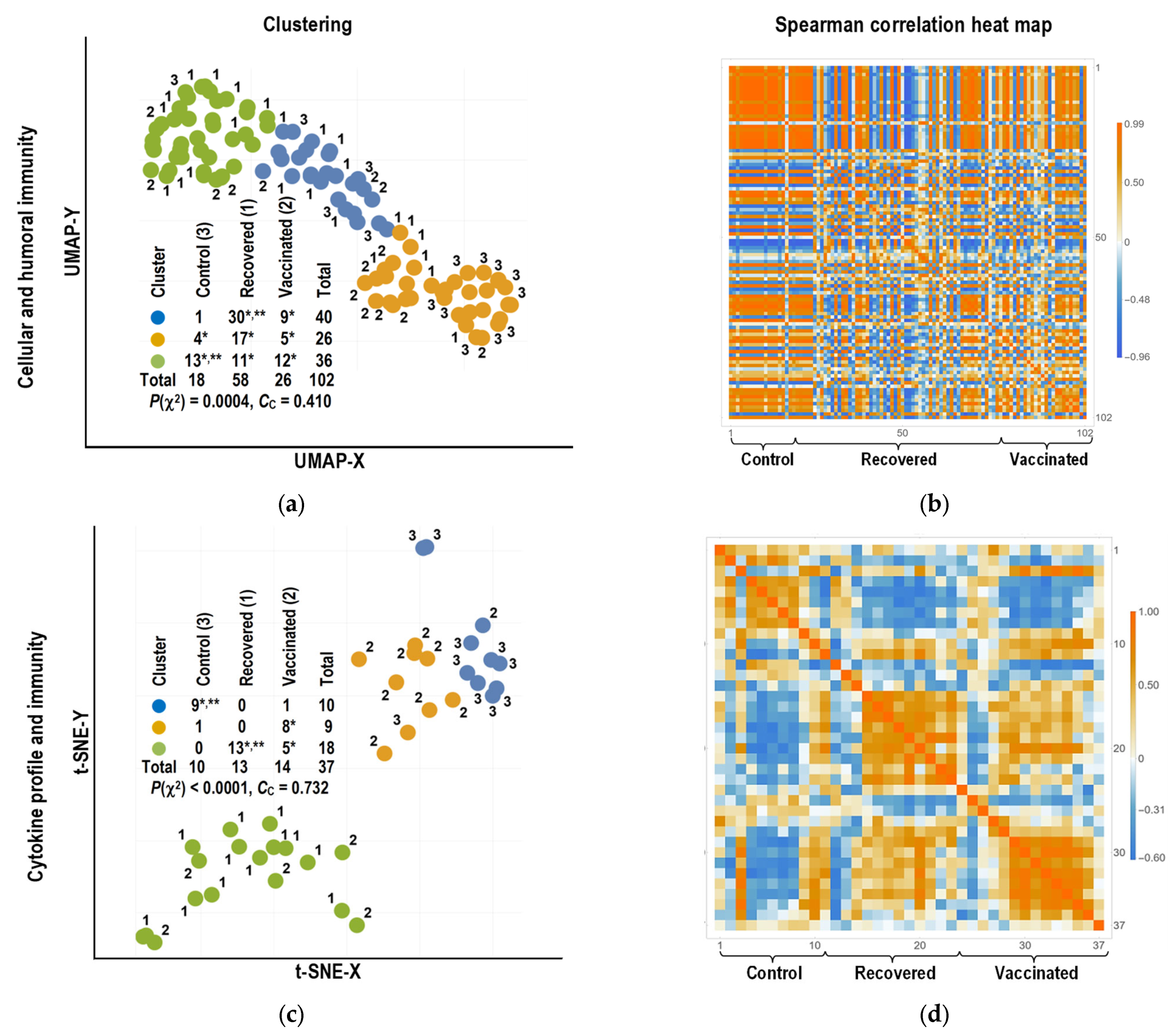
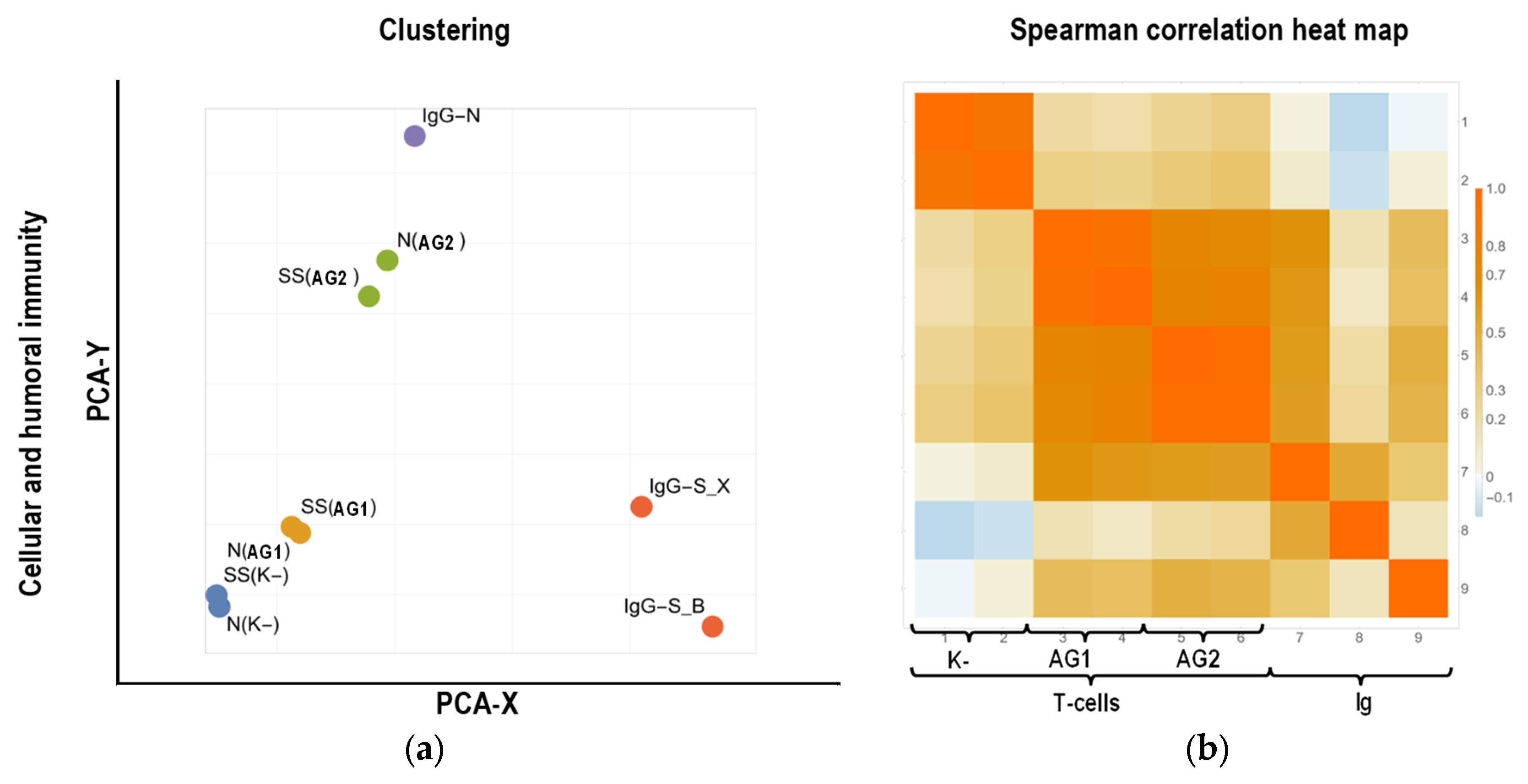
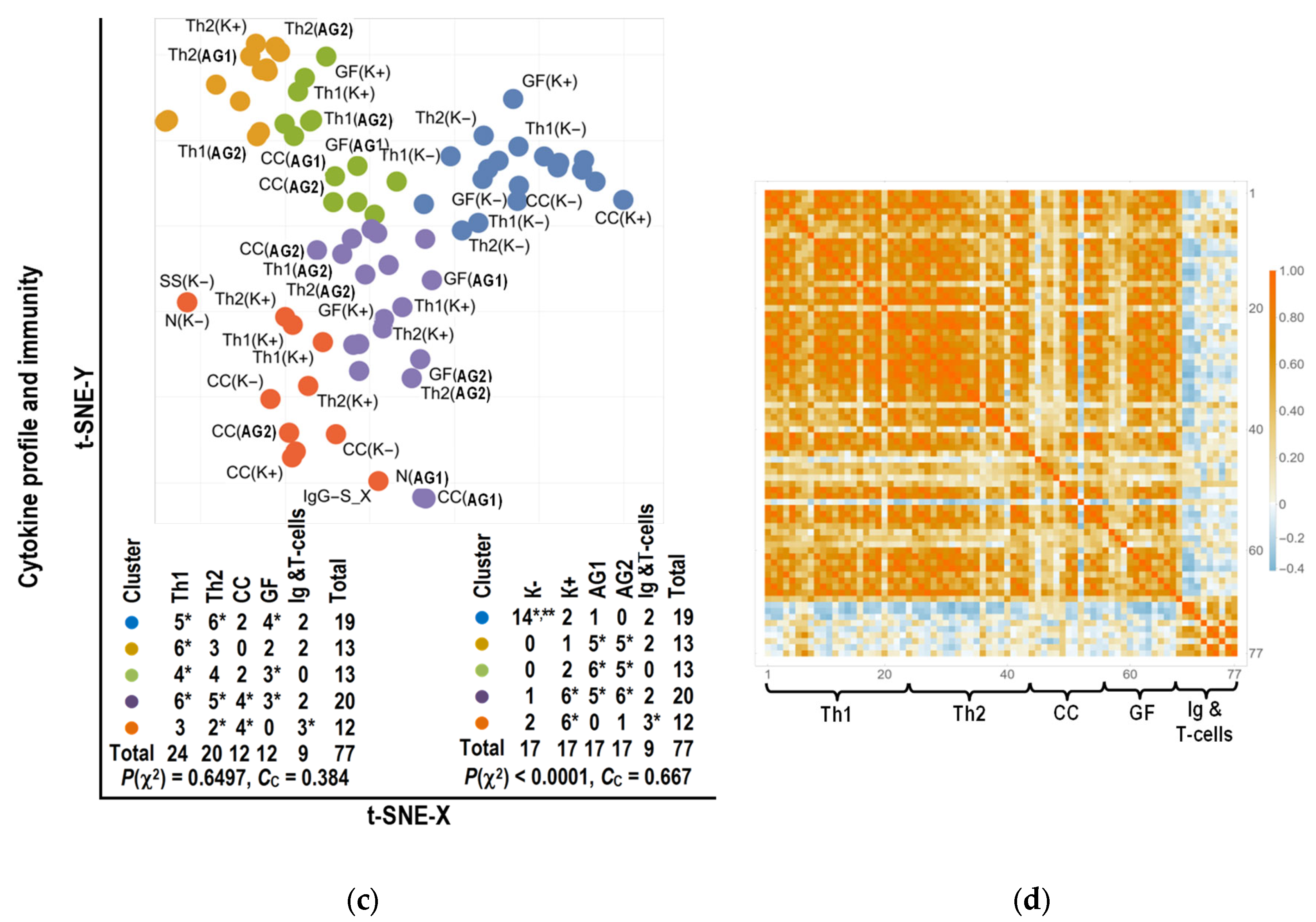
2.4. Bioinformatics Analysis
2.4.1. Clustering of Individuals
2.4.2. Clustering of Parameters
3. Discussion
3.1. Specific T-Cell Immune Response Characteristics during SARS-CoV-2 Infection and after Vaccination
3.2. Cytokine Production by T-Cells Sensitized upon Infection with SARS-CoV-2 and after Vaccination
3.3. Bioinformatics Analysis
4. Materials and Methods
4.1. Samples
4.2. Patients
- signed informed consent;
- age ≥ 18 years;
- negative PCR test during blood sampling for the study;
- absence of concomitant diseases and conditions associated with impaired immunity (autoimmune diseases, malignant neoplasms, HIV infection);
- no treatment with immunomodulatory drugs within three months prior to participation in the study.
- body temperature < 38 °C, cough, fatigue, sore throat;
- absence of criteria for moderate and severe COVID-19.
- body temperature > 38 °C;
- respiratory rate > 22 breaths per minute;
- shortness of breath during physical exertion;
- changes in CT (radiography), typical of a viral lesion;
- SpO2 < 95%;
- serum CRP > 10 mg/L.
4.3. Virus Identification
4.4. Assessment of T-Cell Immune Response
4.5. Determination of Antibody Content
4.6. Determination of Cytokine Concentration
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed on 10 December 2022).
- Lavelle, E.C.; Ward, R.W. Mucosal Vaccines—Fortifying the Frontiers. Nat. Rev. Immunol. 2021, 22, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Crotty, S. Adaptive Immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef]
- Jordan, S.C. Innate and Adaptive Immune Responses to SARS-CoV-2 in Humans: Relevance to Acquired Immunity and Vaccine Responses. Clin. Exp. Immunol. 2021, 204, 310–320. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Wong, B.H.L.; Chan, K.H.; Chu, C.M.; Tsoi, H.W.; Huang, Y.; Peiris, J.S.M.; Yuen, K.Y. Longitudinal Profile of Immunoglobulin G (IgG), IgM, and IgA Antibodies against the Severe Acute Respiratory Syndrome (SARS) Coronavirus Nucleocapsid Protein in Patients with Pneumonia due to the SARS Coronavirus. Clin. Vaccine Immunol. 2004, 11, 665–668. [Google Scholar] [CrossRef]
- Murphy, B.R. Mucosal Immunity to Viruses. In Mucosal Immunology, 3rd ed.; Mestecky, J., Lamm, M.E., Ogra, P., Strober, W., Bienenstock, J., McGhee, J., Mayer, L., Eds.; Academic Press: New York, NY, USA, 2005; pp. 799–813. [Google Scholar] [CrossRef]
- Sheikh-Mohamed, S.; Isho, B.; Chao, G.Y.C.; Zuo, M.; Cohen, C.; Lustig, Y.; Nahass, G.R.; Salomon-Shulman, R.E.; Blacker, G.; Fazel-Zarandi, M.; et al. Systemic and Mucosal IgA Responses Are Variably Induced in Response to SARS-CoV-2 MRNA Vaccination and Are Associated With Protection against Subsequent Infection. Mucosal Immunol. 2022, 15, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Padariya, M.; Kalathiya, U.; Mikac, S.; Dziubek, K.; Fernandez, M.C.T.; Sroka, E.; Fahraeus, R.; Sznarkowska, A. Viruses, Cancer and Non-Self Recognition. Open Biol. 2021, 11, 200348. [Google Scholar] [CrossRef] [PubMed]
- Rijkers, G.T.; Weterings, N.; Obregon-Henao, A.; Lepolder, M.; Dutt, T.S.; Van Overveld, F.J.; Henao-Tamayo, M. Antigen Presentation of MRNA-Based and Virus-Vectored SARS-CoV-2 Vaccines. Vaccines 2021, 9, 848. [Google Scholar] [CrossRef]
- Miyazawa, M. Immunopathogenesis of SARS-CoV-2-Induced Pneumonia: Lessons from Influenza Virus Infection. Inflamm. Regen. 2020, 40, 39. [Google Scholar] [CrossRef]
- Sadarangani, M.; Marchant, A.; Kollmann, T.R. Immunological Mechanisms of Vaccine-Induced Protection against COVID-19 in Humans. Nat. Rev. Immunol. 2021, 21, 475–484. [Google Scholar] [CrossRef]
- Bertoletti, A.; Le Bert, N.; Qui, M.; Tan, A.T. SARS-CoV-2-Specific T cells in Infection and Vaccination. Cell. Mol. Immunol. 2021, 18, 2307–2312. [Google Scholar] [CrossRef]
- Schmidt, A.; Lapuente, D. T Cell Immunity against Influenza: The Long Way from Animal Models towards a Real-Life Universal Flu Vaccine. Viruses 2021, 13, 199. [Google Scholar] [CrossRef] [PubMed]
- Lange, J.; Rivera-Ballesteros, O.; Buggert, M. Human Mucosal Tissue-Resident Memory T Cells in Health and Disease. Mucosal Immunol. 2021, 15, 389–397. [Google Scholar] [CrossRef]
- Crowl, J.T.; Heeg, M.; Ferry, A.; Milner, J.J.; Omilusik, K.D.; Toma, C.; He, Z.; Chang, J.T.; Goldrath, A.W. Tissue-Resident Memory CD8+ T Cells Possess Unique Transcriptional, Epigenetic and Functional Adaptations to Different Tissue Environments. Nat. Immunol. 2022, 23, 1121–1131. [Google Scholar] [CrossRef]
- Paik, D.H.; Farber, D.L. Anti-Viral Protective Capacity of Tissue Resident Memory T Cells. Curr. Opin. Virol. 2021, 46, 20–26. [Google Scholar] [CrossRef]
- Chentoufi, A.A.; Dhanushkodi, N.R.; Srivastava, R.; Prakash, S.; Coulon, P.-G.A.; Zayou, L.; Vahed, H.; Chentoufi, H.A.; Hormi-Carver, K.K.; BenMohamed, L. Combinatorial Herpes Simplex Vaccine Strategies: From Bedside to Bench and Back. Front. Immunol. 2022, 13, 1606. [Google Scholar] [CrossRef] [PubMed]
- Ong, D.S.; Fragkou, P.C.; Schweitzer, V.A.; Chemaly, R.F.; Moschopoulos, C.D.; Skevaki, C. How to Interpret and Use COVID-19 Serology and Immunology Tests. Clin. Microbiol. Infect. 2021, 27, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Wirsching, S.; Harder, L.; Heymanns, M.; Gröndahl, B.; Hilbert, K.; Kowalzik, F.; Meyer, C.; Gehring, S. Long-Term, CD4+ Memory T Cell Response to SARS-CoV-2. Front. Immunol. 2022, 13, 800070. [Google Scholar] [CrossRef]
- Liu, J.; Jaijyan, D.K.; Tang, Q.; Zhu, H. Promising Cytomegalovirus-Based Vaccine Vector Induces Robust CD8+ T-Cell Response. Int. J. Mol. Sci. 2019, 20, 4457. [Google Scholar] [CrossRef]
- Struzik, J.; Szulc-Dąbrowska, L. NF-κB As an Important Factor in Optimizing Poxvirus-Based Vaccines against Viral Infections. Pathogens 2020, 9, 1001. [Google Scholar] [CrossRef]
- Kudlay, D.; Kofiadi, I.; Khaitov, M. Peculiarities of the T Cell Immune Response in COVID-19. Vaccines 2022, 10, 242. [Google Scholar] [CrossRef]
- De Araújo-Souza, P.S.; Hanschke, S.C.H.; Viola, J.P.B. Epigenetic Control of Interferon-Gamma Expression in CD8 T Cells. J. Immunol. Res. 2015, 2015, 849573. [Google Scholar] [CrossRef] [PubMed]
- Nash, G.; Paidimuddala, B.; Zhang, L. Structural Aspects of the MHC Expression Control System. Biophys. Chem. 2022, 284, 106781. [Google Scholar] [CrossRef] [PubMed]
- Schoenborn, J.R.; Wilson, C.B. Regulation of Interferon-γ during Innate and Adaptive Immune Responses. Adv. Immunol. 2007, 96, 41–101. [Google Scholar] [CrossRef]
- De Benedetti, F.; Prencipe, G.; Bracaglia, C.; Marasco, E.; Grom, A.A. Targeting Interferon-Γ in Hyperinflammation: Opportunities and Challenges. Nat. Rev. Rheumatol. 2021, 17, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Pannus, P.; Vanham, G. Viral Inhibitory Activity of CD8+ T Cells in HIV Infection. AIDS Rev. 2019, 21, 115–125. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, L.; Zhang, M.; Zhang, L.; Zhu, C.; He, Q.; Ye, L.; Zhao, C.; Li, Z.; Xu, J.; et al. Prompt Antiviral Action of Pulmonary CD8+ TRM Cells Is Mediated by Rapid IFN-Γ Induction and Its Downstream ISGs in the Lung. Front. Immunol. 2022, 13, 839455. [Google Scholar] [CrossRef] [PubMed]
- Tipton, T.R.W.; Hall, Y.; Bore, J.A.; White, A.; Sibley, L.S.; Sarfas, C.; Yuki, Y.; Martin, M.; Longet, S.; Mellors, J.; et al. Characterisation of the T-Cell Response to Ebola Virus Glycoprotein amongst Survivors of the 2013–16 West Africa Epidemic. Nat. Commun. 2021, 12, 1153. [Google Scholar] [CrossRef]
- Richert, L.; Lelièvre, J.-D.; Lacabaratz, C.; Hardel, L.; Hocini, H.; Wiedemann, A.; Lucht, F.; Poizot-Martin, I.; Bauduin, C.; Diallo, A.; et al. T Cell Immunogenicity, Gene Expression Profile, and Safety of Four Heterologous Prime-Boost Combinations of HIV Vaccine Candidates in Healthy Volunteers: Results of the Randomized Multi-Arm Phase I/II ANRS VRI01 Trial. J. Immunol. 2022, 208, 2663–2674. [Google Scholar] [CrossRef]
- Zepp, F. Principles of Vaccine design—Lessons from Nature. Vaccines 2010, 28, 14–24. [Google Scholar] [CrossRef]
- Kudlay, D.; Svistunov, A.; Satyshev, O. COVID-19 Vaccines: An Updated Overview of Different Platforms. Bioengineering 2022, 9, 714. [Google Scholar] [CrossRef]
- Korenkov, D.; Isakova-Sivak, I.; Rudenko, L. Basics of CD8 T-Cell Immune Responses after Influenza Infection and Vaccination with Inactivated or Live Attenuated Influenza Vaccine. Expert Rev. Vaccines 2018, 17, 977–987. [Google Scholar] [CrossRef] [PubMed]
- Coughlan, L. Factors which Contribute to the Immunogenicity of Non-Replicating Adenoviral Vectored Vaccines. Front. Immunol. 2020, 11, 909. [Google Scholar] [CrossRef] [PubMed]
- Hobernik, D.; Bros, M. DNA Vaccines—How Far from Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Kwon, M.; Im, S.; Lee, K.; Lee, H. MRNA Vaccines: The Most Recent Clinical Applications of Synthetic MRNA. Arch. Pharmacal Res. 2022, 45, 245–262. [Google Scholar] [CrossRef]
- Ghattas, M.; Dwivedi, G.; Lavertu, M.; Alameh, M.-G. Vaccine Technologies and Platforms for Infectious Diseases: Current Progress, Challenges, and Opportunities. Vaccines 2021, 9, 1490. [Google Scholar] [CrossRef]
- Samaranayake, L.P.; Seneviratne, C.J.; Fakhruddin, K.S. Coronavirus Disease 2019 (COVID-19) Vaccines: A Concise Review. Oral Dis. 2021, 28, 2326–2336. [Google Scholar] [CrossRef]
- McCann, N.; O’Connor, D.; Lambe, T.; Pollard, A.J. Viral Vector Vaccines. Curr. Opin. Immunol. 2022, 77, 102210. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V.; Tukhvatulin, A.I.; Zubkova, O.V.; Dzharullaeva, A.S.; Kovyrshina, A.V.; Lubenets, N.L.; Grousova, D.M.; Erokhova, A.S.; et al. Safety and Efficacy of an rAd26 and RAd5 Vector-Based Heterologous Prime-Boost COVID-19 Vaccine: An Interim Analysis of a Randomised Controlled Phase 3 Trial in Russia. Lancet 2021, 39, 671–681. [Google Scholar] [CrossRef]
- Hu, C.; Shen, M.; Han, X.; Chen, Q.; Li, L.; Chen, S.; Zhang, J.; Gao, F.; Wang, W.; Wang, Y.; et al. Identification of Cross-Reactive CD8+ T Cell Receptors with High Functional Avidity to a SARS-CoV-2 Immunodominant Epitope and Its Natural Mutant Variants. Genes Dis. 2022, 9, 216–229. [Google Scholar] [CrossRef]
- Tarke, A.; Sidney, J.; Kidd, C.K.; Dan, J.M.; Ramirez, S.I.; Yu, E.D.; Mateus, J.; Da Silva Antunes, R.; Moore, E.; Rubiro, P.; et al. Comprehensive Analysis of T cell Immunodominance and Immunoprevalence of SARS-CoV-2 Epitopes in COVID-19 Cases. Cell Rep. Med. 2021, 2, 100204. [Google Scholar] [CrossRef]
- Reynolds, C.J.; Swadling, L.; Gibbons, J.M.; Pade, C.; Jensen, M.P.; Diniz, M.O.; Schmidt, N.M.; Butler, D.K.; Amin, O.E.; Bailey, S.N.L.; et al. Discordant Neutralizing Antibody and T Cell Responses in Asymptomatic and Mild SARS-CoV-2 Infection. Sci. Immunol. 2020, 5, 3698. [Google Scholar] [CrossRef] [PubMed]
- Kudlay, D.; Shirobokov, Y.; Gladunova, E.; Borodulina, E. Diagnosis of COVID-19. Methods and Problems of Virus SARS-CoV-2 Detection under Pandemic Conditions. Vrach 2020, 31, 5–10. [Google Scholar] [CrossRef]
- Pandey, S.K.; Mohanta, G.C.; Kumar, V.; Gupta, K. Diagnostic Tools for Rapid Screening and Detection of SARS-CoV-2 Infection. Vaccines 2022, 10, 1200. [Google Scholar] [CrossRef] [PubMed]
- Waldrop, S.L.; Davis, K.A.; Maino, V.C.; Picker, L.J. Normal human CD4+ memory T cells display broad heterogeneity in their activation threshold for cytokine synthesis. J. Immunol. 1998, 161, 5284–5295. [Google Scholar] [CrossRef] [PubMed]
- Farber, D.L. Biochemical Signaling Pathways for Memory T Cell Recall. Semin. Immunol. 2009, 21, 84–91. [Google Scholar] [CrossRef]
- Kelso, A. Cytokines: Principles and Prospects. Immun. Cell Biol. 1998, 76, 300–317. [Google Scholar] [CrossRef]
- Abu-Shah, E.; Trendel, N.; Kruger, P.; Nguyen, J.; Pettmann, J.; Kutuzov, M.; Dushek, O. Human CD8+ T Cells Exhibit a Shared Antigen Threshold for Different Effector Responses. J. Immunol. 2020, 205, 1503–1512. [Google Scholar] [CrossRef]
- Katial, R.K.; Sachanandani, D.; Pinney, C.; Lieberman, M.M. Cytokine Production in Cell Culture by Peripheral Blood Mononuclear Cells from Immunocompetent Hosts. Clin. Vaccine Immunol. 1998, 5, 78–81. [Google Scholar] [CrossRef]
- Hoekstra, M.E.; Vijver, S.V.; Schumacher, T.N. Modulation of the Tumor Micro-Environment by CD8+ T Cell-Derived Cytokines. Curr. Opin. Immunol. 2021, 69, 65–71. [Google Scholar] [CrossRef]
- Morris, G.; Bortolasci, C.C.; Puri, B.K.; Marx, W.; O’Neil, A.; Athan, E.; Walder, K.; Berk, M.; Olive, L.; Carvalho, A.F.; et al. The Cytokine Storms of COVID-19, H1N1 Influenza, CRS and MAS Compared. Can One Sized Treatment Fit All? Cytokine 2021, 144, 155593. [Google Scholar] [CrossRef]
- Wei, F.; Gao, C.; Wang, Y. The Role of Influenza A Virus-Induced Hypercytokinemia. Crit. Rev. Microbiol. 2021, 48, 240–256. [Google Scholar] [CrossRef] [PubMed]
- Chatenoud, L.; Bluestone, J.A. CD3-Specific Antibodies: A Portal to the Treatment of Autoimmunity. Nat. Rev. Immunol. 2007, 7, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Ngoenkam, J.; Schamel, W.W.; Pongcharoen, S. Selected Signalling Proteins Recruited to the T-Cell Receptor-CD3 Complex. Immunology 2017, 153, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Pishesha, N.; Harmand, T.J.; Ploegh, H.L. A Guide to Antigen Processing and Presentation. Nat. Rev. Immunol. 2022, 22, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Clement, L.T. Isoforms of the CD45 Common Leukocyte Antigen Family: Markers for Human T-Cell Differentiation. J. Clin. Immunol. 1992, 12, 1–10. [Google Scholar] [CrossRef]
- Jackola, D.R.; Ruger, J.K.; Miller, R.A. Age-Associated Changes in Human T Cell Phenotype and Function. Aging Clin. Exp. Res. 1994, 6, 25–34. [Google Scholar] [CrossRef]
- Davis, M.M. The Evolutionary and Structural ‘logic’ of Antigen Receptor Diversity. Semin. Immunol. 2004, 16, 239–243. [Google Scholar] [CrossRef]
- Jackson, K.J.L.; Kidd, M.J.; Wang, Y.; Collins, A.M. The Shape of the Lymphocyte Receptor Repertoire: Lessons from the B Cell Receptor. Front. Immunol. 2013, 4, 263. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Y.; Ye, A.Y.; Du, Z.; Xu, M.; Lee, C.-S.; Hwang, J.K.; Kyritsis, N.; Ba, Z.; Neuberg, D.; et al. BCR Selection and Affinity Maturation in Peyer’s Patch Germinal Centres. Nature 2020, 582, 421–425. [Google Scholar] [CrossRef]
- Shi, Z.; Zhang, Q.; Yan, H.; Yang, Y.; Wang, P.; Zhang, Y.; Deng, Z.; Yu, M.; Zhou, W.; Wang, Q.; et al. More Than One Antibody of Individual B Cells Revealed by Single-Cell Immune Profiling. Cell Discov. 2019, 5, 64. [Google Scholar] [CrossRef]
- Martinez, R.J.; Evavold, B.D. Lower Affinity T Cells Are Critical Components and Active Participants of the Immune Response. Front. Immunol. 2015, 6, 468. [Google Scholar] [CrossRef] [PubMed]
- Dessalles, R.; Pan, Y.; Xia, M.; Maestrini, D.; D’Orsogna, M.R.; Chou, T. How Naive T-Cell Clone Counts Are Shaped by Heterogeneous Thymic Output and Homeostatic Proliferation. Front. Immunol. 2022, 12, 07463. [Google Scholar] [CrossRef] [PubMed]
- Kavazović, I.; Polić, B.; Wensveen, F.M. Cheating the Hunger Games; Mechanisms Controlling Clonal Diversity of CD8 Effector and Memory Populations. Front. Immunol. 2018, 9, 2831. [Google Scholar] [CrossRef] [PubMed]
- Mark, M.; Reich-Zeliger, S.; Greenstein, E.; Reshef, D.; Madi, A.; Chain, B.; Friedman, N. A Hierarchy of Selection Pressures Determines the Organization of the T Cell Receptor Repertoire. Front. Immunol. 2022, 13, 4094. [Google Scholar] [CrossRef]
- Merkenschlager, J.; Kassiotis, G. Narrowing the Gap: Preserving Repertoire Diversity despite Clonal Selection during the CD4 T Cell Response. Front. Immunol. 2015, 6, 413. [Google Scholar] [CrossRef]
- Soto, C.; Bombardi, R.G.; Kozhevnikov, M.; Sinkovits, R.S.; Chen, E.C.; Branchizio, A.; Kose, N.; Day, S.B.; Pilkinton, M.; Gujral, M.; et al. High Frequency of Shared Clonotypes in Human T Cell Receptor Repertoires. Cell Rep. 2020, 32, 107882. [Google Scholar] [CrossRef]
- Lavender, P.; Cousins, D.; Lee, T. Regulation of Th2 Cytokine Gene Transcription. Chem. Immunol. 2000, 78, 16–29. [Google Scholar] [CrossRef]
- Sungnak, W.; Wang, C.; Kuchroo, V.K. Multilayer Regulation of CD4 T Cell Subset Differentiation in the Era of Single Cell Genomics. Adv. Immunol. 2019, 141, 1–31. [Google Scholar] [CrossRef]
- Picker, L.; Singh, M.; Zdraveski, Z.; Treer, J.; Waldrop, S.; Bergstresser, P.; Maino, V. Direct Demonstration of Cytokine Synthesis Heterogeneity among Human memory/Effector T Cells by Flow Cytometry. Blood 1995, 86, 1408–1419. [Google Scholar] [CrossRef]
- Andersson, U.; Andersson, J.; Lindfors, A.; Wagner, K.; Möller, G.; Heusser, C.H. Simultaneous Production of Interleukin 2, Interleukin 4 and Interferon-Γ by Activated Human Blood Lymphocytes. Eur. J. Immunol. 1990, 20, 1591–1596. [Google Scholar] [CrossRef]
- Anderson, P. Post-Transcriptional Control of Cytokine Production. Nat. Immunol. 2008, 9, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Gartlan, C.; Tipton, T.; Salguero, F.J.; Sattentau, Q.; Gorringe, A.; Carroll, M.W. Vaccine-Associated Enhanced Disease and Pathogenic Human Coronaviruses. Front. Immunol. 2022, 13, 1504. [Google Scholar] [CrossRef] [PubMed]
- Gousseff, M.; Penot, P.; Gallay, L.; Batisse, D.; Benech, N.; Bouiller, K.; Collarino, R.; Conrad, A.; Slama, D.; Joseph, C.; et al. Clinical Recurrences of COVID-19 Symptoms after Recovery: Viral Relapse, Reinfection or Inflammatory Rebound? J. Infect. 2020, 81, 816–846. [Google Scholar] [CrossRef] [PubMed]
- Mensah, A.A.; Lacy, J.; Stowe, J.; Seghezzo, G.; Sachdeva, R.; Simmons, R.; Bukasa, A.; O’Boyle, S.; Andrews, N.; Ramsay, M.; et al. Disease Severity during SARS-CoV-2 Reinfection: A Nationwide Study. J. Infect. 2022, 84, 542–550. [Google Scholar] [CrossRef]
- Alhusseini, L.B.; Yassen, L.T.; Kouhsari, E.A.; Marjani, M.F. Persistence of SARS-CoV-2: A new paradigm of COVID-19 management. Ann. Ig. 2021, 33, 426–432. [Google Scholar] [CrossRef]
- Bøyum, A. Isolation of Lymphocytes, Granulocytes and Macrophages. Scand. J. Immunol. 1976, 5, 9–15. [Google Scholar] [CrossRef]
- Temporary Guidelines for Prevention, Diagnosis and Treatment of Novel Coronavirus Disease (COVID-19) Version 16.pdf. Available online: https://static-0.minzdrav.gov.ru/system/attachments/attaches/000/060/193/original/%D0%92%D0%9C%D0%A0_COVID-19_V16.pdf (accessed on 10 December 2022).
- Logunov, D.Y.; Dolzhikova, I.V.; Zubkova, O.V.; Tukhvatulin, A.I.; Shcheblyakov, D.V.; Dzharullaeva, A.S.; Grousova, D.M.; Erokhova, A.S.; Kovyrshina, A.V.; Botikov, A.G.; et al. Safety and Immunogenicity of an rAd26 and RAd5 Vector-Based Heterologous Prime-Boost COVID-19 Vaccine in Two Formulations: Two Open, Non-Randomised Phase 1/2 Studies from Russia. Lancet 2020, 396, 887–897. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Wang, X.; Liu, X.; Fu, B.; Lin, Y.; Larsen, L.; Offen, W. Overview of Multiple Testing Methodology and Recent Development in Clinical Trials. Contemp. Clin. Trials 2015, 45, 13–20. [Google Scholar] [CrossRef]

| Group of Individuals | Total Individuals | Number of Women, n (%) | Number of Men,n (%) | Age, Years * | Period of Examination from the Date of the Event, Days * |
|---|---|---|---|---|---|
| Control | 18 | 11 (61.1) | 7 (38.9) | 48.0 (38.0; 55.5) | — |
| Recovered | 58 | 40 (69.0) | 18 (31.0) | 49.0 (42.0; 57.0) | 169 (109; 366) ** |
| Vaccinated | 26 | 20 (76.9) | 6 (23.1) | 49.0 (41.0; 57.0) | 99 (46; 161) *** |
| Clinical Characteristics of the Disease | Characteristic Value |
|---|---|
| Olfaction reduction/anosmia, n (%) | 46 (79.3) |
| Headache, n (%) | 41 (70.7) |
| Myalgia, n (%) | 26 (44.8) |
| Cough, n (%) | 24 (41.3) |
| Lung damage according to CT *, n (%) | 16 (27.6) |
| Weakness, n (%) | 35 (60.3) |
| Duration of mild disease, days ** | 7 (5; 12.8) |
| Duration of moderate disease, days ** | 10 (8; 18.8) |
| Post-Vaccination Complications | Number of Individuals with Post-Vaccination Complications, n (%) | |
|---|---|---|
| After the Administration of Component 1 | After the Administration of Component 2 | |
| Local reactions * | 3 (11.5) | 5 (19.2) |
| Subfebrile fever (37.1–38.0 °C) | 8 (30.8) | 7 (26.9) |
| Febrile fever (38.1–39.0 °C) | 1 (3.8) | 1 (3.8) |
| Systemic reactions ** | 5 (19.2) | 5 (19.2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krechetov, S.P.; Vtorushina, V.V.; Inviyaeva, E.V.; Gorodnova, E.A.; Kolesnik, S.V.; Kudlay, D.A.; Borovikov, P.I.; Krechetova, L.V.; Dolgushina, N.V.; Sukhikh, G.T. T-Cell Immunity in COVID-19-Recovered Individuals and Individuals Vaccinated with the Combined Vector Vaccine Gam-COVID-Vac. Int. J. Mol. Sci. 2023, 24, 1930. https://doi.org/10.3390/ijms24031930
Krechetov SP, Vtorushina VV, Inviyaeva EV, Gorodnova EA, Kolesnik SV, Kudlay DA, Borovikov PI, Krechetova LV, Dolgushina NV, Sukhikh GT. T-Cell Immunity in COVID-19-Recovered Individuals and Individuals Vaccinated with the Combined Vector Vaccine Gam-COVID-Vac. International Journal of Molecular Sciences. 2023; 24(3):1930. https://doi.org/10.3390/ijms24031930
Chicago/Turabian StyleKrechetov, Sergey Petrovich, Valentina Valentinovna Vtorushina, Evgenia Vladimirovna Inviyaeva, Elena Aleksandrovna Gorodnova, Svetlana Vladimirovna Kolesnik, Dmitry Anatolievich Kudlay, Pavel Igorevich Borovikov, Liubov Valentinovna Krechetova, Nataliya Vitalievna Dolgushina, and Gennady Tikhonovich Sukhikh. 2023. "T-Cell Immunity in COVID-19-Recovered Individuals and Individuals Vaccinated with the Combined Vector Vaccine Gam-COVID-Vac" International Journal of Molecular Sciences 24, no. 3: 1930. https://doi.org/10.3390/ijms24031930
APA StyleKrechetov, S. P., Vtorushina, V. V., Inviyaeva, E. V., Gorodnova, E. A., Kolesnik, S. V., Kudlay, D. A., Borovikov, P. I., Krechetova, L. V., Dolgushina, N. V., & Sukhikh, G. T. (2023). T-Cell Immunity in COVID-19-Recovered Individuals and Individuals Vaccinated with the Combined Vector Vaccine Gam-COVID-Vac. International Journal of Molecular Sciences, 24(3), 1930. https://doi.org/10.3390/ijms24031930





