Establishment of Bactrian Camel Induced Pluripotent Stem Cells and Prediction of Their Unique Pluripotency Genes
Abstract
1. Introduction
2. Results
2.1. Identification of BCFFs and Plasmid Preparation
2.2. Acquisition and Passage of bciPSCs
2.3. Pluripotency Gene Detection
2.4. Telomerase Gene Expression in bciPSCs
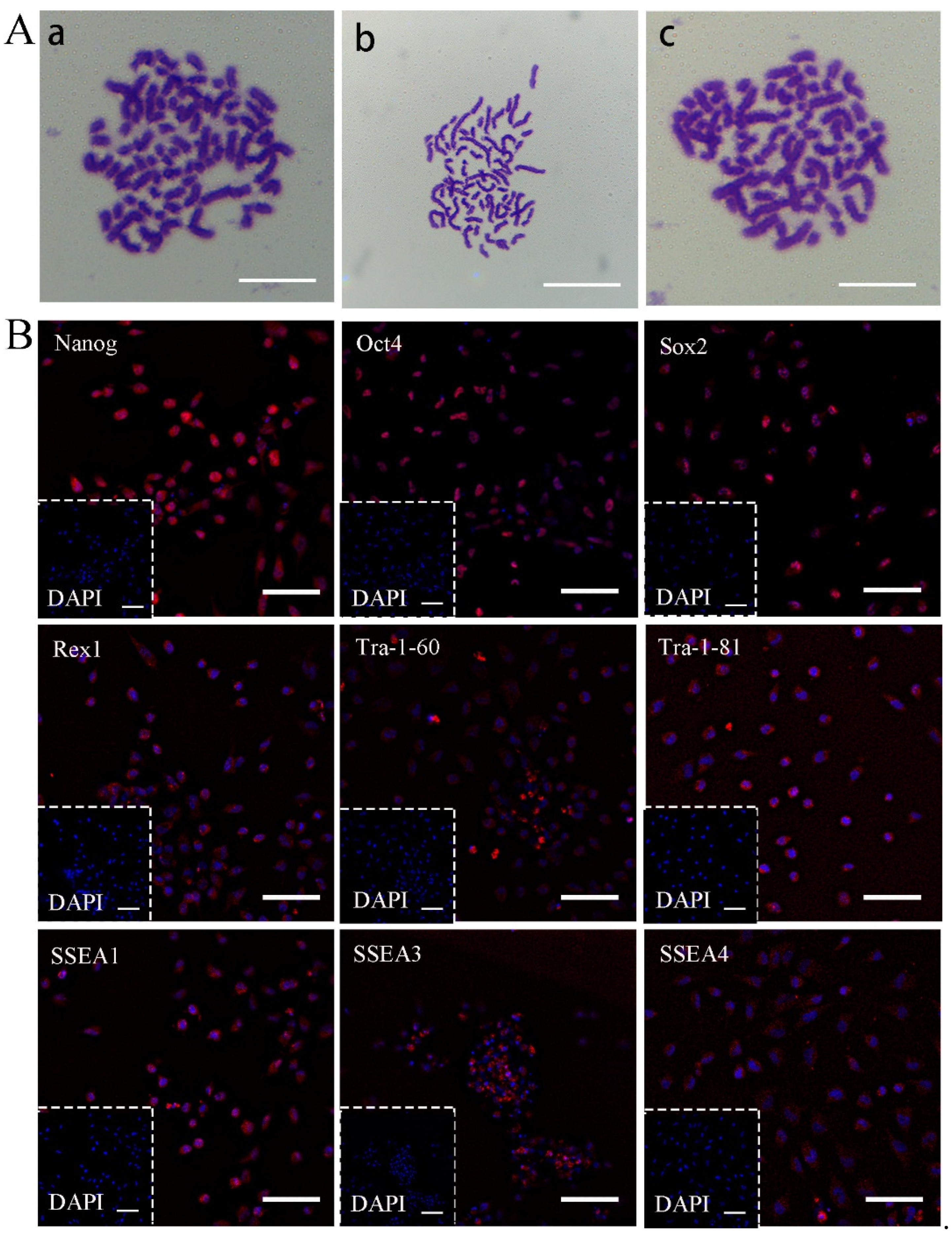
2.5. Karyotype Analysis
2.6. Immunocytochemistry
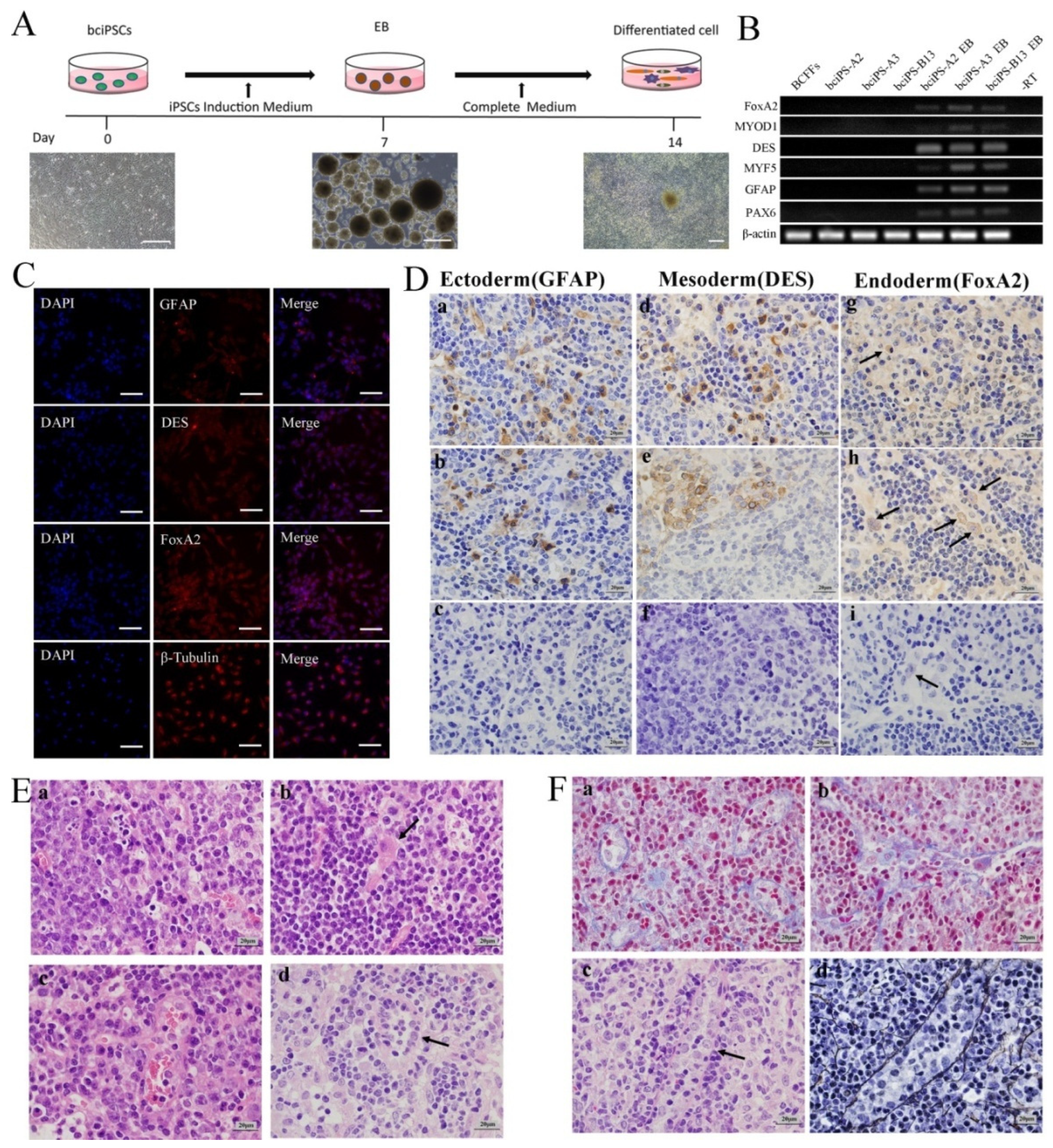
2.7. EB Formation
2.8. Teratoma Formation
2.9. RNA-Seq analysis
2.10. Preliminary Mining of Potentially Pluripotency Genes of bciPSCs
3. Discussion
4. Materials and Methods
4.1. Plasmid Preparation
4.2. Cell Lines and Cell Culture Conditions
4.3. bciPSC Culture Method Analysis
4.4. bciPSC Generation
4.5. Pluripotency Gene Detection
4.6. qRT-PCR
4.7. Alkaline Phosphatase Staining
4.8. Karyotype Analysis
4.9. Immunofluorescence
4.10. In Vitro Differentiation
4.11. Teratoma Formation
4.12. RNA-Seq
4.13. Heatmap Generation and Data Visualization
4.14. Preliminary Mining of Potentially Inducible Genes in bciPSCs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Evans, M.; Kaufman, M. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Ogorevc, J.; Orehek, S.; Dovc, P. Cellular reprogramming in farm animals: An overview of iPSC generation in the mammalian farm animal species. J. Anim. Sci. Biotechnol. 2016, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Han, J.; Ding, F.; Cao, S.; Lim, S.S.; Dai, Y.; Zhang, R.; Zhang, Y.; Lim, B.; Li, N. Generation of induced pluripotent stem cells from bovine embryonic fibroblast cells. Cell Res. 2011, 21, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Cui, C.; Chen, S.; Ren, J.; Chen, J.; Gao, Y.; Li, H.; Jia, N.; Cheng, L.; Xiao, H.; et al. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell 2009, 4, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.; Tian, S.; Nie, J.; Jonsdottir, G.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef]
- Ezashi, T.; Telugu, B.; Alexenko, A.; Sachdev, S.; Sinha, S.; Roberts, R. Derivation of induced pluripotent stem cells from pig somatic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 10993–10998. [Google Scholar] [CrossRef]
- Nagy, K.; Sung, H.; Zhang, P.; Laflamme, S.; Vincent, P.; Agha-Mohammadi, S.; Woltjen, K.; Monetti, C.; Michael, I.P.; Smith, L.C. Induced Pluripotent Stem Cell Lines Derived from Equine Fibroblasts. Stem Cell Rev. 2011, 7, 693–702. [Google Scholar] [CrossRef]
- Liu, J.; Balehosur, D.; Murray, B.; Kelly, J.M.; Sumer, H.; Verma, P.J. Generation and characterization of reprogrammed sheep induced pluripotent stem cells. Theriogenology 2012, 77, 338–346. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, F.; Yong, J.; Zhang, P.; Hou, P.; Li, H.; Wei, J.; Cai, J.; Meng, L.; Kai, C. Generation of Induced Pluripotent Stem Cells from Adult Rhesus Monkey Fibroblasts. Cell Stem Cell 2008, 3, 587–590. [Google Scholar] [CrossRef]
- Buehr, M.; Meek, S.; Blair, K.; Yang, J.; Ure, J.; Silva, J.; Mclay, R.; Hall, J.; Ying, Q.L.; Smith, A. Capture of authentic embryonic stem cells from rat blastocysts. Cell 2008, 135, 1287–1298. [Google Scholar] [CrossRef]
- Lee, A.S.; Xu, D.; Plews, J.R.; Nguyen, P.K.; Nag, D.; Lyons, J.K.; Han, L.; Hu, S.; Lan, F.; Liu, J.; et al. Preclinical derivation and imaging of autologously transplanted canine induced pluripotent stem cells. J. Biol. Chem. 2011, 286, 32697–32704. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Pak, Y.; He, L.; Qian, L.; Gu, Y.; Li, H.; Rao, L.; Liao, J.; Cui, C.; Xu, X.; et al. Generation of hircine-induced pluripotent stem cells by somatic cell reprogramming. Cell Res. 2011, 21, 849–853. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Takizawa, N.; Narita, M.; Lchisaka, T.; Yamanaka, S. Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl. Acad. Sci. USA 2010, 107, 14152–14157. [Google Scholar] [CrossRef]
- Maekawa, M.; Yamaguchi, K.; Nakamura, T.; Shibukawa, R.; Kodanaka, I.; Ichisaka, T.; Kawamura, Y.; Mochizuki, H.; Goshima, N.; Yamanaka, S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 2011, 474, 225–229. [Google Scholar] [CrossRef]
- Lee, B.C.; Kim, M.K.; Jang, G.; Oh, H.J.; Yuda, F.; Kim, H.J.; Shamim, M.H.; Kim, J.J.; Kang, S.K.; Schatten, G. Dogs cloned from adult somatic cells. Nature 2005, 436, 641. [Google Scholar] [CrossRef]
- Bo, F.; Jiang, J.; Kraus, P.; Ng, J.H.; Ng, H.H. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 2009, 11, 197–203. [Google Scholar]
- Chen, J.; Liu, J.; Yang, J.; Chen, Y.; Pei, D. BMPs functionally replace Klf4 and support efficient reprogramming of mouse fibroblasts by Oct4 alone. Cell Res. 2011, 21, 205–212. [Google Scholar] [CrossRef]
- Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Okita, K.; Mochiduki, Y.; Takizawa, N.; Yamanaka, S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008, 26, 101–106. [Google Scholar] [CrossRef]
- Yang, P.; Wang, Y.; Chen, J.; Li, H.; Kang, L.; Zhang, Y.; Chen, S.; Zhu, B.; Gao, S. RCOR2 Is a Subunit of the LSD1 Complex That Regulates ESC Property and Substitutes for SOX2 in Reprogramming Somatic Cells to Pluripotency. STEM CELLS 2011, 29, 791–801. [Google Scholar] [CrossRef]
- Wu, L.; Wu, Y.; Peng, B.; Hou, Z.; Dong, Y.; Chen, K.; Guo, M.; Li, H.; Chen, X.; Kou, X. Oocyte-Specific Homeobox 1, Obox1, Facilitates Reprogramming by Promoting Mesenchymal-to-Epithelial Transition and Mitigating Cell Hyperproliferation. Stem Cell Rep. 2017, 9, 1692–1705. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Wu, C.; Wu, Y.; Li, Z.; Shao, S.; Zhao, W.; Tang, X.; Yang, H.; Shen, L.; Zuo, X. Induction of Pluripotency in Mouse Somatic Cells with Lineage Specifiers. Cell 2015, 161, 1229. [Google Scholar] [CrossRef] [PubMed]
- Shinagawa, T.; Takagi, T.; Tsukamoto, D.; Tomaru, C.; Huynh, L.M.; Sivaraman, P.; Kumarevel, T.; Inoue, K.; Nakato, R.; Katou, Y.; et al. Histone Variants Enriched in Oocytes Enhance Reprogramming to Induced Pluripotent Stem Cells. Cell Stem Cell 2014, 14, 217–227. [Google Scholar] [CrossRef]
- Heng, J.; Feng, B.; Han, J.; Jiang, J.; Kraus, P.; Ng, J.H.; Orlov, Y.L.; Huss, M.; Lin, Y.; Lufkin, T. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell 2010, 6, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Redmer, T.; Diecke, S.; Grigoryan, T.; Quiroga-Negreira, A.; Birchmeier, W.; Besser, D. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011, 12, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Picano-Castro, V.; Russo-Carbolante, E.; Reis, L.; Fraga, A.M.; Covas, D.T. Pluripotent reprogramming of fibroblasts by lentiviral mediated insertion of SOX2, C-MYC, and TCL-1A. Stem Cells Dev. 2011, 20, 169–180. [Google Scholar] [CrossRef]
- Bar-Nur, O.; Verheul, C.; Sommer, A.G.; Brumbaugh, J.; Schwarz, B.A.; Lipchina, I.; Huebner, A.J.; Mostoslavsky, G.; Hochedlinger, K. Lineage conversion induced by pluripotency factors involves transient passage through an iPSC stage. Nat. Biotechnol. 2015, 33, 761–768. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, J.; Li, K.; Wu, T.; Huang, B.; Liu, W.; Kou, X.; Zhang, Y.; Huang, H.; Jiang, Y. Replacement of Oct4 by Tet1 during iPSC Induction Reveals an Important Role of DNA Methylation and Hydroxymethylation in Reprogramming. Cell Stem Cell 2013, 12, 453–469. [Google Scholar] [CrossRef]
- Tulgat, R.; Schaller, G.B. Status and distribution of wild Bactrian camels Camelus bactrianus ferus. Biol. Conserv. 1992, 62, 11–19. [Google Scholar] [CrossRef]
- Bactrian Camels Genome Sequencing and Analysis Consortium; Jirimutu; Wang, Z.; Ding, G.; Chen, G.; Sun, Y.; Sun, Z. Genome sequences of wild and domestic bactrian camels. Nat. Commun 2012, 3, 1202. [Google Scholar]
- Alnohair, S.F. Medical benefits of Camel’s Milk: A comprehensive review. J. Pak. Med. Assoc. 2021, 71, 933–937. [Google Scholar]
- Hussen, J.; Schuberth, H.J. Recent Advances in Camel Immunology. Front. Immunol. 2021, 11, 614150. [Google Scholar] [CrossRef]
- Yu, X.; Xu, Z.; Wang, X.; Xu, Q.; Chen, J. Bactrian camel serum albumins-based nanocomposite as versatile biocargo for drug delivery, biocatalysis and detection of hydrogen peroxide—ScienceDirect. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 109, 110627. [Google Scholar] [CrossRef] [PubMed]
- Mihic, T.; Rainkie, D.; Wilby, K.; Pawluk, S. The Therapeutic Effects of Camel Milk: A Systematic Review of Animal and Human Trials. J. Evid. Based Complement. Altern. Med. 2016, 21, NP110–NP126. [Google Scholar] [CrossRef]
- Mirmiran, P.; Ejtahed, H.S.; Angoorani, P.; Eslami, F.; Azizi, F. Camel Milk Has Beneficial Effects on Diabetes Mellitus: A Systematic Review. Int. J. Endocrinol. Metab. 2017, 15, e42150. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, R.P.; Jain, S.; Shah, S.; Chopra, A.; Agarwal, V. Effect of camel milk on glycemic control and insulin requirement in patients with type 1 diabetes: 2-years randomized controlled trial. Eur. J. Clin. Nutr. 2011, 65, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Wani, N.; Vettical, B.; Hong, S. First cloned Bactrian camel (Camelus bactrianus) calf produced by interspecies somatic cell nuclear transfer: A step towards preserving the critically endangered wild Bactrian camels. PLoS ONE 2017, 12, e0177800. [Google Scholar] [CrossRef]
- Balmus, G.; Trifonov, V.; Biltueva, L.; O’Brien, P.; Alkalaeva, E.; Fu, B.; Skidmore, J.; Allen, T.; Graphodatsky, A.; Yang, F.; et al. Cross-species chromosome painting among camel, cattle, pig and human: Further insights into the putative Cetartiodactyla ancestral karyotype. Chromosome Res. 2007, 15, 499–515. [Google Scholar] [CrossRef]
- Lluis, F.; Pedone, E.; Pepe, S.; Cosma, M.P. Periodic activation of Wnt/beta-catenin signaling enhances somatic cell reprogramming mediated by cell fusion. Cell Stem Cell 2008, 3, 493–507. [Google Scholar] [CrossRef]
- Zhu, S.; Li, W.; Zhou, H.; Wei, W.; Ambasudhan, R.; Lin, T.; Kim, J.; Zhang, K.; Ding, S. Reprogramming of Human Primary Somatic Cells by OCT4 and Chemical Compounds. Cell Stem Cell 2010, 7, 651–655. [Google Scholar] [CrossRef]
- Ichida, J.; Blanchard, J.; Lam, K.; Son, E.; Chung, J.; Egli, D.; Loh, K.; Carter, A.; Di Giorgio, F.; Koszka, K.; et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 2009, 5, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, B.; Xu, J.; Wang, J.; Wu, J.; Shi, C.; Xu, Y.; Dong, J.; Wang, C.; Lai, W. Derivation of Pluripotent Stem Cells with InVivo Embryonic and Extraembryonic Potency. Cell 2017, 169, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ryan, D.J.; Wang, W.; Tsang, C.H.; Lan, G.; Masaki, H.; Gao, X.; Antunes, L.; Yu, Y.; Zhu, Z. Establishment of mouse expanded potential stem cells. Nature 2017, 550, 393. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.; Yin, X.; Yang, W.; Du, Y.; Hou, P.; Ge, J.; Liu, C.; Zhang, W.; Zhang, X. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011, 21, 196–204. [Google Scholar] [CrossRef]
- Hou, P.; Li, Y.; Zhang, X.; Liu, C.; Guan, J.; Li, H.; Zhao, T.; Ye, J.; Yang, W.; Liu, K.; et al. Pluripotent Stem Cells Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science 2013, 341, 651–654. [Google Scholar] [CrossRef]
- Omole, A.E.; Fakoya, A.O.J. Ten years of progress and promise of induced pluripotent stem cells: Historical origins, characteristics, mechanisms, limitations, and potential applications. Peerj 2018, 6, e4370. [Google Scholar] [CrossRef]
- Esteban, M.A.; Wang, T.; Qin, B.; Yang, J.; Qin, D.; Cai, J.; Li, W.; Weng, Z.; Chen, J.; Ni, S. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 2010, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, J.; Yang, X.; Zhou, C.; Guo, J.; Wu, C.; Qin, Y.; Guo, L.; He, J.; Yu, S. Chromatin Accessibility Dynamics During Reprogramming. Cell Stem Cell 2017, 21, 819–833. [Google Scholar] [CrossRef]
- Hong, H.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Kanagawa, O.; Nakagawa, M.; Okita, K.; Yamanaka, S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009, 460, 1132–1135. [Google Scholar] [CrossRef]
- Ba Nito, A.; Rashid, S.T.; Acosta, J.C.; Li, S.D.; Gil, J. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009, 23, 2134–2139. [Google Scholar] [CrossRef]
- Jian, S.; Chen, W.; Wu, Y.; Li, Z.; Deng, H. Induction of Pluripotency in Mouse Somatic Cells with Lineage Specifiers. Cell 2013, 153, 963–975. [Google Scholar]
- Liu, G.; Qu, J.; Suzuki, K.; Nivet, E.; Li, M.; Montserrat, N.; Yi, F.; Xu, X.; Ruiz, S.; Zhang, W.; et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 2012, 491, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Eckersley-Maslin, M.A.; Alda-Catalinas, C.; Reik, W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 2018, 19, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Matoba, S.; Liu, Y.; Lu, F.; Iwabuchi, K.; Shen, L.; Inoue, A.; Zhang, Y. Embryonic Development following Somatic Cell Nuclear Transfer Impeded by Persisting Histone Methylation. Cell 2014, 159, 884–895. [Google Scholar] [CrossRef]
- Wang, T.; Chen, K.; Zeng, X.; Yang, J.; Wu, Y.; Shi, X.; Qin, B.; Zeng, L.; Esteban, M.A.; Pan, G. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 2011, 9, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, H.; Liu, J.; Qi, J.; Wei, B.; Yang, J.; Liang, H.; Chen, Y.; Chen, J.; Wu, Y. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 2013, 45, 34–42. [Google Scholar] [CrossRef]
- Gao, X.; Nowak-Imialek, M.; Chen, X.; Chen, D.; Herrmann, D.; Ruan, D.; Chen, A.; Eckersley-Maslin, M.; Ahmad, S.; Lee, Y.; et al. Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 2019, 21, 687–699. [Google Scholar] [CrossRef]
- Honda, A.; Hirose, M.; Inoue, K.; Ogonuki, N.; Miki, H.; Shimozawa, N.; Hatori, M.; Shimizu, N.; Murata, T.; Hirose, M.; et al. Stable embryonic stem cell lines in rabbits: Potential small animal models for human research. Reprod. Biomed. Online 2008, 17, 706–715. [Google Scholar] [CrossRef]
- Armstrong, L.; Lako, M.; Lincoln, J.; Cairns, P.M.; Hole, N. mTert expression correlates with telomerase activity during the differentiation of murine embryonic stem cells. Mech. Dev. 2000, 97, 109–116. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Brambrink, T.; Foreman, R.; Welstead, G.; Lengner, C.; Wernig, M.; Suh, H.; Jaenisch, R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2008, 2, 151–159. [Google Scholar] [CrossRef]
- Li, X.; Martinez-Fernandez, A.; Hartjes, K.; Kocher, J.; Olson, T.; Terzic, A.; Nelson, T. Transcriptional atlas of cardiogenesis maps congenital heart disease interactome. Physiol. Genom. 2014, 46, 482–495. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, X.; Zheng, Y.; Wang, Z.; Li, X. Establishment of bovine expanded potential stem cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2018505118. [Google Scholar] [CrossRef]
- Ong, D.; Hu, B.; Ho, Y.; Sauvé, C.; Bristow, C.; Wang, Q.; Multani, A.; Chen, P.; Nezi, L.; Jiang, S.; et al. PAF promotes stemness and radioresistance of glioma stem cells. Proc. Natl. Acad. Sci. USA 2017, 114, E9086–E9095. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, L.; Yue, W.; Ouyang, Y.; Wang, Z.; Hou, Y.; Schatten, H.; Sun, Q. CENP-W regulates kinetochore-microtubule attachment and meiotic progression of mouse oocytes. Biochem Biophys Res Commun 2020, 527, 8–14. [Google Scholar] [CrossRef]
- Chen, Q.; Wan, X.; Chen, Y.; Liu, C.; Gu, M.; Wang, Z. SGO1 induces proliferation and metastasis of prostate cancer through AKT-mediated signaling pathway. Am J Cancer Res 2019, 9, 2693–2705. [Google Scholar]
- Serafim, R.B.; Cardoso, C.; Di Cristofaro, L.F.; Pienna Soares, C.; Araújo Silva, W., Jr.; Espreafico, E.M.; Paçó-Larson, M.L.; Price, B.D.; Valente, V. HJURP knockdown disrupts clonogenic capacity and increases radiation-induced cell death of glioblastoma cells. Cancer Gene 2020, 27, 319–329. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, L.; Feng, J.; Tan, D.; Zhu, Y.; Hou, J.; Li, W.; Lv, K.; Wang, W.; Jiang, L.; et al. ASF1B is a Promising Prognostic Biomarker and Correlates With Immunotherapy Efficacy in Hepatocellular Carcinoma. Front. Genet. 2022, 13, 842351. [Google Scholar] [CrossRef]
- Zhang, Y.; Tseng, J.; Lien, I.; Li, F.; Wu, W.; Li, H. mRNAsi Index: Machine Learning in Mining Lung Adenocarcinoma Stem Cell Biomarkers. Genes 2020, 11, 257. [Google Scholar] [CrossRef]
- Fujii-Yamamoto, H.; Kim, J.; Arai, K.; Masai, H. Cell cycle and developmental regulations of replication factors in mouse embryonic stem cells. J. Biol. Chem. 2005, 280, 12976–12987. [Google Scholar] [CrossRef]
- Wen, M.; Wang, H.; Zhang, X.; Long, J.; Lv, Z.; Kong, Q.; An, Y. Cytokine-like 1 is involved in the growth and metastasis of neuroblastoma cells. Int. J. Oncol. 2012, 41, 1419–1424. [Google Scholar] [CrossRef] [PubMed]
- Stoll, S.; Stuart, P.; Swindell, W.; Tsoi, L.; Li, B.; Gandarillas, A.; Lambert, S.; Johnston, A.; Nair, R.; Elder, J. The EGF receptor ligand amphiregulin controls cell division via FoxM1. Oncogene 2016, 35, 2075–2086. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Feng, Z.; Zhou, M.; Ren, Z.; Zhang, F.; Li, Y. Bioinformatic Evidence Reveals that Cell Cycle Correlated Genes Drive the Communication between Tumor Cells and the Tumor Microenvironment and Impact the Outcomes of Hepatocellular Carcinoma. Biomed. Res. Int. 2021, 2021, 4092635. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, L.; Ran, W.; Li, G.; Xiao, Y.; Wang, X.; Zhao, H.; Xing, X. Overexpression of SAPCD2 correlates with proliferation and invasion of colorectal carcinoma cells. Cancer Cell Int. 2020, 20, 43. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Cong, M.; Li, J.; He, D.; Wu, Q.; Tian, P.; Wang, Y.; Yang, S.; Liang, C.; Liang, Y.; et al. Cathepsin C promotes breast cancer lung metastasis by modulating neutrophil infiltration and neutrophil extracellular trap formation. Cancer Cell 2021, 39, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, H.; Zhang, R.; Luo, B.; Xu, B.; Zhu, Z.; Lin, P. MEOX2 serves as a novel biomarker associated with macrophage infiltration in oesophageal squamous cell carcinoma and other digestive system carcinomas. Autoimmunity 2021, 54, 373–383. [Google Scholar] [CrossRef]
- Hoffmann, C.; Mao, X.; Brown-Clay, J.; Moreau, F.; Al, A.A.; Wurzer, H.; Sousa, B.; Schmitt, F.; Berchem, G.; Janji, B.; et al. Hypoxia promotes breast cancer cell invasion through HIF-1α-mediated up-regulation of the invadopodial actin bundling protein CSRP2. Sci. Rep. 2018, 8, 10191. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Hu, C.; Ning, Z.; Huang, J.; Zhu, Z. Identification of key genes controlling cancer stem cell characteristics in gastric cancer. World J. Gastrointest. Surg. 2020, 12, 442–459. [Google Scholar] [CrossRef]
- Pan, S.; Zhan, Y.; Chen, X.; Wu, B.; Liu, B. Identification of Biomarkers for Controlling Cancer Stem Cell Characteristics in Bladder Cancer by Network Analysis of Transcriptome Data Stemness Indices. Front. Oncol. 2019, 9, 613. [Google Scholar] [CrossRef]
- Luo, W.; Gao, F.; Li, S.; Liu, L. FoxM1 Promotes Cell Proliferation, Invasion, and Stem Cell Properties in Nasopharyngeal Carcinoma. Front. Oncol. 2018, 8, 483. [Google Scholar] [CrossRef]
- Hasegawa, D.; Hasegawa, K.; Kaneko, H.; Yoshida, S.; Mitarai, H.; Arima, M.; Tomokiyo, A.; Hamano, S.; Sugii, H.; Wada, N.; et al. MEST Regulates the Stemness of Human Periodontal Ligament Stem Cells. Stem. Cells Int. 2020, 2020, 9672673. [Google Scholar] [CrossRef] [PubMed]
- Katoh, Y.; Katoh, M. Comparative genomics on PROM1 gene encoding stem cell marker CD133. Int. J. Mol. Med. 2007, 19, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Becker-Herman, S.; Rozenberg, M.; Hillel-Karniel, C.; Gil-Yarom, N.; Kramer, M.P.; Barak, A.; Sever, L.; David, K.; Radomir, L.; Lewinsky, H.; et al. CD74 is a regulator of hematopoietic stem cell maintenance. PLoS Biol. 2021, 19, e3001121. [Google Scholar] [CrossRef]
- Dormeyer, W.; van Hoof, D.; Braam, S.R.; Heck, A.J.R.; Mummery, C.L.; Krijgsveld, J. Plasma membrane proteomics of human embryonic stem cells and human embryonal carcinoma cells. J. Proteome Res. 2008, 7, 2936–2951. [Google Scholar] [CrossRef] [PubMed]
- Zubakov, D.; Stupar, Z.; Kovacs, G. Differential expression of a new isoform of DLG2 in renal oncocytoma. BMC Cancer 2006, 6, 106. [Google Scholar] [CrossRef] [PubMed]
- Iino, K.; Mitobe, Y.; Ikeda, K.; Takayama, K.I.; Suzuki, T.; Kawabata, H.; Suzuki, Y.; Horie-Inoue, K.; Inoue, S. RNA-binding protein NONO promotes breast cancer proliferation by post-transcriptional regulation of SKP2 and E2F8. Cancer Sci. 2020, 111, 148–159. [Google Scholar] [CrossRef]
- Hong, S.H.; Son, K.H.; Ha, S.Y.; Wee, T.I.; Choi, S.K.; Won, J.E.; Han, H.D.; Ro, Y.; Park, Y.M.; Eun, J.W.; et al. Nucleoporin 210 Serves a Key Scaffold for SMARCB1 in Liver Cancer. Cancer Res. 2021, 81, 356–370. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, Z.; Wei, S.; Li, B.; Zhao, Z. Genomic features of Chinese small cell lung cancer. BMC Med. Genom. 2022, 15, 117. [Google Scholar] [CrossRef]
- Zhang, L.; Fan, B.; Zheng, Y.; Lou, Y.; Cui, Y.; Wang, K.; Zhang, T.; Tan, X. Identification SYT13 as a novel biomarker in lung adenocarcinoma. J. Cell Biochem. 2020, 121, 963–973. [Google Scholar] [CrossRef]
- Frank, D.A. Culture of Animal Cells: A Manual of Basic Technique. J. R. Soc. Med. 1984, 57, 247–248. [Google Scholar]
- Liu, J.H.Q.; Peng, T.; Peng, M.; Wei, B.; Li, D. The oncogene c-Jun impedes somatic cell reprogramming. Nat. Cell Biol. 2015, 17, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.H.; Kim, J.S.; Jun, E.K.; Lee, J.H.; Kim, A. Reprogramming fibroblasts into induced pluripotent stem cells with Bmi1. Cell Res. 2011, 21, 1305–1315. [Google Scholar] [CrossRef]
- Buganim, Y.; Faddah, D.; Cheng, A. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell 2012, 150, 1209–1222. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Li, Y.; Zhang, Q.; Ge, W.; Zhang, Y.; Zhao, X.; Hu, J.; Yuan, L.; Zhang, W. Establishment of Bactrian Camel Induced Pluripotent Stem Cells and Prediction of Their Unique Pluripotency Genes. Int. J. Mol. Sci. 2023, 24, 1917. https://doi.org/10.3390/ijms24031917
Li Z, Li Y, Zhang Q, Ge W, Zhang Y, Zhao X, Hu J, Yuan L, Zhang W. Establishment of Bactrian Camel Induced Pluripotent Stem Cells and Prediction of Their Unique Pluripotency Genes. International Journal of Molecular Sciences. 2023; 24(3):1917. https://doi.org/10.3390/ijms24031917
Chicago/Turabian StyleLi, Zongshuai, Yina Li, Qiran Zhang, Wenbo Ge, Yong Zhang, Xingxu Zhao, Junjie Hu, Ligang Yuan, and Wangdong Zhang. 2023. "Establishment of Bactrian Camel Induced Pluripotent Stem Cells and Prediction of Their Unique Pluripotency Genes" International Journal of Molecular Sciences 24, no. 3: 1917. https://doi.org/10.3390/ijms24031917
APA StyleLi, Z., Li, Y., Zhang, Q., Ge, W., Zhang, Y., Zhao, X., Hu, J., Yuan, L., & Zhang, W. (2023). Establishment of Bactrian Camel Induced Pluripotent Stem Cells and Prediction of Their Unique Pluripotency Genes. International Journal of Molecular Sciences, 24(3), 1917. https://doi.org/10.3390/ijms24031917






