Identification of Potent Zika Virus NS5 RNA-Dependent RNA Polymerase Inhibitors Combining Virtual Screening and Biological Assays
Abstract
1. Introduction
2. Results
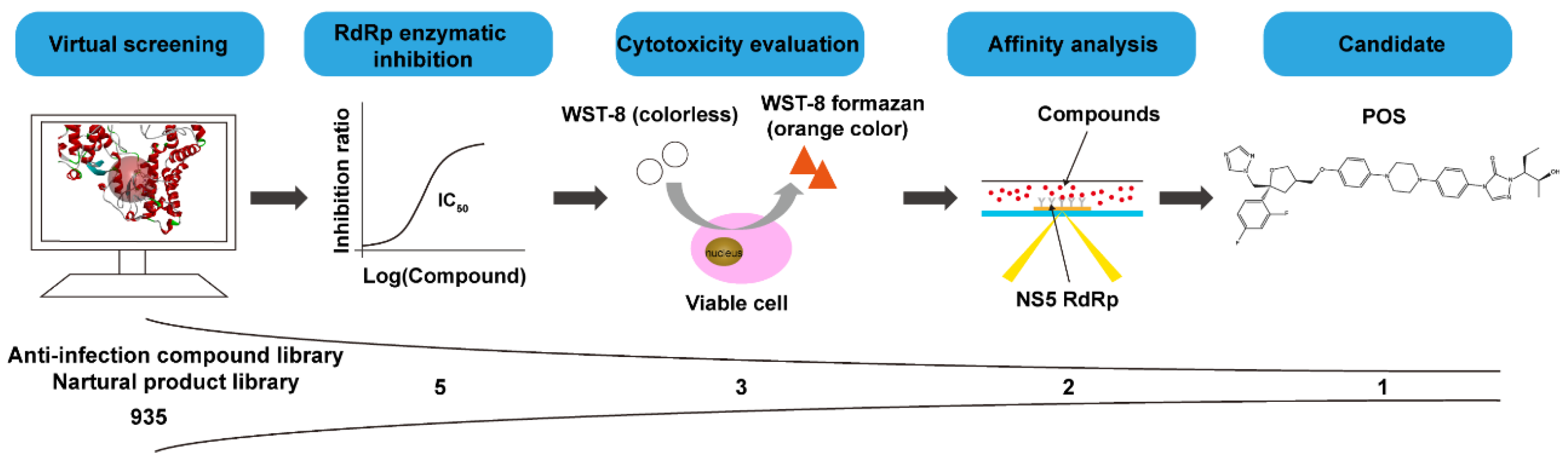
2.1. Virtual Screening of NS5 RdRp Inhibitors
2.2. Biological Assays of NS5 RdRp Inhibitors
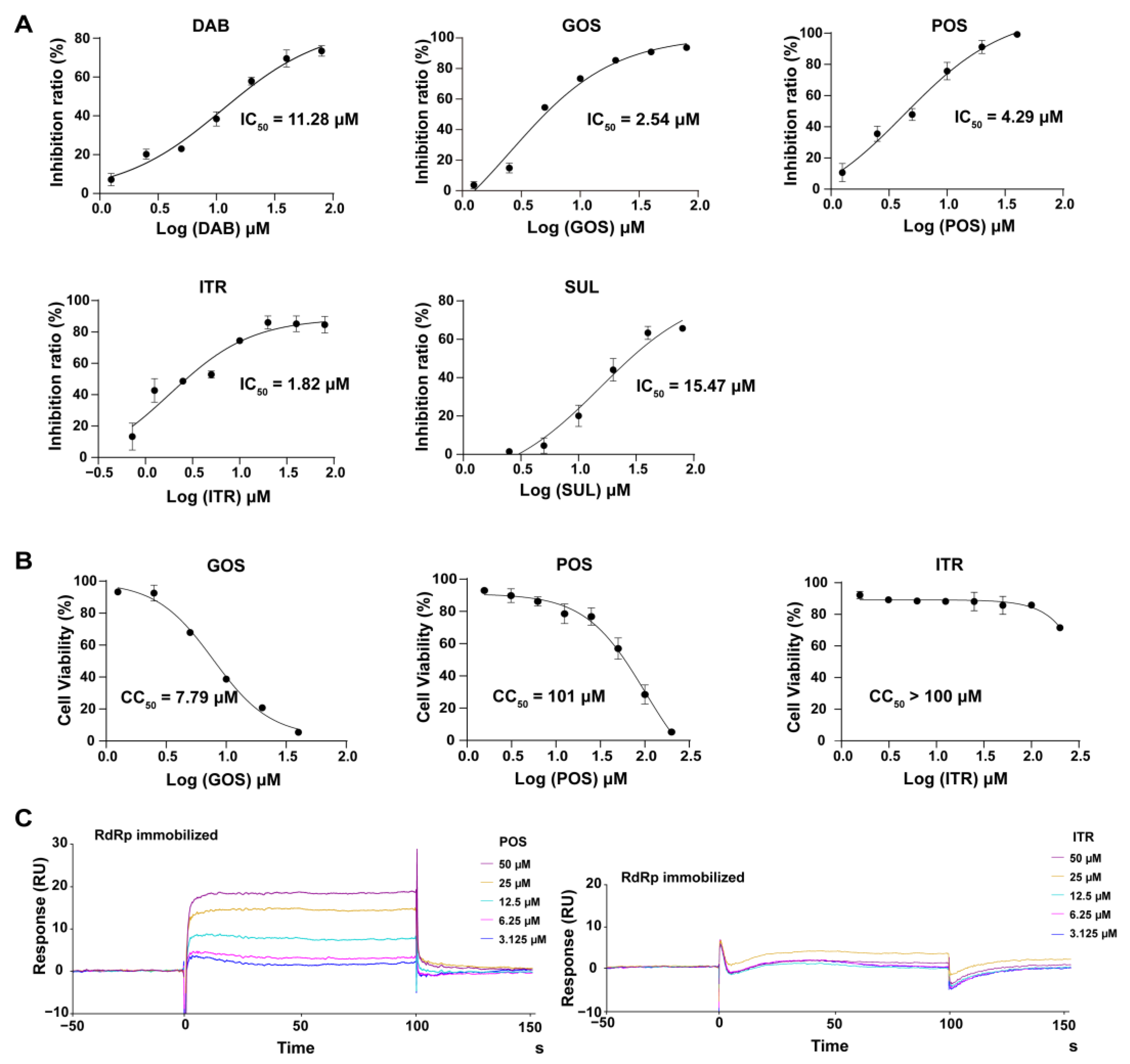
2.2.1. The Inhibitory Effect of Compounds on NS5 RdRp Activity
2.2.2. Cytotoxicity of Compounds
2.2.3. Interaction of Compounds with NS5 RdRp by SPR
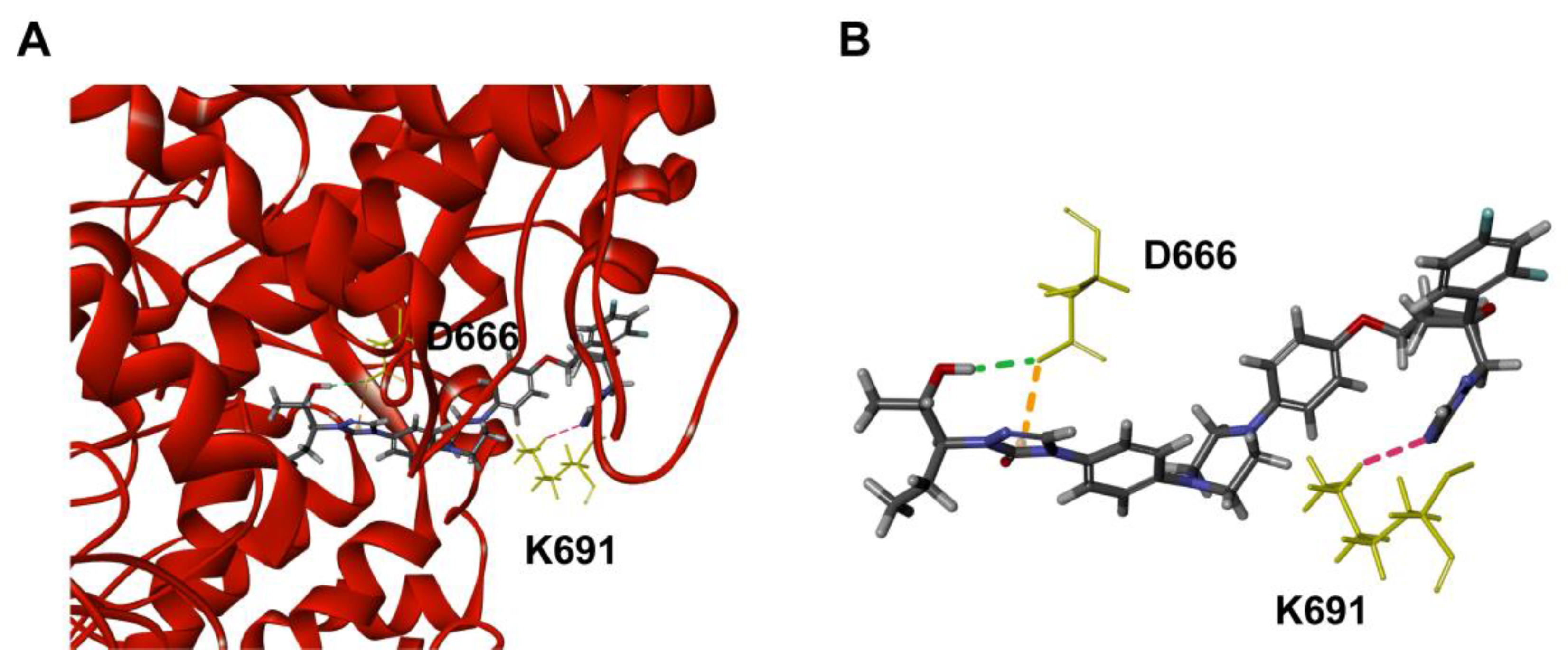
2.3. The Mechanism of POS against NS5 RdRp
2.3.1. Molecular Docking of POS and NS5 RdRp
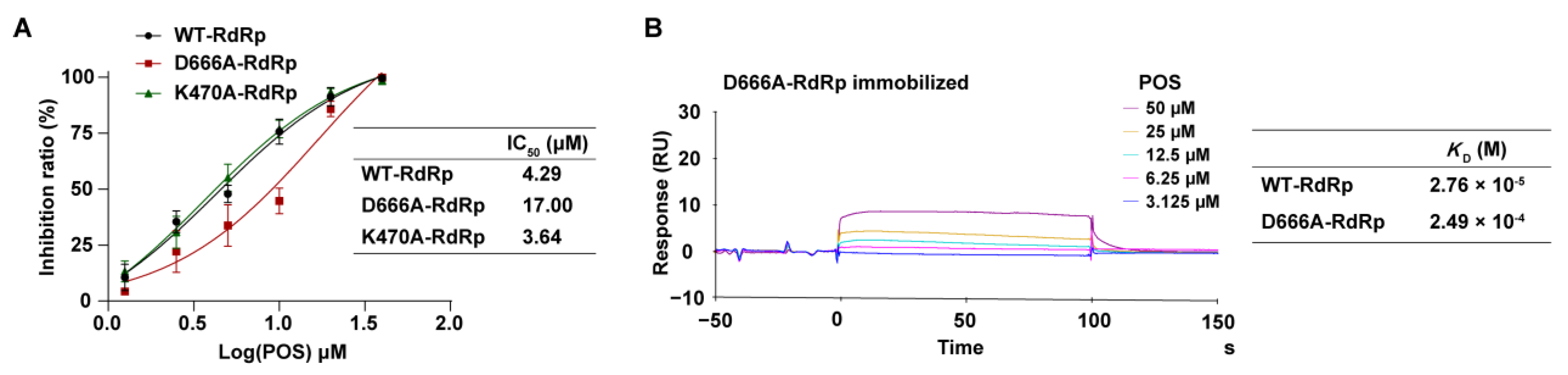
2.3.2. The Inhibitory Effect of POS on NS5 RdRp Mutants
2.3.3. Interaction Detected by SPR between POS and D666A-RdRp
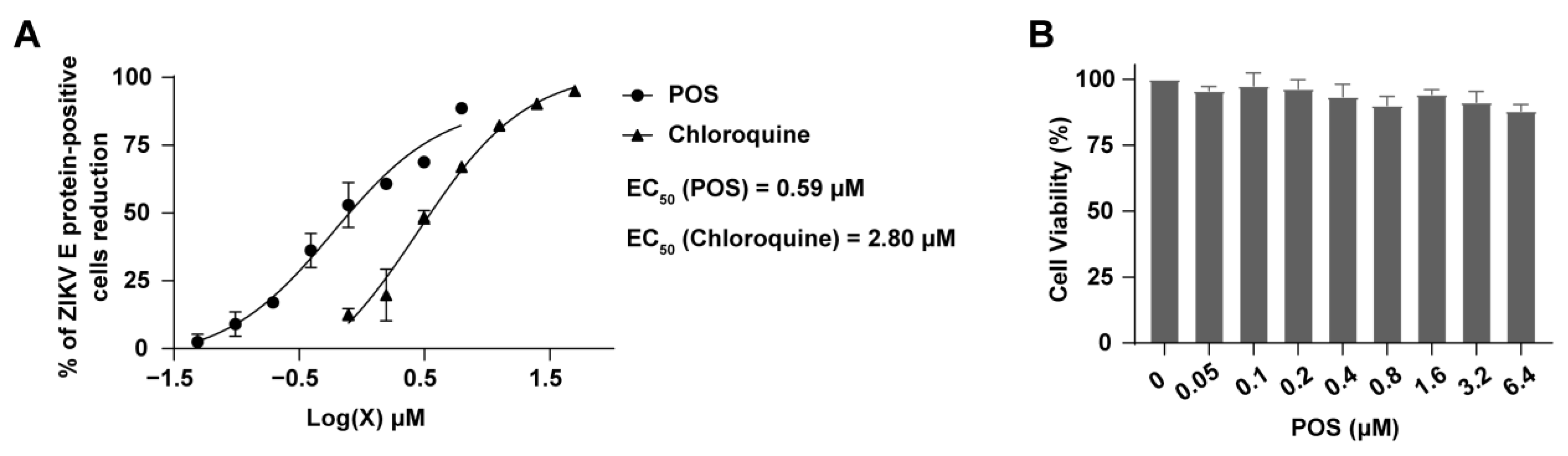
2.4. Anti-ZIKV Activity of POS
3. Discussion
4. Materials and Methods
4.1. Cells and Virus
4.2. Chemicals and Antibodies
4.3. Virtual Screening
4.4. NS5 RdRp Enzymatic Inhibition Assay
4.5. Cell Viability Assay
4.6. Surface Plasmon Resonance (SPR) Assay
4.7. Molecular Docking
4.8. Anti-ZIKV Activity
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bernatchez, J.A.; Tran, L.T.; Li, J.; Luan, Y.; Siqueira-Neto, J.L.; Li, R. Drugs for the Treatment of Zika Virus Infection. J. Med. Chem. 2020, 63, 470–489. [Google Scholar] [CrossRef]
- Shan, C.; Xie, X.; Shi, P.Y. Zika Virus Vaccine: Progress and Challenges. Cell Host Microb. 2018, 24, 12–17. [Google Scholar] [CrossRef]
- Ikejezie, J.; Shapiro, C.N.; Kim, J.; Chiu, M.; Almiron, M.; Ugarte, C.; Espinal, M.A.; Aldighieri, S. Zika Virus Transmission—Region of the Americas, 15 May–15 December, 2016. MMWR Morb. Mortal. Wkly Rep. 2017, 66, 329–334. [Google Scholar] [CrossRef]
- Gulland, A. Zika virus is a global public health emergency, declares WHO. BMJ 2016, 352, i657. [Google Scholar] [CrossRef]
- Gasco, S.; Muñoz-Fernández, M. A Review on the Current Knowledge on ZIKV Infection and the Interest of Organoids and Nanotechnology on Development of Effective Therapies against Zika Infection. Int. J. Mol. Sci. 2020, 22, 35. [Google Scholar] [CrossRef] [PubMed]
- Sirohi, D.; Kuhn, R.J. Zika Virus Structure, Maturation, and Receptors. J. Infect. Dis. 2017, 216, S935–S944. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Yi, G.; Du, F.; Chuang, Y.C.; Vaughan, R.C.; Sankaran, B.; Kao, C.C.; Li, P. Structure and function of the Zika virus full-length NS5 protein. Nat. Commun. 2017, 8, 14762. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, I.; Santos-Júnior, P.; Aquino, T.M.; Araújo-Júnior, J.X.; Silva-Júnior, E.F.D. Insights on Dengue and Zika NS5 RNA-dependent RNA polymerase (RdRp) inhibitors. Eur. J. Med. Chem. 2021, 224, 113698. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.P.; Noble, C.G.; Shi, P.Y. The dengue virus NS5 protein as a target for drug discovery. Antivir. Res. 2015, 119, 57–67. [Google Scholar] [CrossRef]
- Ramaswamy, K.; Rashid, M.; Ramasamy, S.; Jayavelu, T.; Venkataraman, S. Revisiting Viral RNA-Dependent RNA Polymerases: Insights from Recent Structural Studies. Viruses 2022, 14, 2200. [Google Scholar] [CrossRef] [PubMed]
- Kattakuzhy, S.; Levy, R.; Kottilil, S. Sofosbuvir for treatment of chronic hepatitis C. Hepatol. Int. 2015, 9, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Boccuto, A.; Dragoni, F.; Picarazzi, F.; Lai, A.; Della Ventura, C.; Veo, C.; Giammarino, F.; Saladini, F.; Zehender, G.; Zazzi, M.; et al. Sofosbuvir Selects for Drug-Resistant Amino Acid Variants in the Zika Virus RNA-Dependent RNA-Polymerase Complex In Vitro. Int. J. Mol. Sci. 2021, 22, 2670. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.R.; Yang, Q.H.; Luo, R.H.; Peng, Y.M.; Dai, S.X.; Zhang, X.J.; Chen, H.; Cui, X.Q.; Liu, Y.J.; Huang, J.F.; et al. Azvudine, a novel nucleoside reverse transcriptase inhibitor showed good drug combination features and better inhibition on drug-resistant strains than lamivudine in vitro. PLoS ONE 2014, 9, e105617. [Google Scholar] [CrossRef]
- Zhang, J.L.; Li, Y.H.; Wang, L.L.; Liu, H.Q.; Lu, S.Y.; Liu, Y.; Li, K.; Liu, B.; Li, S.Y.; Shao, F.M.; et al. Azvudine is a thymus-homing anti-SARS-CoV-2 drug effective in treating COVID-19 patients. Signal Transduct. Target. Ther. 2021, 6, 414. [Google Scholar] [CrossRef]
- Rao, S.N.; Head, M.S.; Kulkarni, A.; LaLonde, J.M. Validation studies of the site-directed docking program LibDock. J. Chem. Inf. Model. 2007, 47, 2159–2171. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Xu, J.; Qi, M.; Wang, D.; Xu, W.; Qiu, L.; Li, Y.; Cao, Y. A strategy of screening and binding analysis of bioactive components from traditional Chinese medicine based on surface plasmon resonance biosensor. J. Pharm. Anal. 2022, 12, 500–508. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, H.; Song, W.; Si, S.; Han, Y.; Jiang, J. Identification and characterization of Zika virus NS5 RNA-dependent RNA polymerase inhibitors. Int. J. Antimicrob. Agents 2019, 54, 502–506. [Google Scholar] [CrossRef]
- Niyomrattanakit, P.; Abas, S.N.; Lim, C.C.; Beer, D.; Shi, P.Y.; Chen, Y.L. A fluorescence-based alkaline phosphatase-coupled polymerase assay for identification of inhibitors of dengue virus RNA-dependent RNA polymerase. J. Biomol. Screen. 2011, 16, 201–210. [Google Scholar] [CrossRef]
- Jason-Moller, L.; Murphy, M.; Bruno, J. Overview of Biacore systems and their applications. Curr. Protoc. Protein Sci. 2006, 45, 19.13.1–19.13.14. [Google Scholar]
- Li, C.; Zhu, X.; Ji, X.; Quanquin, N.; Deng, Y.Q.; Tian, M.; Aliyari, R.; Zuo, X.; Yuan, L.; Afridi, S.K.; et al. Chloroquine, a FDA-approved Drug, Prevents Zika Virus Infection and its Associated Congenital Microcephaly in Mice. EBioMedicine 2017, 24, 189–194. [Google Scholar]
- Zhang, S.; Yi, C.; Li, C.; Zhang, F.; Peng, J.; Wang, Q.; Liu, X.; Ye, X.; Li, P.; Wu, M.; et al. Chloroquine inhibits endosomal viral RNA release and autophagy-dependent viral replication and effectively prevents maternal to fetal transmission of Zika virus. Antivir. Res. 2019, 169, 104547. [Google Scholar] [CrossRef] [PubMed]
- Delvecchio, R.; Higa, L.M.; Pezzuto, P.; Valadão, A.L.; Garcez, P.P.; Monteiro, F.L.; Loiola, E.C.; Dias, A.A.; Silva, F.J.; Aliota, M.T.; et al. Chloroquine, an Endocytosis Blocking Agent, Inhibits Zika Virus Infection in Different Cell Models. Viruses 2016, 8, 322. [Google Scholar] [CrossRef]
- Mizui, T.; Yamashina, S.; Tanida, I.; Takei, Y.; Ueno, T.; Sakamoto, N.; Ikejima, K.; Kitamura, T.; Enomoto, N.; Sakai, T.; et al. Inhibition of hepatitis C virus replication by chloroquine targeting virus-associated autophagy. J. Gastroenterol. 2010, 45, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G. Virtual screening: An endless staircase? Nat. Rev. Drug Discov. 2010, 9, 273–276. [Google Scholar] [CrossRef]
- de Sousa Luis, J.A.; Barros, R.P.C.; de Sousa, N.F.; Muratov, E.; Scotti, L.; Scotti, M.T. Virtual Screening of Natural Products Database. Mini Rev. Med. Chem. 2021, 21, 2657–2730. [Google Scholar] [CrossRef] [PubMed]
- Kumarasiri, M.; Teo, T.; Yu, M.; Philip, S.; Basnet, S.K.; Albrecht, H.; Sykes, M.J.; Wang, P.; Wang, S. In Search of Novel CDK8 Inhibitors by Virtual Screening. J. Chem. Inf. Model. 2017, 57, 413–416. [Google Scholar] [CrossRef]
- Liu, G.; Jiao, Y.; Lin, Y.; Hao, H.; Dou, Y.; Yang, J.; Jiang, C.S.; Chang, P. Discovery and Biological Evaluation of New Selective Acetylcholinesterase Inhibitors with Anti-Aβ Aggregation Activity through Molecular Docking-Based Virtual Screening. Chem. Pharm. Bull. 2020, 68, 161–166. [Google Scholar] [CrossRef]
- Unoh, Y.; Uehara, S.; Nakahara, K.; Nobori, H.; Yamatsu, Y.; Yamamoto, S.; Maruyama, Y.; Taoda, Y.; Kasamatsu, K.; Suto, T.; et al. Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19. J. Med. Chem. 2022, 65, 6499–6512. [Google Scholar] [CrossRef] [PubMed]
- Pirzada, R.H.; Haseeb, M.; Batool, M.; Kim, M.; Choi, S. Remdesivir and Ledipasvir among the FDA-Approved Antiviral Drugs Have Potential to Inhibit SARS-CoV-2 Replication. Cells 2021, 10, 1052. [Google Scholar] [CrossRef] [PubMed]
- Power, H.; Wu, J.; Turville, S.; Aggarwal, A.; Valtchev, P.; Schindeler, A.; Dehghani, F. Virtual screening and in vitro validation of natural compound inhibitors against SARS-CoV-2 spike protein. Bioorg. Chem. 2022, 119, 105574. [Google Scholar] [CrossRef]
- Shin, H.J.; Kim, M.H.; Lee, J.Y.; Hwang, I.; Yoon, G.Y.; Kim, H.S.; Kwon, Y.C.; Ahn, D.G.; Kim, K.D.; Kim, B.T.; et al. Structure-Based Virtual Screening: Identification of a Novel NS2B-NS3 Protease Inhibitor with Potent Antiviral Activity against Zika and Dengue Viruses. Microorganisms 2021, 9, 545. [Google Scholar] [CrossRef]
- Pathak, N.; Kuo, Y.P.; Chang, T.Y.; Huang, C.T.; Hung, H.C.; Hsu, J.T.; Yu, G.Y.; Yang, J.M. Zika Virus NS3 Protease Pharmacophore Anchor Model and Drug Discovery. Sci. Rep. 2020, 10, 8929. [Google Scholar] [CrossRef] [PubMed]
- Pattnaik, A.; Palermo, N.; Sahoo, B.R.; Yuan, Z.; Hu, D.; Annamalai, A.S.; Vu, H.L.X.; Correas, I.; Prathipati, P.K.; Destache, C.J.; et al. Discovery of a non-nucleoside RNA polymerase inhibitor for blocking Zika virus replication through in silico screening. Antivir. Res. 2018, 151, 78–86. [Google Scholar] [CrossRef]
- Zhu, W.; Chen, C.Z.; Gorshkov, K.; Xu, M.; Lo, D.C.; Zheng, W. RNA-Dependent RNA Polymerase as a Target for COVID-19 Drug Discovery. SLAS Discov. 2020, 25, 1141–1151. [Google Scholar] [CrossRef]
- Eltahla, A.A.; Lackovic, K.; Marquis, C.; Eden, J.S.; White, P.A. A fluorescence-based high-throughput screen to identify small compound inhibitors of the genotype 3a hepatitis C virus RNA polymerase. J. Biomol. Screen. 2013, 18, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Gong, E.Y.; Kenens, H.; Ivens, T.; Dockx, K.; Vermeiren, K.; Vandercruyssen, G.; Devogelaere, B.; Lory, P.; Kraus, G. Expression and purification of dengue virus NS5 polymerase and development of a high-throughput enzymatic assay for screening inhibitors of dengue polymerase. Methods Mol. Biol. 2013, 1030, 237–247. [Google Scholar]
- Su, C.Y.; Cheng, T.J.; Lin, M.I.; Wang, S.Y.; Huang, W.I.; Lin-Chu, S.Y.; Chen, Y.H.; Wu, C.Y.; Lai, M.M.; Cheng, W.C.; et al. High-throughput identification of compounds targeting influenza RNA-dependent RNA polymerase activity. Proc. Natl. Acad. Sci. USA 2010, 107, 19151–19156. [Google Scholar] [CrossRef] [PubMed]
- Cubitt, B.; Ortiz-Riano, E.; Cheng, B.Y.; Kim, Y.J.; Yeh, C.D.; Chen, C.Z.; Southall, N.O.E.; Zheng, W.; Martinez-Sobrido, L.; de la Torre, J.C. A cell-based, infectious-free, platform to identify inhibitors of lassa virus ribonucleoprotein (vRNP) activity. Antivir. Res. 2020, 173, 104667. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Tseng, C.K.; Chen, K.J.; Huang, K.J.; Lin, C.K.; Lin, Y.T. A cell-based reporter assay for inhibitor screening of hepatitis C virus RNA-dependent RNA polymerase. Anal. Biochem. 2010, 403, 52–62. [Google Scholar] [CrossRef]
- Smith, S.A.; Lynch, K.W. Cell-based splicing of minigenes. In Spliceosomal Pre-mRNA Splicing; Springer: Berlin/Heidelberg, Germany; Totowa: New York, NJ, USA, 2014; pp. 243–255. [Google Scholar]
- Godoy, A.S.; Lima, G.M.; Oliveira, K.I.; Torres, N.U.; Maluf, F.V.; Guido, R.V.; Oliva, G. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat. Commun. 2017, 8, 14764. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zou, J.; Shan, C.; Yang, Y.; Kum, D.B.; Dallmeier, K.; Neyts, J.; Shi, P.Y. Zika Virus Replicons for Drug Discovery. EBioMedicine 2016, 12, 156–160. [Google Scholar] [CrossRef]
- Chen, L.; Krekels, E.H.J.; Verweij, P.E.; Buil, J.B.; Knibbe, C.A.J.; Brüggemann, R.J.M. Pharmacokinetics and Pharmacodynamics of Posaconazole. Drugs 2020, 80, 671–695. [Google Scholar] [CrossRef]
- Chen, B.; Trang, V.; Lee, A.; Williams, N.S.; Wilson, A.N.; Epstein, E.H., Jr.; Tang, J.Y.; Kim, J. Posaconazole, a Second-Generation Triazole Antifungal Drug, Inhibits the Hedgehog Signaling Pathway and Progression of Basal Cell Carcinoma. Mol. Cancer Ther. 2016, 15, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Mast, N.; Lin, J.B.; Pikuleva, I.A. Marketed Drugs Can Inhibit Cytochrome P450 27A1, a Potential New Target for Breast Cancer Adjuvant Therapy. Mol. Pharmacol. 2015, 88, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Varghese, F.S.; Meutiawati, F.; Teppor, M.; Jacobs, S.; de Keyzer, C.; Taşköprü, E.; van Woudenbergh, E.; Overheul, G.J.; Bouma, E.; Smit, J.M.; et al. Posaconazole inhibits multiple steps of the alphavirus replication cycle. Antivir. Res. 2022, 197, 105223. [Google Scholar] [CrossRef]
- Mercorelli, B.; Luganini, A.; Celegato, M.; Palù, G.; Gribaudo, G.; Lepesheva, G.I.; Loregian, A. The Clinically Approved Antifungal Drug Posaconazole Inhibits Human Cytomegalovirus Replication. Antimicrob. Agents Chemother. 2020, 64, e00056-20. [Google Scholar] [CrossRef]
- Meutiawati, F.; Bezemer, B.; Strating, J.; Overheul, G.J.; Žusinaite, E.; van Kuppeveld, F.J.M.; van Cleef, K.W.R.; van Rij, R.P. Posaconazole inhibits dengue virus replication by targeting oxysterol-binding protein. Antivir. Res. 2018, 157, 68–79. [Google Scholar] [CrossRef] [PubMed]
| No. | Compound Name | Structure | LibDock Score | Absolute Energy | Relative Energy | Conf Number |
|---|---|---|---|---|---|---|
| 1 | Dabigatran etexilate (DAB) | 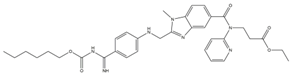 | 164.77 | 99.27 | 6.00 | 10 |
| 2 | Gossypol (acetic acid) (GOS) | 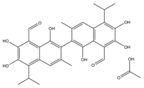 | 158.62 | 93.92 | 19.20 | 40 |
| 3 | Posaconazole (POS) | 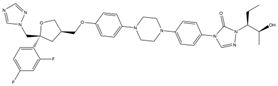 | 147.89 | 115.52 | 16.73 | 137 |
| 4 | Itraconazole (ITR) | 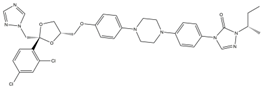 | 147.33 | 117.48 | 14.03 | 131 |
| 5 | Sulconazole (nitrate) (SUL) | 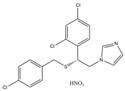 | 146.89 | 55.28 | 8.26 | 28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Chi, X.; Zhang, H.; Zhang, Y.; Qiao, L.; Ding, J.; Han, Y.; Lin, Y.; Jiang, J. Identification of Potent Zika Virus NS5 RNA-Dependent RNA Polymerase Inhibitors Combining Virtual Screening and Biological Assays. Int. J. Mol. Sci. 2023, 24, 1900. https://doi.org/10.3390/ijms24031900
Chen Y, Chi X, Zhang H, Zhang Y, Qiao L, Ding J, Han Y, Lin Y, Jiang J. Identification of Potent Zika Virus NS5 RNA-Dependent RNA Polymerase Inhibitors Combining Virtual Screening and Biological Assays. International Journal of Molecular Sciences. 2023; 24(3):1900. https://doi.org/10.3390/ijms24031900
Chicago/Turabian StyleChen, Ying, Xiangyin Chi, Hongjuan Zhang, Yu Zhang, Luyao Qiao, Jinwen Ding, Yanxing Han, Yuan Lin, and Jiandong Jiang. 2023. "Identification of Potent Zika Virus NS5 RNA-Dependent RNA Polymerase Inhibitors Combining Virtual Screening and Biological Assays" International Journal of Molecular Sciences 24, no. 3: 1900. https://doi.org/10.3390/ijms24031900
APA StyleChen, Y., Chi, X., Zhang, H., Zhang, Y., Qiao, L., Ding, J., Han, Y., Lin, Y., & Jiang, J. (2023). Identification of Potent Zika Virus NS5 RNA-Dependent RNA Polymerase Inhibitors Combining Virtual Screening and Biological Assays. International Journal of Molecular Sciences, 24(3), 1900. https://doi.org/10.3390/ijms24031900




