Genome-Wide Identification, Evolutionary Analysis, and Functional Studies of APX Genes in Melon (Cucuis melo L.)
Abstract
:1. Introduction
2. Results
2.1. Identification of APX Genes in the Melon Genome
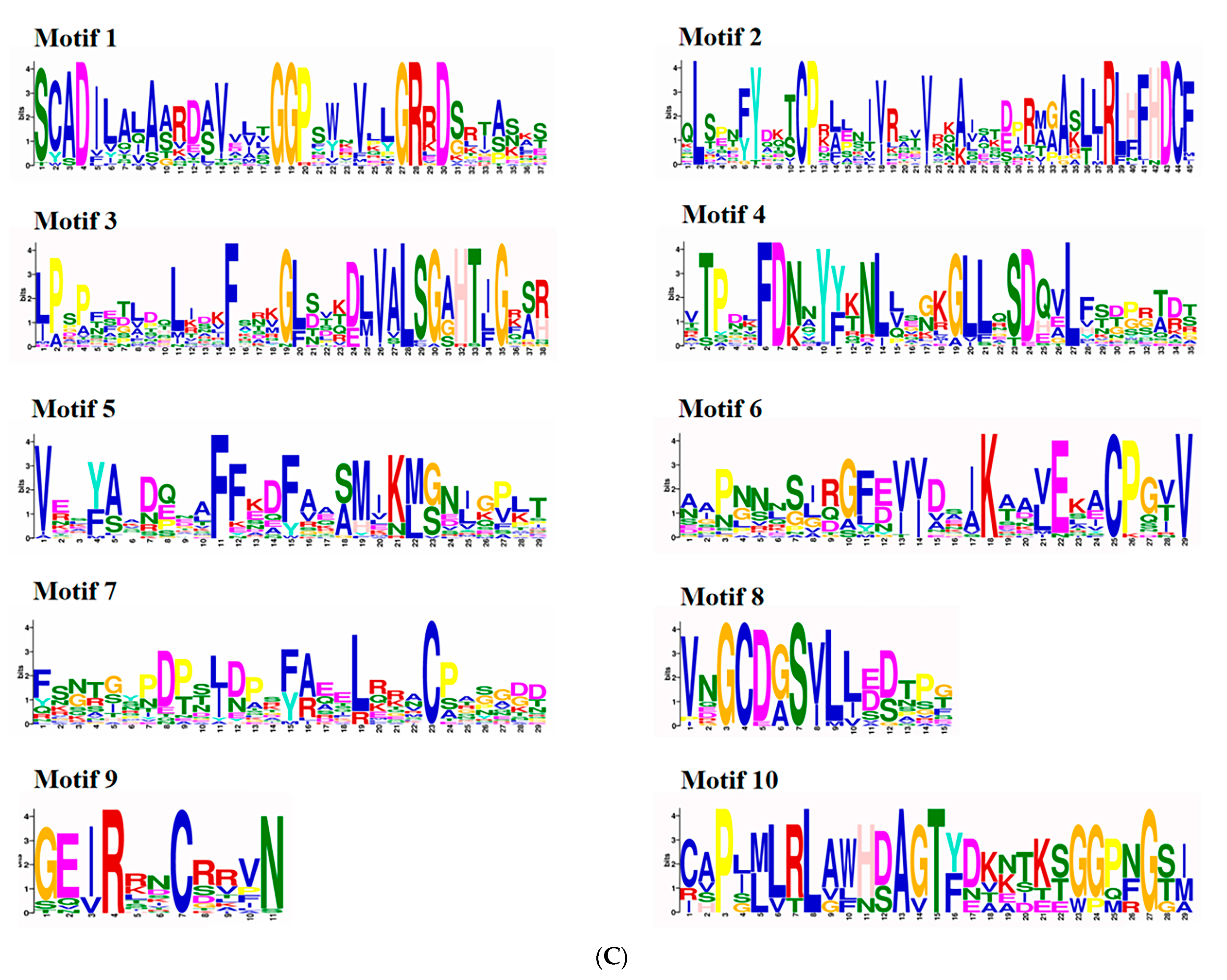
2.2. Structure and Conserved Motif Analyses for the CmAPX Genes
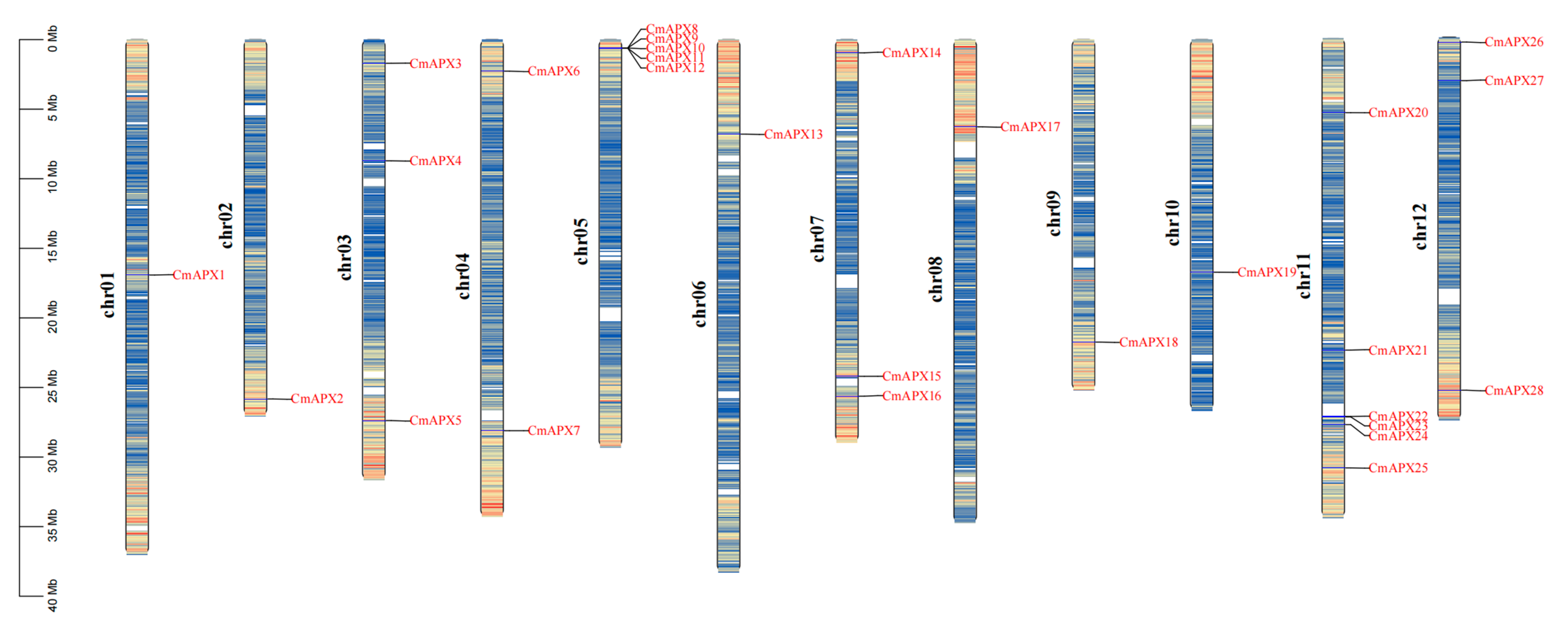
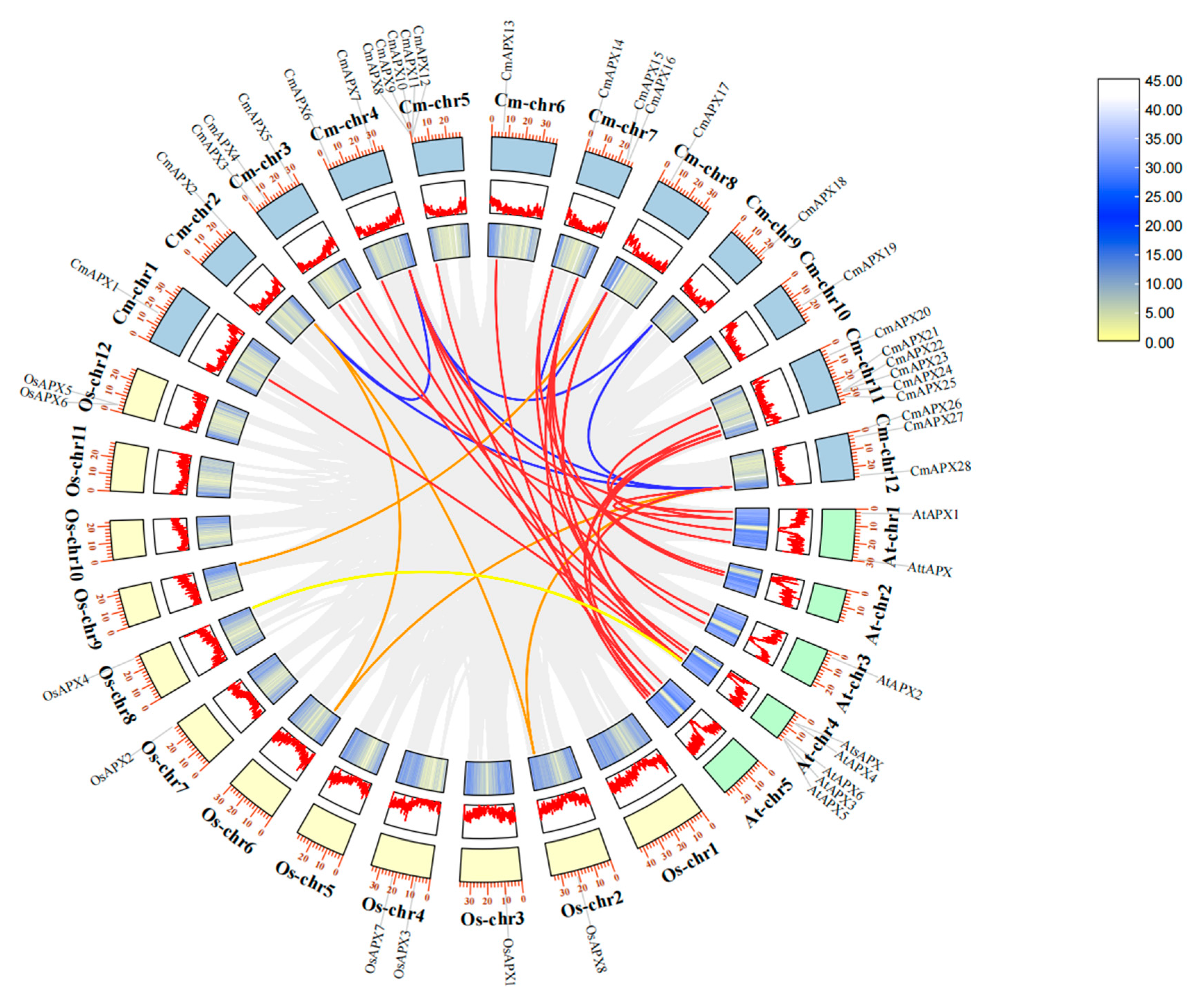
2.3. Chromosomal Localization of CmAPX Genes and Collinearity Analysis of APX Genes
2.4. Tissue Expression Profiles of CmAPX Genes
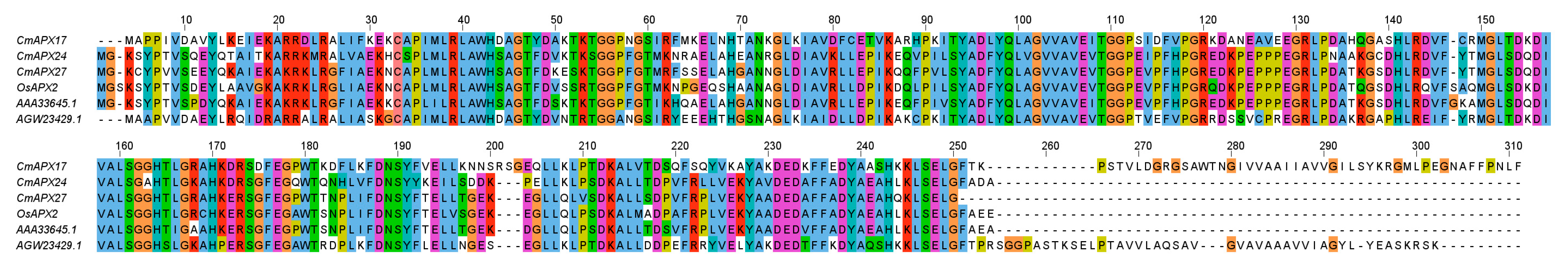
2.5. Phylogenetic Analysis and Gene Cloning
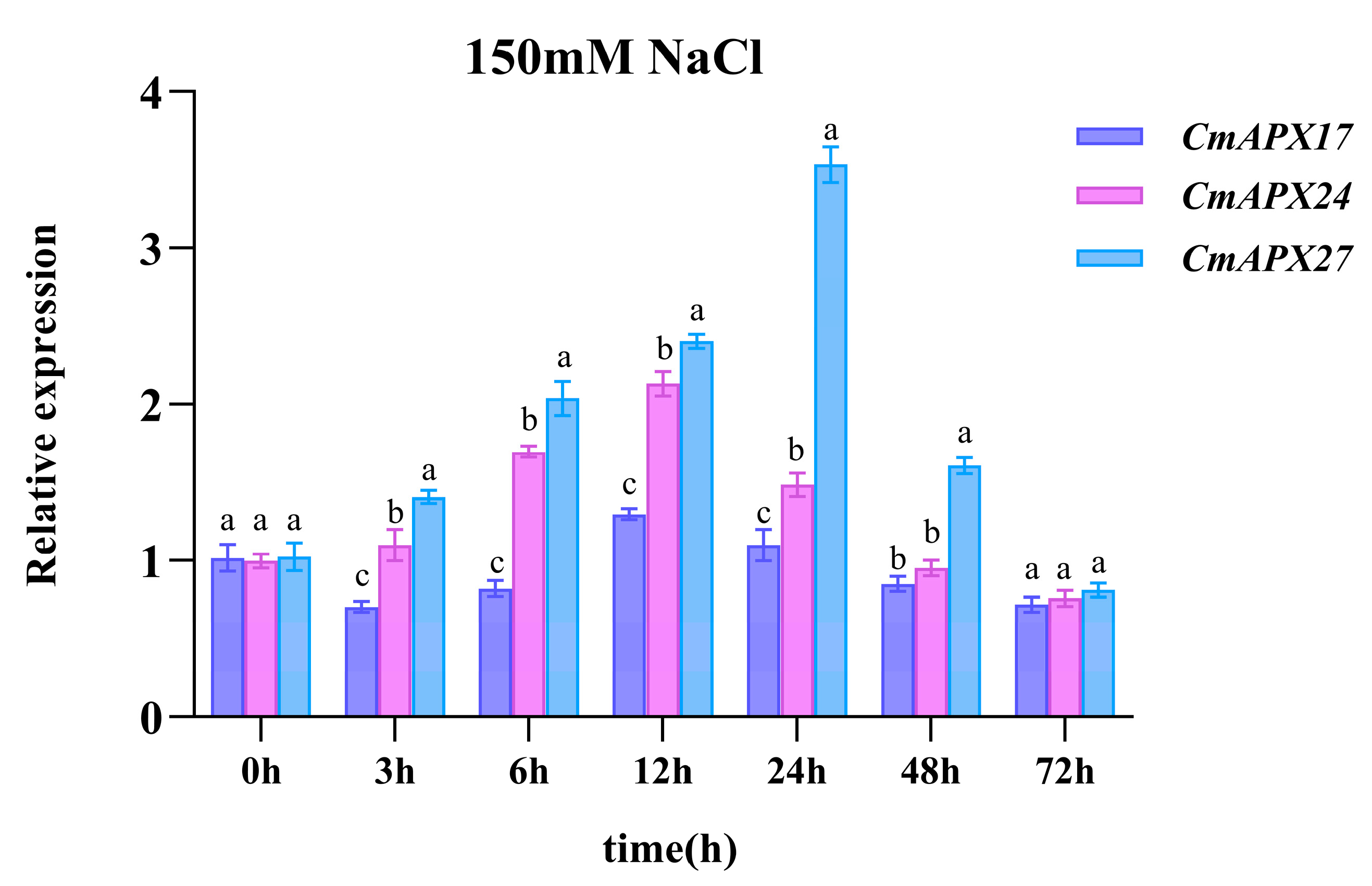
2.6. Expression Patterns of the Three Candidate Genes in the Context of Salt Stress
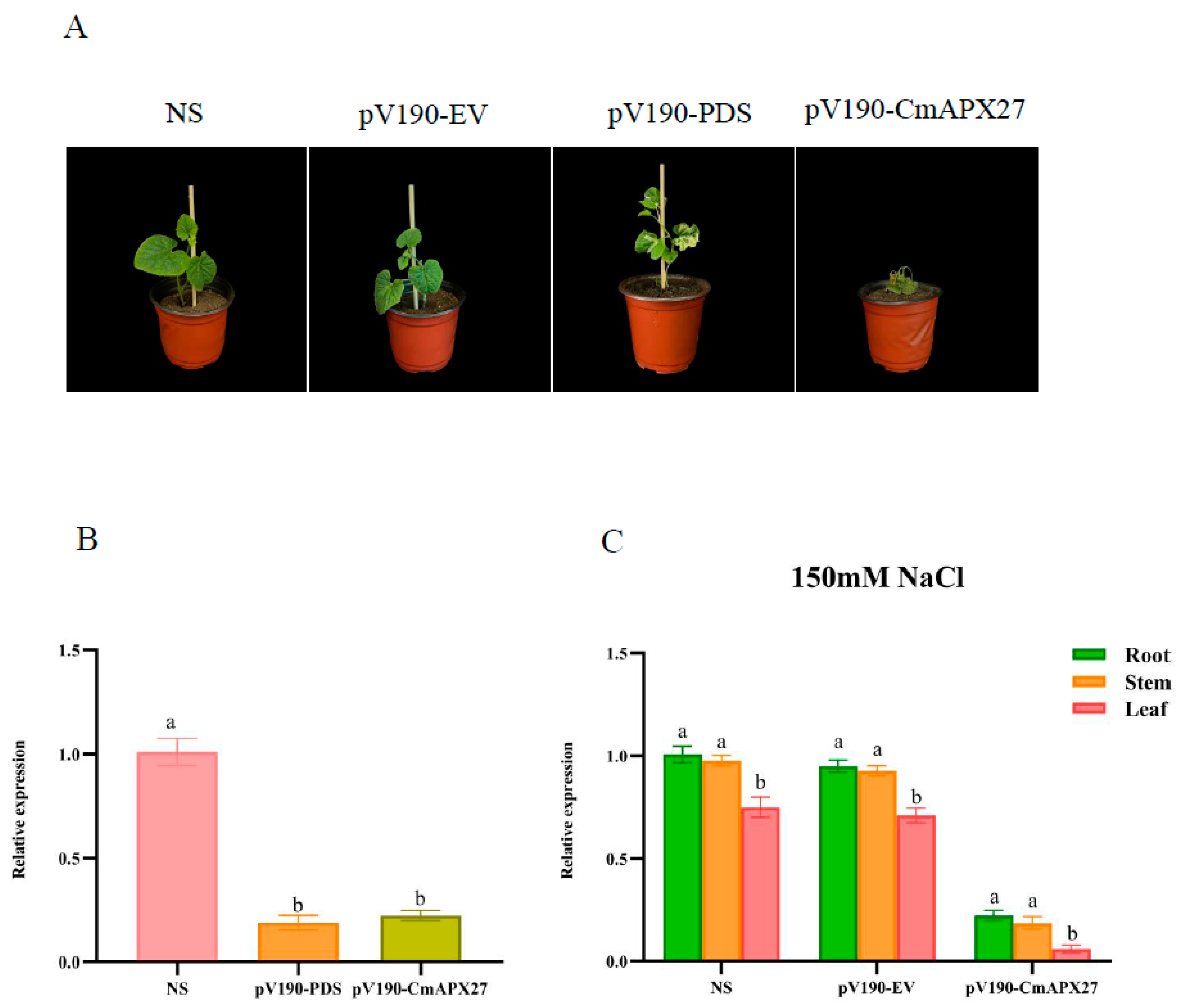
2.7. Functional Validation of CmAPX27
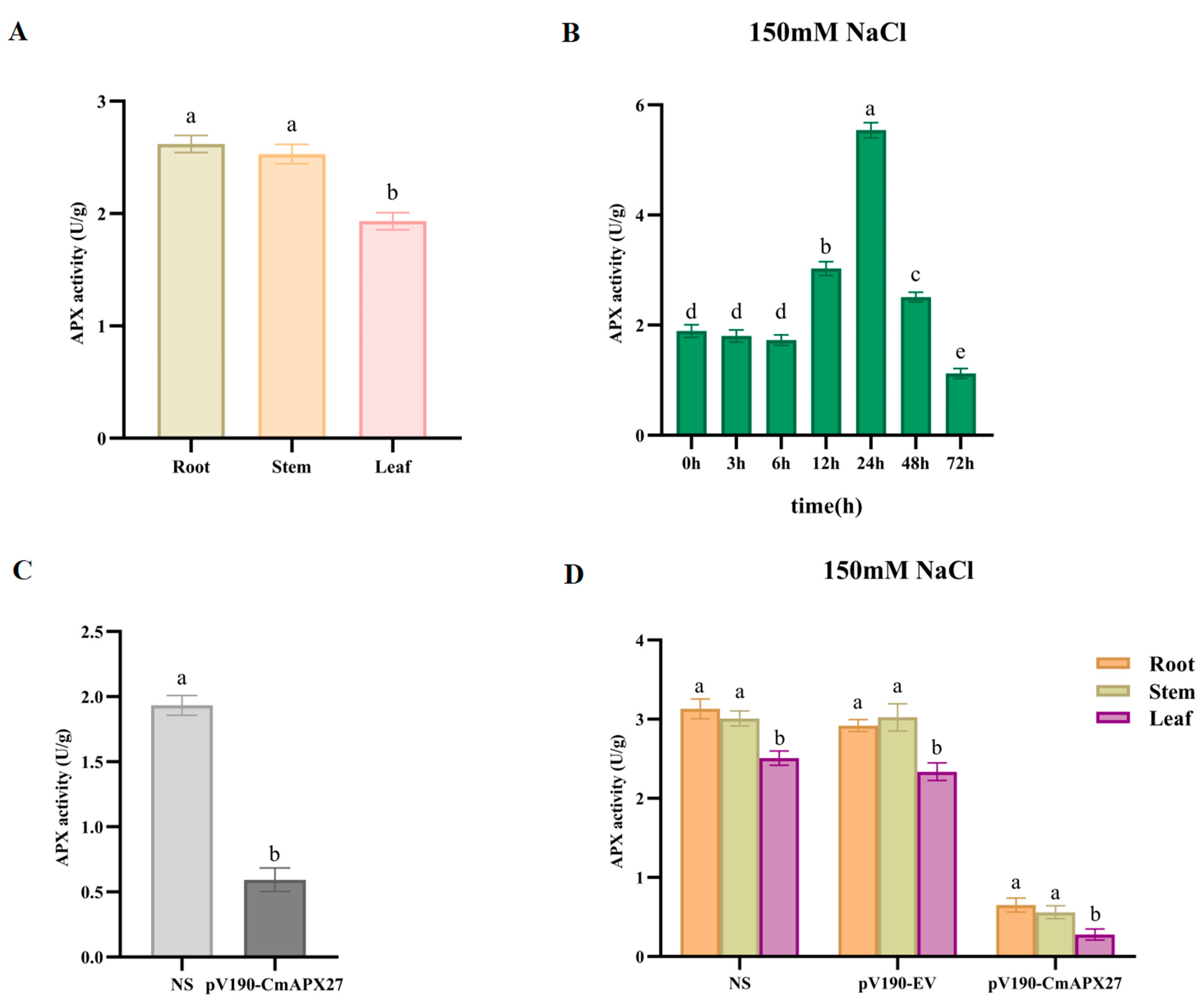
2.8. APX Activity Assay
3. Discussion
4. Materials and Methods
4.1. Identification of APX Genes in the Melon Genome
4.2. Structure and Conserved Motif Analyses for the CmAPX Genes
4.3. Phylogenetic Relationships, Chromosomal Localization, and Collinearity Analyses of APX Genes in Melon
4.4. RNA Sequencing (RNA-Seq) Analysis of CmAPX Genes
4.5. Plant Material and Salt Stress Treatment
4.6. Ascorbate Peroxidase (APX) Activity Assay
4.7. Cloning of APX Genes from Melon
4.8. CmAPX Gene Expression Analysis
4.9. Functional Characterization of the Candidate Gene CmAPX27
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amanullah, S.; Gao, P.; Osae, B.A.; Saroj, A.; Yang, T.; Liu, S.; Weng, Y.; Luan, F. Genetic linkage mapping and QTLs identification for morphology and fruit quality related traits of melon by SNP based CAPS markers. Sci. Hortic. 2021, 278, 109849. [Google Scholar] [CrossRef]
- Amanullah, S.; Liu, S.; Gao, P.; Zhu, Z.; Zhu, Q.; Fan, C.; Luan, F. QTL mapping for melon (Cucumis melo L.) fruit traits by assembling and utilization of novel SNPs based CAPS markers. Sci. Hortic. 2018, 236, 18–29. [Google Scholar] [CrossRef]
- Wang, L.M.; Zhang, L.D.; Chen, J.B.; Huang, D.F.; Zhang, Y.D. Physiological analysis and transcriptome comparison of two muskmelon (Cucumis melo L.) cultivars in response to salt stress. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Cabello, M.J.; Castellanos, M.T.; Romojaro, F.; Martínez-Madrid, C.; Ribas, F. Yield and quality of melon grown under different irrigation and nitrogen rates. Agric. Water Manag. 2009, 96, 866–874. [Google Scholar] [CrossRef]
- Rodríguez-López, J.N.; Espín, J.C.; Del Amor, F.; Tudela, J.; Martínez, V.; Cerdá, A.; García-Cánovas, F. Purification and kinetic characterization of an anionic peroxidase from melon (Cucumis melo L.) cultivated under different salinity conditions. J. Agric. Food Chem. 2000, 48, 1537–1541. [Google Scholar] [CrossRef] [PubMed]
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Adams, P.; Thomas, J.C.; Vernon, D.M.; Bohnert, H.J.; Jensen, R.G. Distinct Cellular and Organismic Responses to Salt Stress. Plant Cell Physiol. 1992, 33, 1215–1223. [Google Scholar]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Fahad, S.; Nie, L.; Chen, Y.; Wu, C.; Xiong, D.; Saud, S.; Hongyan, L.; Cui, K.; Huang, J. Crop Plant Hormones and Environmental Stress. In Sustainable Agriculture Reviews; Springer: Berlin/Heidelberg, Germany, 2015; pp. 371–400. [Google Scholar]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Hernández, J.A.; Ferrer, M.A.; Jiménez, A.; Barceló, A.R.; Sevilla, F. Antioxidant systems and O2−/H2O2 production in the apoplast of pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiol. 2001, 127, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Gilroy, S.; Bialasek, M.; Suzuki, N.; Górecka, M.; Devireddy, A.R.; Karpiński, S.; Mittler, R. ROS, Calcium, and Electric Signals: Key Mediators of Rapid Systemic Signaling in Plants. Plant Physiol. 2016, 171, 1606–1615. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Qi, W.; Bai, J.; Li, H.; Fang, Y.; Xu, J.; Xu, Y.; Zeng, X.; Pu, Y.; Wang, W.; et al. Genome-Wide Identification and Analysis of the Ascorbate Peroxidase (APX) Gene Family of Winter Rapeseed (Brassica rapa L.) Under Abiotic Stress. Front. Genet. 2021, 12, 753624. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. THE WATER-WATER CYCLE IN CHLOROPLASTS: Scavenging of Active Oxygens and Dissipation of Excess Photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.M.; Muramoto, Y.; Ueda, A.; Takabe, T. Cloning of peroxisomal ascorbate peroxidase gene from barley and enhanced thermotolerance by overexpressing in Arabidopsis thaliana. Gene 2001, 273, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Panchuk, I.I.; Volkov, R.A.; Schöffl, F. Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 2002, 129, 838–853. [Google Scholar] [CrossRef] [PubMed]
- Panchuk, I.I.; Zentgraf, U.; Volkov, R.A. Expression of the Apx gene family during leaf senescence of Arabidopsis thaliana. Planta 2005, 222, 926–932. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Menezes-Benavente, L.; Margis, R.; Margis-Pinheiro, M. Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: Inferences from the rice genome. J. Mol. Evol. 2004, 59, 761–770. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.; Wu, J.; Zheng, X.; Zheng, S.; Sun, X.; Qiu, Q.; Lu, T. Gene knockout study reveals that cytosolic ascorbate peroxidase 2(OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 2013, 8, e57472. [Google Scholar] [CrossRef]
- Tao, C.; Jin, X.; Zhu, L.; Xie, Q.; Wang, X.; Li, H. Genome-wide investigation and expression profiling of APX gene family in Gossypium hirsutum provide new insights in redox homeostasis maintenance during different fiber development stages. Mol. Genet. Genom. 2018, 293, 685–697. [Google Scholar] [CrossRef]
- Akbudak, M.A.; Filiz, E.; Vatansever, R.; Kontbay, K. Genome-Wide Identification and Expression Profiling of Ascorbate Peroxidase (APX) and Glutathione Peroxidase (GPX) Genes Under Drought Stress in Sorghum (Sorghum bicolor L.). J. Plant Growth Regul. 2018, 37, 925–936. [Google Scholar] [CrossRef]
- Tyagi, S.; Shumayla; Verma, P.C.; Singh, K.; Upadhyay, S.K. Molecular characterization of ascorbate peroxidase (APX) and APX-related (APX-R) genes in Triticum aestivum L. Genomics 2020, 112, 4208–4223. [Google Scholar] [CrossRef] [PubMed]
- Leng, X.; Wang, H.; Zhang, S.; Qu, C.; Yang, C.; Xu, Z.; Liu, G. Identification and Characterization of the APX Gene Family and Its Expression Pattern under Phytohormone Treatment and Abiotic Stress in Populus trichocarpa. Genes 2021, 12, 334. [Google Scholar] [CrossRef] [PubMed]
- Najami, N.; Janda, T.; Barriah, W.; Kayam, G.; Tal, M.; Guy, M.; Volokita, M. Ascorbate peroxidase gene family in tomato: Its identification and characterization. Mol. Genet. Genom. 2008, 279, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.L.; Liu, Q.; Li, Y.Q.; Zhong, M.; Huang, C.H.; Jia, D.F.; Xu, X.B. Identification and expression profiling analysis of ascorbate peroxidase gene family in Actinidia chinensis (Hongyang). J. Plant Res. 2020, 133, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Sharif, Y.; Chen, K.; Wang, L.; Fu, H.; Zhuang, Y.; Chitikineni, A.; Chen, H.; Zhang, C.; Varshney, R.K.; et al. Genome-Wide Characterization of Ascorbate Peroxidase Gene Family in Peanut (Arachis hypogea L.) Revealed Their Crucial Role in Growth and Multiple Stress Tolerance. Front. Plant Sci. 2022, 13, 962182. [Google Scholar] [CrossRef] [PubMed]
- Malambane, G.; Tsujimoto, H.; Akashi, K. The cDNA Structures and Expression Profile of the Ascorbate Peroxidase Gene Family During Drought Stress in Wild Watermelon. J. Agric. Sci. 2018, 10, 56. [Google Scholar] [CrossRef]
- Yan, H.; Li, Q.; Park, S.C.; Wang, X.; Liu, Y.J.; Zhang, Y.G.; Tang, W.; Kou, M.; Ma, D.F. Overexpression of CuZnSOD and APX enhance salt stress tolerance in sweet potato. Plant Physiol. Biochem. 2016, 109, 20–27. [Google Scholar] [CrossRef]
- Guan, Q.; Wang, Z.; Wang, X.; Takano, T.; Liu, S. A peroxisomal APX from Puccinellia tenuiflora improves the abiotic stress tolerance of transgenic Arabidopsis thaliana through decreasing of H2O2 accumulation. J. Plant Physiol. 2015, 175, 183–191. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Serrano, R.; Mulet, J.M.; Rios, G.; Marquez, J.A.; De Larrinoa, I.F.; Leube, M.P.; Mendizabal, I.; Pascual-Ahuir, A.; Proft, M.; Ros, R.; et al. A glimpse of the mechanisms of ion homeostasis during salt stress. J. Exp. Bot. 1999, 50, 1023–1036. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Shukla, P.S.; Gupta, K.; Jha, B. Bioengineering for salinity tolerance in plants: State of the art. Mol. Biotechnol. 2013, 54, 102–123. [Google Scholar] [CrossRef] [PubMed]
- Chevilly, S.; Dolz-Edo, L.; Martínez-Sánchez, G.; Morcillo, L.; Vilagrosa, A.; López-Nicolás, J.M.; Blanca, J.; Yenush, L.; Mulet, J.M. Distinctive Traits for Drought and Salt Stress Tolerance in Melon (Cucumis melo L.). Front. Plant Sci. 2021, 12, 777060. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Storozhenko, S.; De Pauw, P.; Van Montagu, M.; Inzé, D.; Kushnir, S. The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol. 1998, 118, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Suda, Y.; Kondo, N.; Sugahara, K. O3 Tolerance and the Ascorbate-Dependent H2O2 Decomposing System in Chloroplasts. Plant Cell Physiol. 1985, 26, 1425–1431. [Google Scholar]
- Gupta, A.S.; Webb, R.P.; Holaday, A.S.; Allen, R.D. Overexpression of Superoxide Dismutase Protects Plants from Oxidative Stress (Induction of Ascorbate Peroxidase in Superoxide Dismutase-Overexpressing Plants). Plant Physiol. 1993, 103, 1067–1073. [Google Scholar] [CrossRef]
- Bender, J.; Weigel, H.J.; Wegner, U.; Jäger, H.J. Response of cellular antioxidants to ozone in wheat flag leaves at different stages of plant development. Environ. Pollut. 1994, 84, 15–21. [Google Scholar] [CrossRef]
- Guan, Q.; Takano, T.; Liu, S. Genetic transformation and analysis of rice OsAPx2 gene in Medicago sativa. PLoS ONE 2012, 7, e41233. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Salehi, H.; Rahman, M.A.; Zahid, Z.; Madadkar Haghjou, M.; Najafi-Kakavand, S.; Charagh, S.; Osman, H.S.; Albaqami, M.; Zhuang, Y.; et al. Plant hormones and neurotransmitter interactions mediate antioxidant defenses under induced oxidative stress in plants. Front. Plant Sci. 2022, 13, 961872. [Google Scholar] [CrossRef] [PubMed]
- Arasimowicz-Jelonek, M.; Floryszak-Wieczorek, J.; Suarez, S.; Doctorovich, F.; Sobieszczuk-Nowicka, E.; Bruce King, S.; Milczarek, G.; Rębiś, T.; Gajewska, J.; Jagodzik, P.; et al. Discovery of endogenous nitroxyl as a new redox player in Arabidopsis thaliana. Nat. Plants 2023, 9, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Veljovic-Jovanovic, S.; Foyer, C.H. Peroxide processing in photosynthesis: Antioxidant coupling and redox signalling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1465–1475. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G. Metabolic signalling in defence and stress: The central roles of soluble redox couples. Plant Cell Environ. 2006, 29, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Shigeoka, S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011, 155, 93–100. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Beavis, W.; Berardini, T.Z.; Chen, G.; Dixon, D.; Doyle, A.; Garcia-Hernandez, M.; Huala, E.; Lander, G.; Montoya, M.; et al. The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003, 31, 224–228. [Google Scholar] [CrossRef]
- Yu, T.; Cen, Q.; Kang, L.; Mou, W.; Zhang, X.; Fang, Y.; Zhang, X.; Tian, Q.; Xue, D. Identification and expression pattern analysis of the OsSnRK2 gene family in rice. Front. Plant Sci. 2022, 13, 1088281. [Google Scholar] [CrossRef]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Chung, B.Y.; Hardcastle, T.J.; Jones, J.D.; Irigoyen, N.; Firth, A.E.; Baulcombe, D.C.; Brierley, I. The use of duplex-specific nuclease in ribosome profiling and a user-friendly software package for Ribo-seq data analysis. RNA 2015, 21, 1731–1745. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, J.; Amanullah, S.; Ding, Z.; Cui, H.; Luan, F.; Gao, P. Fine Mapping of Cla015407 Controlling Plant Height in Watermelon. J. Am. Soc. Hortic. Sci. 2021, 146, 196–205. [Google Scholar] [CrossRef]
- Zhang, T.; Cui, H.; Luan, F.; Liu, H.; Ding, Z.; Amanullah, S.; Zhang, M.; Ma, T.; Gao, P. A recessive gene Cmpmr2F confers powdery mildew resistance in melon (Cucumis melo L.). Theor. Appl. Genet. 2023, 136, 4. [Google Scholar] [CrossRef]
- Liu, M.; Liang, Z.; Aranda, M.A.; Hong, N.; Liu, L.; Kang, B.; Gu, Q. A cucumber green mottle mosaic virus vector for virus-induced gene silencing in cucurbit plants. Plant Methods 2020, 16, 9. [Google Scholar] [CrossRef]




| Gene Name | Gene ID | Genomic Region | CDS Length (bp) | Exon | Intron | Protein Length (aa) | PI | Molecular Weight/kDa | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|
| CmAPX1 | MELO3C013362.2 | chr01 (16914932, 16920143) | 471 | 5 | 4 | 156 | 5.82 | 17.26 | −0.245 | Cytoplasmic |
| CmAPX2 | MELO3C017120.2 | chr02 (25825071, 25826275) | 984 | 1 | 0 | 327 | 8.47 | 35.64 | −0.084 | Extracellular |
| CmAPX3 | MELO3C008186.2 | chr03 (1701410, 1704553) | 1008 | 3 | 2 | 335 | 6.15 | 36.09 | −0.124 | Chloroplast |
| CmAPX4 | MELO3C010627.2 | chr03 (8704812, 8712281) | 1251 | 12 | 11 | 416 | 8.45 | 45.04 | −0.411 | Chloroplast |
| CmAPX5 | MELO3C011261.2 | chr03 (27375748, 27380146) | 1008 | 10 | 9 | 335 | 7.61 | 36.65 | −0.079 | Chloroplast |
| CmAPX6 | MELO3C003559.2 | chr04 (2265163, 2269072) | 819 | 9 | 8 | 272 | 8.67 | 29.51 | −0.251 | Chloroplast |
| CmAPX7 | MELO3C009924.2 | chr04 (28104983, 28106495) | 1107 | 1 | 0 | 368 | 8.74 | 41.49 | −0.077 | Plasma membrane |
| CmAPX8 | MELO3C014658.2 | chr05 (595378, 597928) | 957 | 4 | 3 | 318 | 6.43 | 34.67 | −0.125 | Extracellular |
| CmAPX9 | MELO3C014657.2 | chr05 (603858, 605423) | 1038 | 4 | 3 | 345 | 6.01 | 37.11 | −0.182 | Chloroplast |
| CmAPX10 | MELO3C014656.2 | chr05 (606283, 608027) | 1038 | 4 | 3 | 345 | 5.69 | 37.19 | −0.234 | Chloroplast |
| CmAPX11 | MELO3C014655.2 | chr05 (613307, 615280) | 990 | 3 | 2 | 329 | 5.92 | 36.67 | −0.336 | Extracellular |
| CmAPX12 | MELO3C014654.2 | chr05 (618721, 620283) | 1002 | 3 | 2 | 333 | 8.09 | 36.36 | −0.060 | Extracellular |
| CmAPX13 | MELO3C006862.2 | chr06 (6793499, 6795861) | 951 | 3 | 2 | 316 | 7.56 | 34.69 | −0.101 | Extracellular |
| CmAPX14 | MELO3C016943.2 | chr07 (948078, 951116) | 984 | 3 | 2 | 327 | 5.32 | 34.92 | 0.040 | Chloroplast |
| CmAPX15 | MELO3C016405.2 | chr07 (24195633, 24198118) | 1032 | 4 | 3 | 343 | 9.12 | 37.37 | −0.296 | Extracellular |
| CmAPX16 | MELO3C017603.2 | chr07 (25625229, 25628495) | 759 | 2 | 1 | 252 | 5.36 | 28.14 | −0.261 | Cytoplasmic |
| CmAPX17 | MELO3C007923.2 | chr08 (6265300, 6269688) | 990 | 10 | 9 | 329 | 8.75 | 36.59 | −0.242 | Cytoplasmic |
| CmAPX18 | MELO3C005456.2 | chr09 (21737263, 21739391) | 1014 | 4 | 3 | 337 | 4.53 | 35.93 | −0.046 | Extracellular |
| CmAPX19 | MELO3C022604.2 | chr10 (16715480, 16716883) | 951 | 3 | 2 | 316 | 8.07 | 34.27 | −0.079 | Plasma membrane |
| CmAPX20 | MELO3C021914.2 | chr11 (5335837, 5338322) | 1008 | 3 | 2 | 335 | 5.04 | 37.20 | −0.140 | Extracellular |
| CmAPX21 | MELO3C034836.2 | chr11 (22401880, 22403778) | 975 | 4 | 3 | 324 | 8.03 | 35.43 | −0.170 | Extracellular |
| CmAPX22 | MELO3C025681.2 | chr11 (27168191, 27169996) | 978 | 4 | 3 | 325 | 9.15 | 35.43 | −0.169 | Chloroplast |
| CmAPX23 | MELO3C025683.2 | chr11 (27200212, 27202002) | 969 | 3 | 2 | 322 | 6.78 | 34.93 | −0.166 | Extracellular |
| CmAPX24 | MELO3C025724.2 | chr11 (27753082, 27756921) | 750 | 9 | 8 | 249 | 5.86 | 27.63 | −0.374 | Cytoplasmic |
| CmAPX25 | MELO3C021259.2 | chr11 (30864215, 30866563) | 993 | 4 | 3 | 330 | 4.92 | 35.39 | 0.011 | Extracellular |
| CmAPX26 | MELO3C020501.2 | chr12 (349119, 351142) | 960 | 3 | 2 | 319 | 5.51 | 34.41 | −0.048 | Extracellular |
| CmAPX27 | MELO3C020719.2 | chr12 (3120431, 3122973) | 738 | 8 | 7 | 245 | 5.32 | 26.99 | −0.383 | Cytoplasmic |
| CmAPX28 | MELO3C002242.2 | chr12 (25369164, 25370763) | 1005 | 2 | 1 | 334 | 7.59 | 37.07 | −0.052 | Extracellular |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Zhu, Z.; Zhang, T.; Meng, X.; Zhang, W.; Gao, P. Genome-Wide Identification, Evolutionary Analysis, and Functional Studies of APX Genes in Melon (Cucuis melo L.). Int. J. Mol. Sci. 2023, 24, 17571. https://doi.org/10.3390/ijms242417571
Song J, Zhu Z, Zhang T, Meng X, Zhang W, Gao P. Genome-Wide Identification, Evolutionary Analysis, and Functional Studies of APX Genes in Melon (Cucuis melo L.). International Journal of Molecular Sciences. 2023; 24(24):17571. https://doi.org/10.3390/ijms242417571
Chicago/Turabian StyleSong, Jiayan, Zicheng Zhu, Taifeng Zhang, Xiaobing Meng, Wencheng Zhang, and Peng Gao. 2023. "Genome-Wide Identification, Evolutionary Analysis, and Functional Studies of APX Genes in Melon (Cucuis melo L.)" International Journal of Molecular Sciences 24, no. 24: 17571. https://doi.org/10.3390/ijms242417571
APA StyleSong, J., Zhu, Z., Zhang, T., Meng, X., Zhang, W., & Gao, P. (2023). Genome-Wide Identification, Evolutionary Analysis, and Functional Studies of APX Genes in Melon (Cucuis melo L.). International Journal of Molecular Sciences, 24(24), 17571. https://doi.org/10.3390/ijms242417571





