Probiotic Potential and Safety Assessment of Lactiplantibacillus plantarum cqf-43 and Whole-Genome Sequence Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Genomic Sequencing and Taxonomic Identification
2.2. Genomic Features
2.3. Genomic Functional Annotation
2.4. Mobile Genetic Element Analysis
2.5. Virulence Factors
2.6. Probiotic Potential of Strain cqf-43
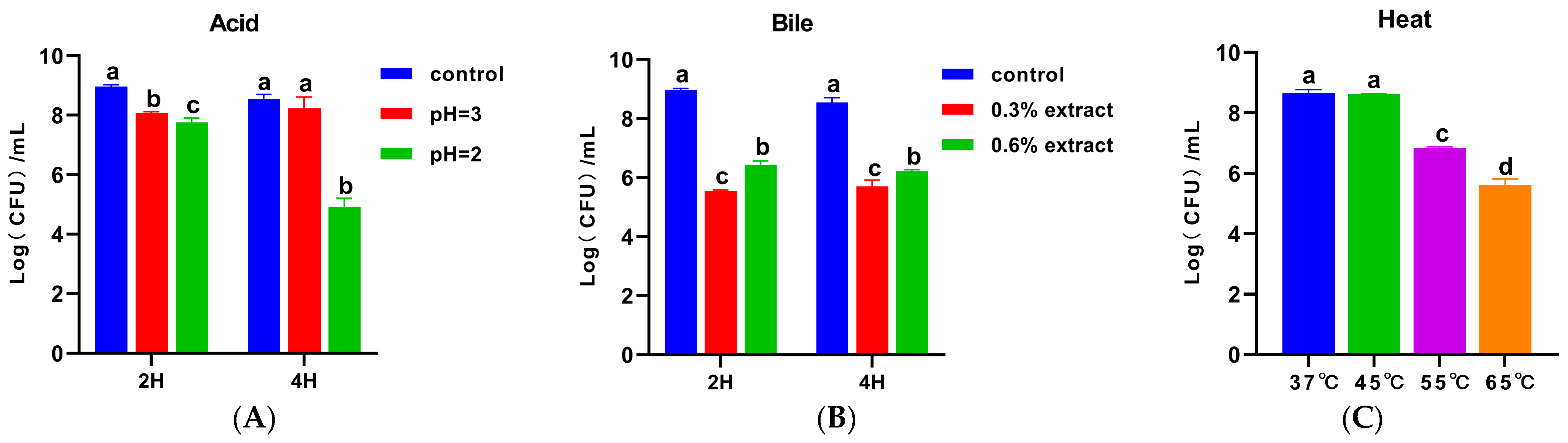
2.6.1. Tolerance to Acid, Bile, and Heat
2.6.2. Antibacterial Activity
2.7. Safety Test
2.7.1. Antibiotic Susceptibility
2.7.2. Hemolytic and Gelatinase Activities and Biogenic Amine-Producing Ability
2.7.3. The 28-Day Oral Feeding Toxicity Test
3. Materials and Methods
3.1. Bacterial Strains, Culture Conditions, and DNA Isolation
3.2. Whole-Genome Sequencing of Strain cqf-43
3.3. Gene Prediction and Functional Annotation
3.4. Phylogenetic and Genome-Related Analyses
3.5. Tolerance of Acid, Bile, and Heat
3.6. Preparation of Pathogenic Bacteria and Antibacterial Activity
3.7. Antibiotic Susceptibility Test
3.8. Phenotypic Tests for Hemolytic and Gelatinase Activities and Biogenic Amine-Producing Ability
3.9. The 28-Day Oral Feeding Toxicity Test
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Fijan, S. Microorganisms with Claimed Probiotic Properties: An Overview of Recent Literature. Int. J. Environ. Res. Public Health 2014, 11, 4745–4767. [Google Scholar] [CrossRef] [PubMed]
- Ezendam, J.; van Loveren, H. Probiotics: Immunomodulation and evaluation of safety and efficacy. Nutr. Rev. 2006, 64, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hou, G.F.; Peng, W.; Wei, L.K.; Li, R.; Huang, X.G.; Yin, Y.L. Probiotics and Achyranthes bidentata Polysaccharides Improve Growth Performance via Promoting Intestinal Nutrient Utilization and Enhancing Immune Function of Weaned Pigs. Animals 2021, 11, 2617. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, N.; Battista, N.; Prete, R.; Corsetti, A. Health-Promoting Role of Lactiplantibacillus plantarum Isolated from Fermented Foods. Microorganisms 2021, 9, 349. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Gao, Y.; Zhao, Y.; Gao, L.; Zhao, Z.; He, Z.; Li, S. Lactiplantibacillus plantarum ELF051 Alleviates Antibiotic-Associated Diarrhea by Regulating Intestinal Inflammation and Gut Microbiota. Probiotics Antimicrob. Proteins 2023. [Google Scholar] [CrossRef]
- Paswan, V.K.; Singh, P.; Pandey, R.K.; Yadav, S.P.; Bhinchhar, B.K.; Singh, C.S. Effect of supplementation of L. plantarum and L. casei based probiotic milk powder on hematology, blood biochemistry and lipid profile of Charles Foster rats. Indian J. Anim. Res. 2019, 53, 332–335. [Google Scholar] [CrossRef]
- Wang, W.; Chen, J.; Zhou, H.; Wang, L.; Ding, S.; Wang, Y.; Song, D.; Li, A. Effects of microencapsulated Lactobacillus plantarum and fructooligosaccharide on growth performance, blood immune parameters, and intestinal morphology in weaned piglets. Food Agric. Immunol. 2017, 29, 84–94. [Google Scholar] [CrossRef]
- Tartrakoon, W.; Charoensook, R.; Incharoen, T.; Numthuam, S.; Pechrkong, T.; Onoda, S.; Shoji, G.; Brenig, B. Effects of Heat-Killed Lactobacillus plantarum L-137 Supplementation on Growth Performance, Blood Profiles, Intestinal Morphology, and Immune Gene Expression in Pigs. Vet. Sci. 2023, 10, 87. [Google Scholar] [CrossRef]
- Echegaray, N.; Yilmaz, B.; Sharma, H.; Kumar, M.; Pateiro, M.; Ozogul, F.; Lorenzo, J.M. A novel approach to Lactiplantibacillus plantarum: From probiotic properties to the omics insights. Microbiol. Res. 2023, 268, 127289. [Google Scholar] [CrossRef]
- Zheng, M.; Zhang, R.J.; Tian, X.C.; Zhou, X.; Pan, X.T.; Wong, A. Assessing the Risk of Probiotic Dietary Supplements in the Context of Antibiotic Resistance. Front. Microbiol. 2017, 8, 908. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.Q.; Liu, L.M.; Fu, T.Y.; Li, C.H.; Jin, N.Y.; Zhang, H.P.; Li, C.; Liu, Y.W.; Zhao, C.Q. Genome Sequence and Evaluation of Safety and Probiotic Potential of Lactiplantibacillus plantarum LPJZ-658. Microorganisms 2023, 11, 1620. [Google Scholar] [CrossRef] [PubMed]
- Syrokou, M.K.; Paramithiotis, S.; Drosinos, E.H.; Bosnea, L.; Mataragas, M. A Comparative Genomic and Safety Assessment of Six Lactiplantibacillus plantarum subsp. argentoratensis Strains Isolated from Spontaneously Fermented Greek Wheat Sourdoughs for Potential Biotechnological Application. Int. J. Mol. Sci. 2022, 23, 2487. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Yang, S.M.; Kim, D.; Kim, H.Y. Complete Genome Sequencing and Comparative Genomics of Three Potential Probiotic Strains, Lacticaseibacillus casei FBL6, Lacticaseibacillus chiayiensis FBL7, and Lacticaseibacillus zeae FBL8. Front. Microbiol. 2022, 12, 794315. [Google Scholar] [CrossRef] [PubMed]
- Florez, A.B.; Vazquez, L.; Rodriguez, J.; Mayo, B. Phenotypic and Safety Assessment of the Cheese Strain Lactiplantibacillus plantarum LL441, and Sequence Analysis of its Complete Genome and Plasmidome. Int. J. Mol. Sci. 2022, 24, 605. [Google Scholar] [CrossRef] [PubMed]
- Fujimori, S.; Washio, T.; Tomita, M. GC-compositional strand bias around transcription start sites in plants and fungi. BMC Genom. 2005, 6, 26. [Google Scholar] [CrossRef]
- Kandasamy, S.; Yoo, J.; Yun, J.; Lee, K.H.; Kang, H.B.; Kim, J.E.; Oh, M.H.; Ham, J.S. Probiogenomic In-Silico Analysis and Safety Assessment of Lactiplantibacillus plantarum DJF10 Strain Isolated from Korean Raw Milk. Int. J. Mol. Sci. 2022, 23, 14494. [Google Scholar] [CrossRef]
- Kwon, Y.J.; Chun, B.H.; Jung, H.S.; Chu, J.; Joung, H.; Park, S.Y.; Kim, B.K.; Jeon, C.O. Safety Assessment of Lactiplantibacillus (formerly Lactobacillus) plantarum Q180. J. Microbiol. Biotechnol. 2021, 31, 1420–1429. [Google Scholar] [CrossRef]
- Renaux, A.; UniProt, C. UniProt: The universal protein knowledgebase (vol 45, pg D158, 2017). Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; Selengut, J.D.; White, O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003, 31, 371–373. [Google Scholar] [CrossRef] [PubMed]
- Ou, D.; Ling, N.; Wang, X.; Zou, Y.; Dong, J.; Zhang, D.; Shen, Y.; Ye, Y. Safety Assessment of One Lactiplantibacillus plantarum Isolated from the Traditional Chinese Fermented Vegetables-Jiangshui. Foods 2022, 11, 2177. [Google Scholar] [CrossRef]
- Katiku, M.M.; Matofari, J.W.; Nduko, J.M. Preliminary evaluation of probiotic properties and safety profile of Lactiplantibacillus plantarum isolated from spontaneously fermented milk, Amabere amaruranu. Heliyon 2022, 8, e10342. [Google Scholar] [CrossRef]
- Angmo, K.; Kumari, A.; Savitri; Bhalla, T.C. Probiotic characterization of lactic acid bacteria isolated from fermented foods and beverage of Ladakh. Lwt-Food Sci. Technol. 2016, 66, 428–435. [Google Scholar] [CrossRef]
- Sohn, H.; Chang, Y.H.; Yune, J.H.; Jeong, C.H.; Shin, D.M.; Kwon, H.C.; Kim, D.H.; Hong, S.W.; Hwang, H.; Jeong, J.Y.; et al. Probiotic Properties of Lactiplantibacillus plantarum LB5 Isolated from Kimchi Based on Nitrate Reducing Capability. Foods 2020, 9, 1777. [Google Scholar] [CrossRef]
- Abarquero, D.; Bodelon, R.; Florez, A.B.; Fresno, J.M.; Renes, E.; Mayob, B.; Tornadijoa, M.E. Technological and safety assessment of selected lactic acid bacteria for cheese starter cultures design: Enzymatic and antimicrobial activity, antibiotic resistance and biogenic amine production. Lwt-Food Sci. Technol. 2023, 180, 114709. [Google Scholar] [CrossRef]
- Chikindas, M.L.; Weeks, R.; Drider, D.; Chistyakov, V.A.; Dicks, L.M.T. Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 2018, 49, 23–28. [Google Scholar] [CrossRef]
- Jeong, C.H.; Sohn, H.; Hwang, H.; Lee, H.J.; Kim, T.W.; Kim, D.S.; Kim, C.S.; Han, S.G.; Hong, S.W. Comparison of the Probiotic Potential between Lactiplantibacillus plantarum Isolated from Kimchi and Standard Probiotic Strains Isolated from Different Sources. Foods 2021, 10, 2125. [Google Scholar] [CrossRef] [PubMed]
- Naughton, L.M.; Romano, S.; O’Gara, F.; Dobson, A.D.W. Identification of Secondary Metabolite Gene Clusters in the Pseudovibrio Genus Reveals Encouraging Biosynthetic Potential toward the Production of Novel Bioactive Compounds. Front. Microbiol. 2017, 8, 1494. [Google Scholar] [CrossRef] [PubMed]
- Aziz, T.; Naveed, M.; Sarwar, A.; Makhdoom, S.I.; Mughal, M.S.; Ali, U.; Yang, Z.; Shahzad, M.; Sameeh, M.Y.; Alruways, M.W.; et al. Functional Annotation of Lactiplantibacillus plantarum 13-3 as a Potential Starter Probiotic Involved in the Food Safety of Fermented Products. Molecules 2022, 27, 5399. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.J.; Kim, S.A.; Yang, S.H.; Kim, D.H.; Cheng, Y.Y.; Kang, J.I.; Lee, S.Y.; Han, N.S. Integrated genome-based assessment of safety and probiotic characteristics of Lactiplantibacillus plantarum PMO 08 isolated from kimchi. PLoS ONE 2022, 17, e0273986. [Google Scholar] [CrossRef] [PubMed]
- Tegopoulos, K.; Stergiou, O.S.; Kiousi, D.E.; Tsifintaris, M.; Koletsou, E.; Papageorgiou, A.C.; Argyri, A.A.; Chorianopoulos, N.; Galanis, A.; Kolovos, P. Genomic and Phylogenetic Analysis of Lactiplantibacillus plantarum L125, and Evaluation of Its Anti-Proliferative and Cytotoxic Activity in Cancer Cells. Biomedicines 2021, 9, 1718. [Google Scholar] [CrossRef] [PubMed]
- Das, D.J.; Shankar, A.; Johnson, J.B.; Thomas, S. Critical insights into antibiotic resistance transferability in probiotic Lactobacillus. Nutrition 2020, 69, 110567. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Chae, S.A.; Bang, W.Y.; Lee, M.; Ban, O.H.; Kim, S.J.; Jung, Y.H.; Yang, J. Anti-inflammatory potential of Lactiplantibacillus plantarum IDCC 3501 and its safety evaluation. Braz. J. Microbiol. 2021, 52, 2299–2306. [Google Scholar] [CrossRef]
- Lee, B.S.; Ban, O.H.; Bang, W.Y.; Chae, S.A.; Oh, S.; Park, C.; Lee, M.; Kim, S.J.; Yang, J.; Jung, Y.H. Safety assessment of Lactobacillus reuteri IDCC 3701 based on phenotypic and genomic analysis. Ann. Microbiol. 2021, 71, 10. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.S.; Kim, Y.; Jeong, Y.; Kim, J.E.; Paek, N.S.; Kang, C.H. Antioxidant and Probiotic Properties of Lactobacilli and Bifidobacteria of Human Origins. Biotechnol. Bioprocess Eng. 2020, 25, 421–430. [Google Scholar] [CrossRef]
- Florez, A.B.; Egervarn, M.; Danielsen, M.; Tosi, L.; Morelli, L.; Lindgren, S.; Mayo, B. Susceptibility of Lactobacillus plantarum strains to six antibiotics and definition of new susceptibility-resistance cutoff values. Microb. Drug Resist.-Mech. Epidemiol. Dis. 2006, 12, 252–256. [Google Scholar] [CrossRef]
- Ministry of Agriculture and Rural Affairs of the People’s Republic of China. Guide for Identification and Safety Evaluation of Strains Produced by Direct Feeding Microorganisms and Fermented Products; Documents of the General Office of the Ministry of Agriculture and Village; General Office of the Ministry of Agriculture and Village: Beijing, China, 2021.
- Nakajima, Y.; Ishibashi, J.; Yukuhiro, F.; Asaoka, A.; Taylor, D.; Yamakawa, M. Antibacterial activity and mechanism of action of tick defensin against Gram-positive bacteria. Biochim. Et Biophys. Acta-Gen. Subj. 2003, 1624, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Beecher, D.J.; Schoeni, J.L.; Wong, A.C.L. Enterotoxic activity of hemolysin bl from bacillus-cereus. Infect. Immun. 1995, 63, 4423–4428. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, A.R. Significance of biogenic amines to food safety and human health. Food Res. Int. 1996, 29, 675–690. [Google Scholar] [CrossRef]
- Gao, X.L.; Li, C.; He, R.H.; Zhang, Y.Q.; Wang, B.; Zhang, Z.H.; Ho, C.T. Research advances on biogenic amines in traditional fermented foods: Emphasis on formation mechanism, detection and control methods. Food Chem. 2023, 405, 134911. [Google Scholar] [CrossRef]
- del Pulgar, E.M.G.; Benitez-Paez, A.; Sanz, Y. Safety Assessment of Bacteroides Uniformis CECT 7771, a Symbiont of the Gut Microbiota in Infants. Nutrients 2020, 12, 551. [Google Scholar] [CrossRef] [PubMed]
- Sellers, R.S.; Morton, D.; Michael, B.; Roome, N.; Johnson, J.K.; Yano, B.L.; Perry, R.; Schafer, K. Society of toxicologic pathology position paper: Organ weight recommendations for toxicology studies. Toxicol. Pathol. 2007, 35, 751–755. [Google Scholar] [CrossRef]
- Kim, J.; Han, M.; Jeon, W.K. Acute and Subacute Oral Toxicity of Mumefural, Bioactive Compound Derived from Processed Fruit of Prunus mume Sieb. et Zucc., in ICR Mice. Nutrients 2020, 12, 1328. [Google Scholar] [CrossRef]
- Zhang, C.; Li, C.X.; Shao, Q.; Chen, W.B.; Ma, L.; Xu, W.H.; Li, Y.X.; Huang, S.C.; Ma, Y.B. Effects of Glycyrrhiza polysaccharide in diet on growth performance, serum antioxidant capacity, and biochemistry of broilers. Poult. Sci. 2021, 100, 100927. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.D.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Cuccuru, G.; Orsini, M.; Pinna, A.; Sbardellati, A.; Soranzo, N.; Travaglione, A.; Uva, P.; Zanetti, G.; Fotia, G. Orione, a web-based framework for NGS analysis in microbiology. Bioinformatics 2014, 30, 1928–1929. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Chilton, J.M.; Grüning, B.; Johnson, J.E.; Soranzo, N. NCBI BLAST plus integrated into Galaxy. Gigascience 2015, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.J.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Auke, J.; Anne; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.D.; Zhou, S.Y.; Chen, L.H.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Saboktakin-Rizi, M.; Alizadeh Behbahani, B.; Hojjati, M.; Noshad, M. Identification of Lactobacillus plantarum TW29-1 isolated from Iranian fermented cereal-dairy product (Yellow Zabol Kashk): Probiotic characteristics, antimicrobial activity and safety evaluation. J. Food Meas. Charact. 2021, 15, 2615–2624. [Google Scholar] [CrossRef]
- Kim, H.; Lee, Y.S.; Yu, H.Y.; Kwon, M.; Kim, K.K.; In, G.; Hong, S.K.; Kim, S.K. Anti-Inflammatory Effects of Limosilactobacillus fermentum KGC1601 Isolated from Panax ginseng and Its Probiotic Characteristics. Foods 2022, 11, 1707. [Google Scholar] [CrossRef]
- Mu, G.; Zhang, Z.; Wang, J.; Jiang, S.; Wang, H.; Xu, Y.; Li, X.; Chi, L.; Li, Y.; Tuo, Y.; et al. Antigenicity and Safety Evaluation of Lactiplantibacillus plantarum 7-2 Screened to Reduce alpha-Casein Antigen. Foods 2021, 11, 88. [Google Scholar] [CrossRef]
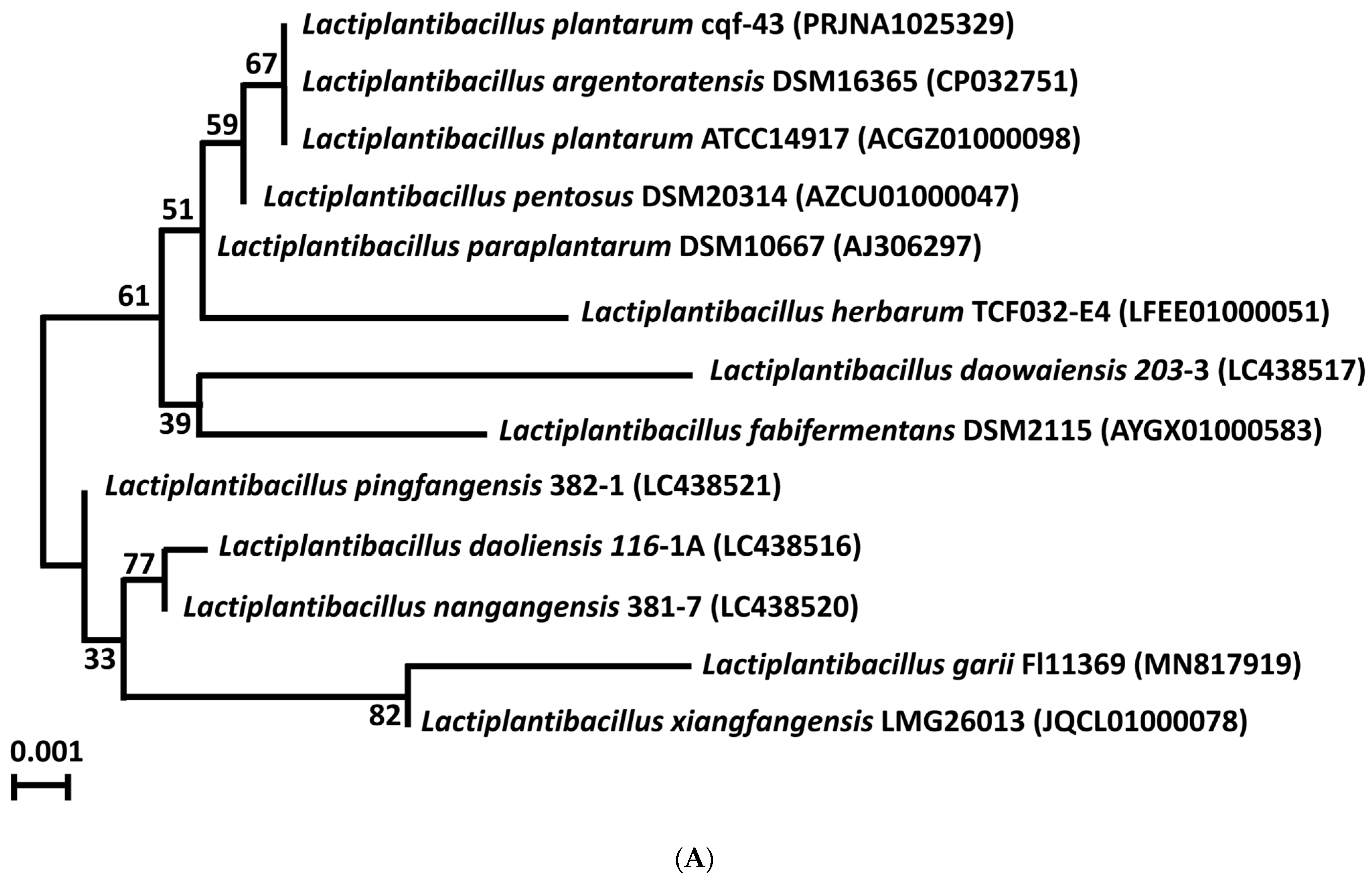





| Features | DSM 16365 (T) | ATCC14917 (T) | cqf-43 |
|---|---|---|---|
| Genome size (bp) | 3,350,338 | 3,198,761 | 3,169,201 |
| GC content (%) | 44.97 | 44.48 | 44.59 |
| No. of counts | 6 | 36 | 5 |
| Number of genes | 2814 | 3061 | 3141 |
| Protein coding sequences | 3191 | 3013 | 2990 |
| tRNA | 84 | 61 | 71 |
| rRNA | 16 | 2 | 16 |
| Proteins with function prediction | 2814 | 2779 | 2973 |
| Item | Count | Percentage |
|---|---|---|
| All | 3141 | 100% |
| Annotation | 2973 | 94.65% |
| Uniprot | 1788 | 56.92% |
| Pfam | 2573 | 81.92% |
| RefSeq | 2950 | 93.92% |
| NR | 2743 | 87.33% |
| Tigrfam | 1749 | 55.68% |
| GO | 1712 | 54.50% |
| KEGG | 908 | 28.91% |
| KEGG Pathway | 868 | 27.63% |
| COG | 1449 | 46.13% |
| Type | Bacteria Name | Zone Diameter 1 (mm) |
|---|---|---|
| ATCC35666 | Streptococcus dysgalactiae | 25.67 ± 0.58 b |
| ATCC25923 | Staphylococcus aureus | 22.67 ± 0.58 c |
| ATCC25922 | Escherichia coli | 23.00 ± 0.50 c |
| CVCC3764 | Listeria monocytogenes | 22.50 ± 0.50 c |
| ATCC27853 | Pseudomonas aeruginosa | 24.67 ± 0.58 b |
| CGMCC1.1869 | Shigella dysenteriae | 25.33 ± 0.58 b |
| CVCC4101 | Bacillus cereus | 19.67 ± 0.58 d |
| CVCC534 | Salmonella | 27.67 ± 0.58 a |
| Gene Name | Antibiotic Resistance Ontology Category | Target Antibiotics | Identify |
|---|---|---|---|
| poxtA | Antibiotic inactivation enzyme | Carbapenem, aminoglycoside, phenicol antibiotic, lincosamide, and streptogramin | 100% |
| Antibiotics | MIC 1 (µg/mL) | Cut-Off Value 2 (µg/mL) |
|---|---|---|
| Ampicillin | 1 | 2 |
| Vancomycin | >128 | nr |
| Gentamicin | 32 | 16 |
| Kanamycin | >128 | 64 |
| Erythromycin | 0.25 | 1 |
| Clindamycin | 0.25 | 4 |
| Tetracycline | 8 | 32 |
| Chloramphenicol | 1 | 8 |
| Item | MH | MM | ML | MC | FH | FM | FL | FC |
|---|---|---|---|---|---|---|---|---|
| Hematological parameters | ||||||||
| Red blood cell (RBC, 1012/L) | 8.45 ± 0.63 | 8.29 ± 0.79 | 6.87 ± 2.51 | 8.13 ± 1.28 | 8.17 ± 0.53 | 7.48 ± 1.68 | 7.03 ± 1.08 | 7.57 ± 0.97 |
| Hemoglobin concentration (HGB, g/L) | 137.33 ± 8.31 | 136.33 ± 14.65 | 110.00 ± 43.49 | 128.33 ± 22.64 | 139.50 ± 8.50 | 126.50 ± 30.65 | 123.67 ± 20.7 | 124.67 ± 21.01 |
| Hematocrit (HCT, %) | 38.17 ± 1.64 | 37.18 ± 3.60 | 31.17 ± 11.82 | 35.55 ± 5.67 | 37.57 ± 1.77 | 33.82 ± 7.46 | 32.98 ± 5.20 | 34.05 ± 4.81 |
| Mean erythrocyte hemoglobin concentration (MCHC, g/L) | 359.83 ± 11.55 ab | 366.00 ± 6.16 a | 351.50 ± 15.37 b | 360.33 ± 9.18 ab | 371.17 ± 6.05 | 372.67 ± 9.83 | 373.83 ± 7.47 | 365.50 ± 21.82 |
| Coefficient of variation of erythrocyte distribution width (RDW-CV, %) | 15.92 ± 1.56 a | 14.87 ± 0.35 ab | 14.5 ± 0.73 b | 15.05 ± 0.5 ab | 15.17 ± 0.57 | 15.10 ± 0.42 | 15.13 ± 0.27 | 14.98 ± 0.56 |
| Standard deviation of red blood cell distribution width (RDW-SD, fL) | 28.73 ± 4.52 | 26.60 ± 1.00 | 26.40 ± 1.67 | 26.27 ± 0.73 | 27.90 ± 2.24 | 27.43 ± 1.36 | 28.72 ± 0.74 | 27.08 ± 1.35 |
| Serum biochemical indicators | ||||||||
| Alanine aminotransferase (ALT, U/L) | 31.80 ± 6.26 | 28.80 ± 5.89 | 41.20 ± 13.44 | 36.00 ± 10.79 | 38.00 ± 5.70 | 36.60 ± 10.64 | 38.40 ± 5.37 | 51.00 ± 19.65 |
| Aspartate aminotransferase (AST, U/L) | 92.40 ± 8.85 | 98.40 ± 16.13 | 88.60 ± 12.28 | 113.60 ± 37.53 | 124.00 ± 43.69 | 97.20 ± 18.91 | 98.40 ± 12.62 | 105.80 ± 10.45 |
| Creatinine (CREA, umol/L) | 35.80 ± 8.04 b | 42.2 ± 18.05 b | 46.20 ± 14.65 b | 66.60 ± 14.57a | 44.80 ± 14.43 | 59.80 ± 21.82 | 42.80 ± 17.14 | 46.60 ± 26.36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Zhong, X.; Liu, Z.; Guan, X.; Wang, Q.; Qi, R.; Zhou, X.; Huang, J. Probiotic Potential and Safety Assessment of Lactiplantibacillus plantarum cqf-43 and Whole-Genome Sequence Analysis. Int. J. Mol. Sci. 2023, 24, 17570. https://doi.org/10.3390/ijms242417570
Liu B, Zhong X, Liu Z, Guan X, Wang Q, Qi R, Zhou X, Huang J. Probiotic Potential and Safety Assessment of Lactiplantibacillus plantarum cqf-43 and Whole-Genome Sequence Analysis. International Journal of Molecular Sciences. 2023; 24(24):17570. https://doi.org/10.3390/ijms242417570
Chicago/Turabian StyleLiu, Baiheng, Xiaoxia Zhong, Zhiyun Liu, Xiaofeng Guan, Qi Wang, Renli Qi, Xiaorong Zhou, and Jinxiu Huang. 2023. "Probiotic Potential and Safety Assessment of Lactiplantibacillus plantarum cqf-43 and Whole-Genome Sequence Analysis" International Journal of Molecular Sciences 24, no. 24: 17570. https://doi.org/10.3390/ijms242417570
APA StyleLiu, B., Zhong, X., Liu, Z., Guan, X., Wang, Q., Qi, R., Zhou, X., & Huang, J. (2023). Probiotic Potential and Safety Assessment of Lactiplantibacillus plantarum cqf-43 and Whole-Genome Sequence Analysis. International Journal of Molecular Sciences, 24(24), 17570. https://doi.org/10.3390/ijms242417570






