The Role of Transcription Factors in the Loss of Inter-Chromosomal Co-Expression for Breast Cancer Subtypes
Abstract
1. Introduction
1.1. Breast Cancer Heterogeneity and Molecular Subtypes
1.2. Gene Co-Expression Networks
1.3. Loss of Long-Range Co-Expression in Cancer
1.4. Transcription Factors Influence in GCNs
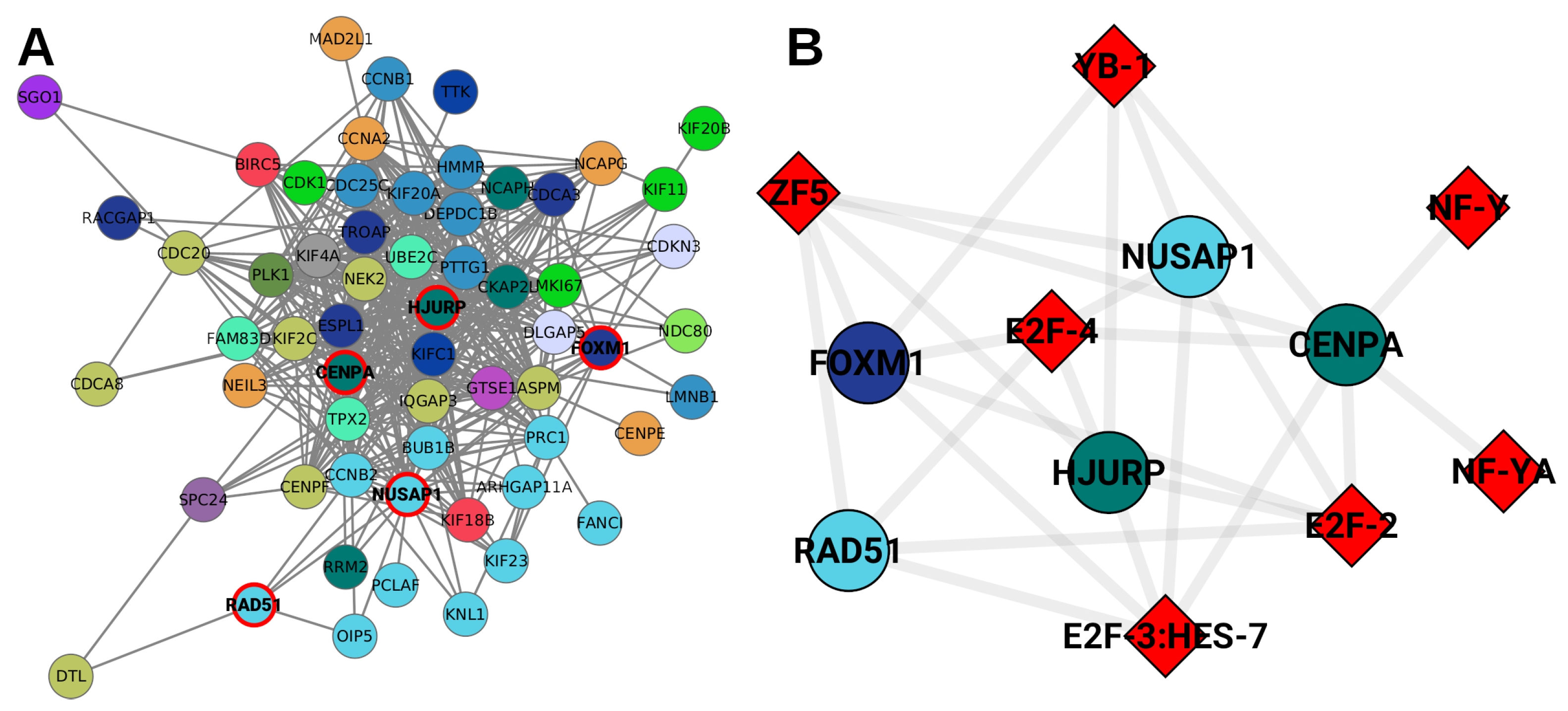
2. Results
2.1. Gene Co-Expression Networks
2.2. TF-Target Network Topology Is Similar between Cancer Subtypes
2.3. TFs Are Strongly Shared between Cancer Subtypes
2.4. Gene Targets for the Cancer Subtypes Share Regulation Patterns with the Non-Cancer GRN
2.5. Transcriptional Targets in Cancer Have More Regulators than in Control
2.6. Unique TFs for Each Subtype Regulate Chromosome-Specific Target Genes in Cancer
- All unique TFs in cancer regulate genes from the same chromosome, except NF-Y and YB-1 for Luminal A GRN.
- In the control GRN, the TFs regulate genes from any chromosome.
3. Discussion
4. Materials and Methods
4.1. Gene Expression Data
4.2. Co-Expression Network Inference and Network Modularity Calculations
4.3. Transcription Factor Binding Motif Analysis and TF-Target Network Construction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sorlie, T.; Perou, C.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Raj-Kumar, P.K.; Liu, J.; Hooke, J.A.; Kovatich, A.J.; Kvecher, L.; Shriver, C.D.; Hu, H. PCA-PAM50 improves consistency between breast cancer intrinsic and clinical subtyping reclassifying a subset of luminal A tumors as luminal B. Sci. Rep. 2019, 9, 7956. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Jiang, G.; Zhang, S.; Yazdanparast, A.; Li, M.; Pawar, A.V.; Liu, Y.; Inavolu, S.M.; Cheng, L. Comprehensive comparison of molecular portraits between cell lines and tumors in breast cancer. BMC Genom. 2016, 17, 281–301. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Burstein, M.D.; Tsimelzon, A.; Poage, G.M.; Covington, K.R.; Contreras, A.; Fuqua, S.A.; Savage, M.I.; Osborne, C.K.; Hilsenbeck, S.G.; Chang, J.C.; et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015, 21, 1688–1698. [Google Scholar] [CrossRef]
- Prat, A.; Pineda, E.; Adamo, B.; Galván, P.; Fernández, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Muñoz, M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015, 24, S26–S35. [Google Scholar] [CrossRef]
- Dai, X.; Xiang, L.; Li, T.; Bai, Z. Cancer hallmarks, biomarkers and breast cancer molecular subtypes. J. Cancer 2016, 7, 1281. [Google Scholar] [CrossRef]
- Garcia-Cortes, D.; Hernandez-Lemus, E.; Espinal-Enriquez, J. Loss of long-range co-expression is a common trait in cancer. bioRxiv 2022. [Google Scholar] [CrossRef]
- Ovens, K.; Eames, B.F.; McQuillan, I. Comparative analyses of gene co-expression networks: Implementations and applications in the study of evolution. Front. Genet. 2021, 12, 695399. [Google Scholar] [CrossRef]
- Serin, E.A.; Nijveen, H.; Hilhorst, H.W.; Ligterink, W. Learning from co-expression networks: Possibilities and challenges. Front. Plant Sci. 2016, 7, 444. [Google Scholar] [CrossRef] [PubMed]
- Stuart, J.M.; Segal, E.; Koller, D.; Kim, S.K. A gene-coexpression network for global discovery of conserved genetic modules. Science 2003, 302, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Sonawane, A.R.; Weiss, S.T.; Glass, K.; Sharma, A. Network medicine in the age of biomedical big data. Front. Genet. 2019, 10, 294. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Han, L.; Yuan, Y.; Li, J.; Hei, N.; Liang, H. Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types. Nat. Commun. 2014, 5, 3231. [Google Scholar] [CrossRef]
- Tang, J.; Kong, D.; Cui, Q.; Wang, K.; Zhang, D.; Gong, Y.; Wu, G. Prognostic genes of breast cancer identified by gene co-expression network analysis. Front. Oncol. 2018, 8, 374. [Google Scholar] [CrossRef]
- Yin, W.; Mendoza, L.; Monzon-Sandoval, J.; Urrutia, A.O.; Gutierrez, H. Emergence of co-expression in gene regulatory networks. PLoS ONE 2021, 16, e0247671. [Google Scholar] [CrossRef]
- Mao, L.; Van Hemert, J.L.; Dash, S.; Dickerson, J.A. Arabidopsis gene co-expression network and its functional modules. BMC Bioinform. 2009, 10, 346. [Google Scholar] [CrossRef]
- Liu, X.; Hu, A.X.; Zhao, J.L.; Chen, F.L. Identification of key gene modules in human osteosarcoma by co-expression analysis weighted gene co-expression network analysis (WGCNA). J. Cell. Biochem. 2017, 118, 3953–3959. [Google Scholar] [CrossRef]
- Van Dam, S.; Vosa, U.; van der Graaf, A.; Franke, L.; de Magalhaes, J.P. Gene co-expression analysis for functional classification and gene–disease predictions. Brief. Bioinform. 2018, 19, 575–592. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.G.; Huang, X.L.; Liang, S.Y.; Tang, S.M.; Wu, S.K.; Huang, T.T.; Mo, Z.N.; Wang, Q.Y. Identifying miRNA and gene modules of colon cancer associated with pathological stage by weighted gene co-expression network analysis. OncoTargets Ther. 2018, 11, 2815–2830. [Google Scholar] [CrossRef] [PubMed]
- García-Cortés, D.; Hernández-Lemus, E.; Espinal-Enríquez, J. Luminal a breast cancer co-expression network: Structural and functional alterations. Front. Genet. 2021, 12, 629475. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Gómez, C.; Hernández-Lemus, E.; Espinal-Enríquez, J. CNVs in 8q24. 3 do not influence gene co-expression in breast cancer subtypes. Front. Genet. 2023, 14, 1141011. [Google Scholar] [CrossRef] [PubMed]
- Drago-García, D.; Espinal-Enríquez, J.; Hernández-Lemus, E. Network analysis of EMT and MET micro-RNA regulation in breast cancer. Sci. Rep. 2017, 7, 13534. [Google Scholar] [CrossRef]
- Benetatos, L.; Hatzimichael, E.; Londin, E.; Vartholomatos, G.; Loher, P.; Rigoutsos, I.; Briasoulis, E. The microRNAs within the DLK1-DIO3 genomic region: Involvement in disease pathogenesis. Cell. Mol. Life Sci. 2013, 70, 795–814. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Fuentes, J.M.; Hernández-Lemus, E.; Espinal-Enríquez, J. Oncogenic Role of miR-217 During Clear Cell Renal Carcinoma Progression. Front. Oncol. 2022, 12, 934711. [Google Scholar] [CrossRef]
- Zamora-Fuentes, J.M.; Hernández-Lemus, E.; Espinal-Enríquez, J. Methylation-related genes involved in renal carcinoma progression. Front. Genet. 2023, 14, 1225158. [Google Scholar] [CrossRef]
- Rondón-Lagos, M.; Verdun Di Cantogno, L.; Rangel, N.; Mele, T.; Ramírez-Clavijo, S.R.; Scagliotti, G.; Marchiò, C.; Sapino, A. Unraveling the chromosome 17 patterns of FISH in interphase nuclei: An in-depth analysis of the HER2amplicon and chromosome 17 centromere by karyotyping, FISH and M-FISH in breast cancer cells. BMC Cancer 2014, 14, 922. [Google Scholar] [CrossRef]
- Pärssinen, J.; Kuukasjärvi, T.; Karhu, R.; Kallioniemi, A. High-level amplification at 17q23 leads to coordinated overexpression of multiple adjacent genes in breast cancer. Br. J. Cancer 2007, 96, 1258–1264. [Google Scholar] [CrossRef][Green Version]
- Thennavan, A.; Beca, F.; Xia, Y.; Garcia-Recio, S.; Allison, K.; Collins, L.C.; Gary, M.T.; Chen, Y.Y.; Schnitt, S.J.; Hoadley, K.A.; et al. Molecular analysis of TCGA breast cancer histologic types. Cell Genom. 2021, 1, 100067. [Google Scholar] [CrossRef] [PubMed]
- Nueda, M.J.; Ferrer, A.; Conesa, A. ARSyN: A method for the identification and removal of systematic noise in multifactorial time course microarray experiments. Biostatistics 2012, 13, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Pietro, A.D.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef] [PubMed]
- Risso, D.; Schwartz, K.; Sherlock, G.; Dudoit, S. GC-content normalization for RNA-Seq data. BMC Bioinform. 2011, 12, 480. [Google Scholar] [CrossRef]
- Margolin, A.A.; Nemenman, I.; Basso, K.; Wiggins, C.; Stolovitzky, G.; Favera, R.D.; Califano, A. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform. 2006, 7, S7. [Google Scholar] [CrossRef]
- Basso, K.; Margolin, A.A.; Stolovitzky, G.; Klein, U.; Dalla-Favera, R.; Califano, A. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 2005, 37, 382–390. [Google Scholar] [CrossRef]
- Margolin, A.A.; Wang, K.; Lim, W.K.; Kustagi, M.; Nemenman, I.; Califano, A. Reverse engineering cellular networks. Nat. Protoc. 2006, 1, 662–671. [Google Scholar] [CrossRef]
- Blondel, V.D.; Guillaume, J.L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat.Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Matys, V.; Kel-Margoulis, O.V.; Fricke, E.; Liebich, I.; Land, S.; Barre-Dirrie, A.; Reuter, I.; Chekmenev, D.; Krull, M.; Hornischer, K.; et al. TRANSFAC® and its module TRANSCompel®: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006, 34, D108–D110. [Google Scholar] [CrossRef]
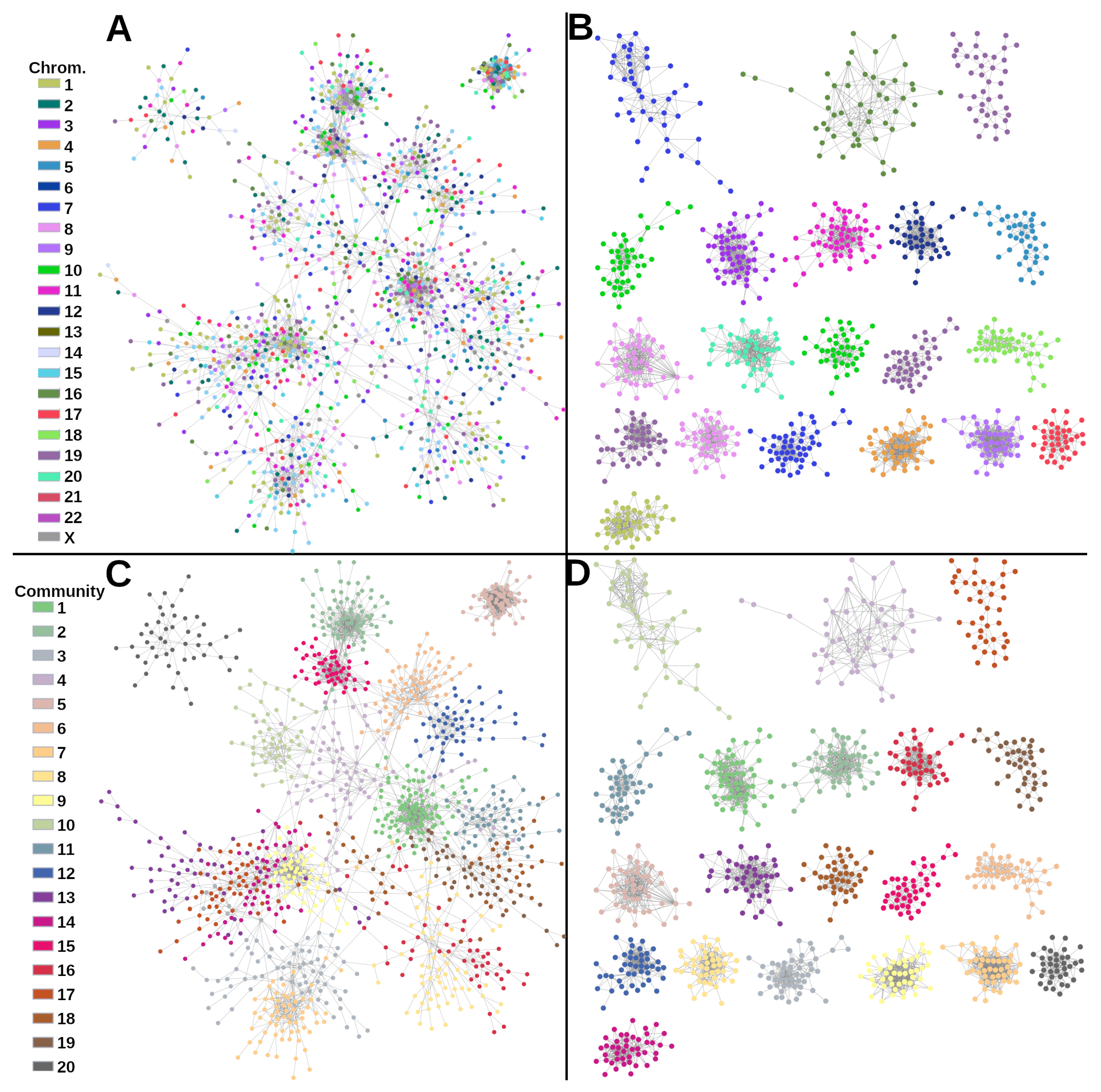

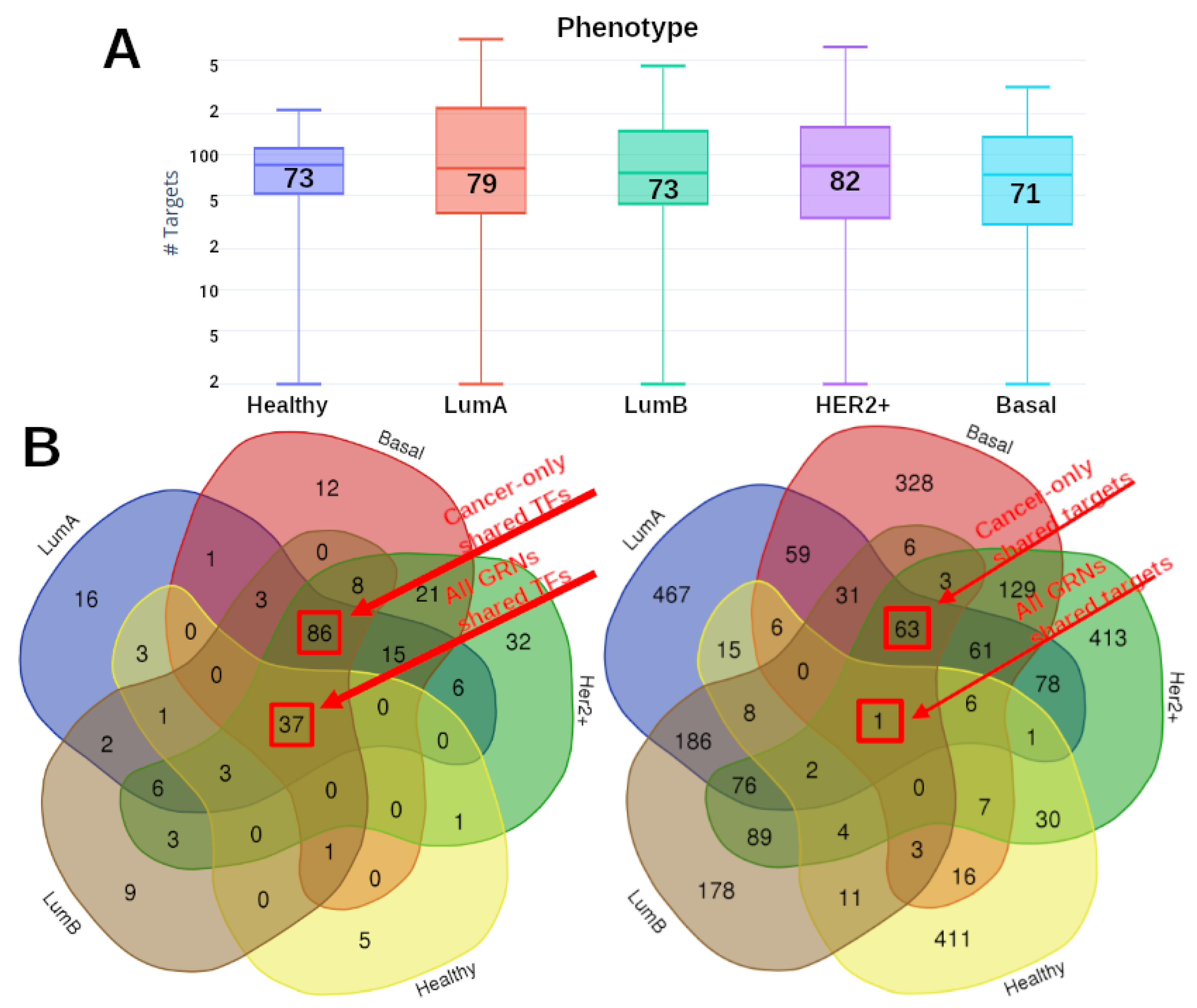
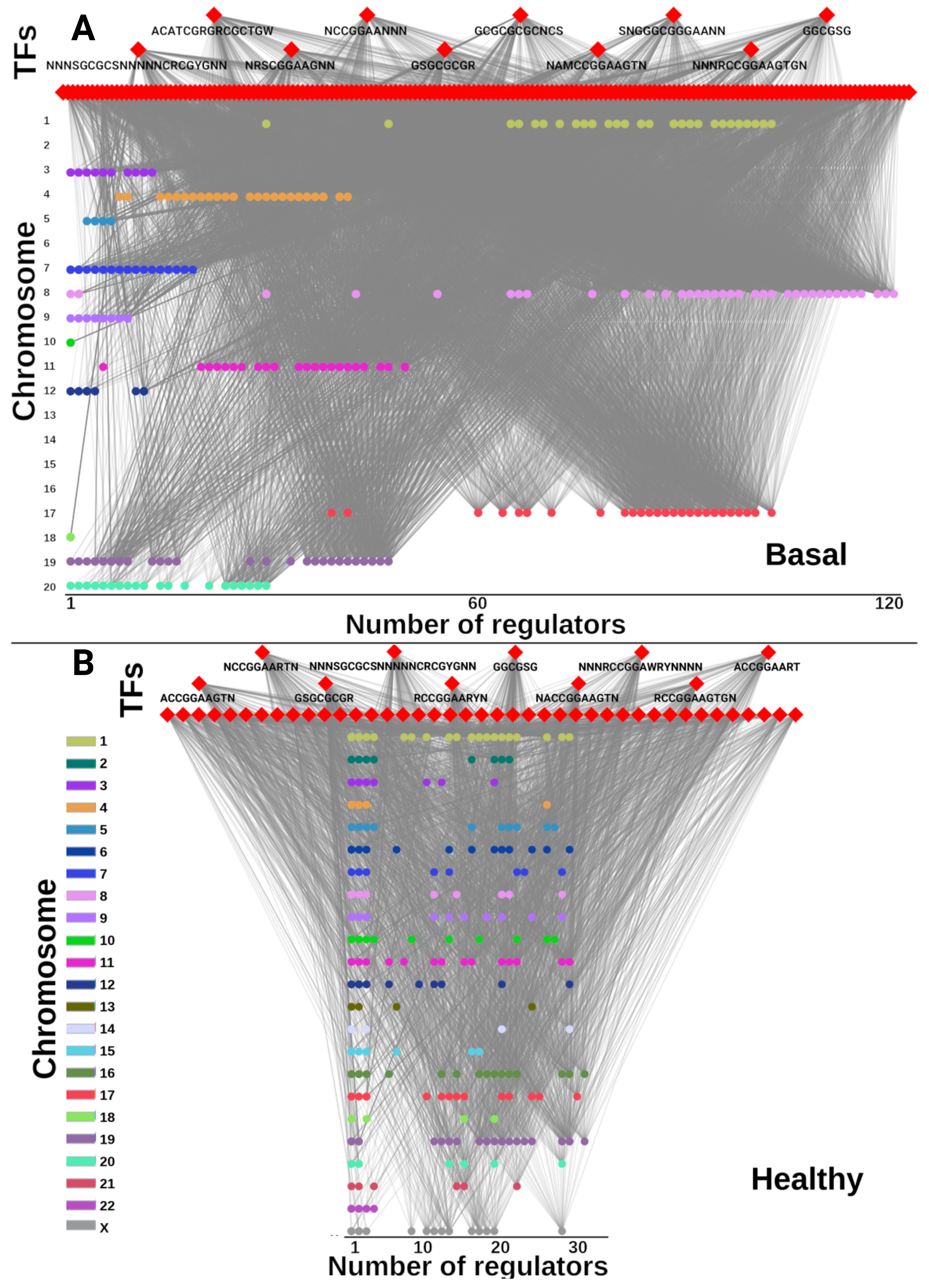
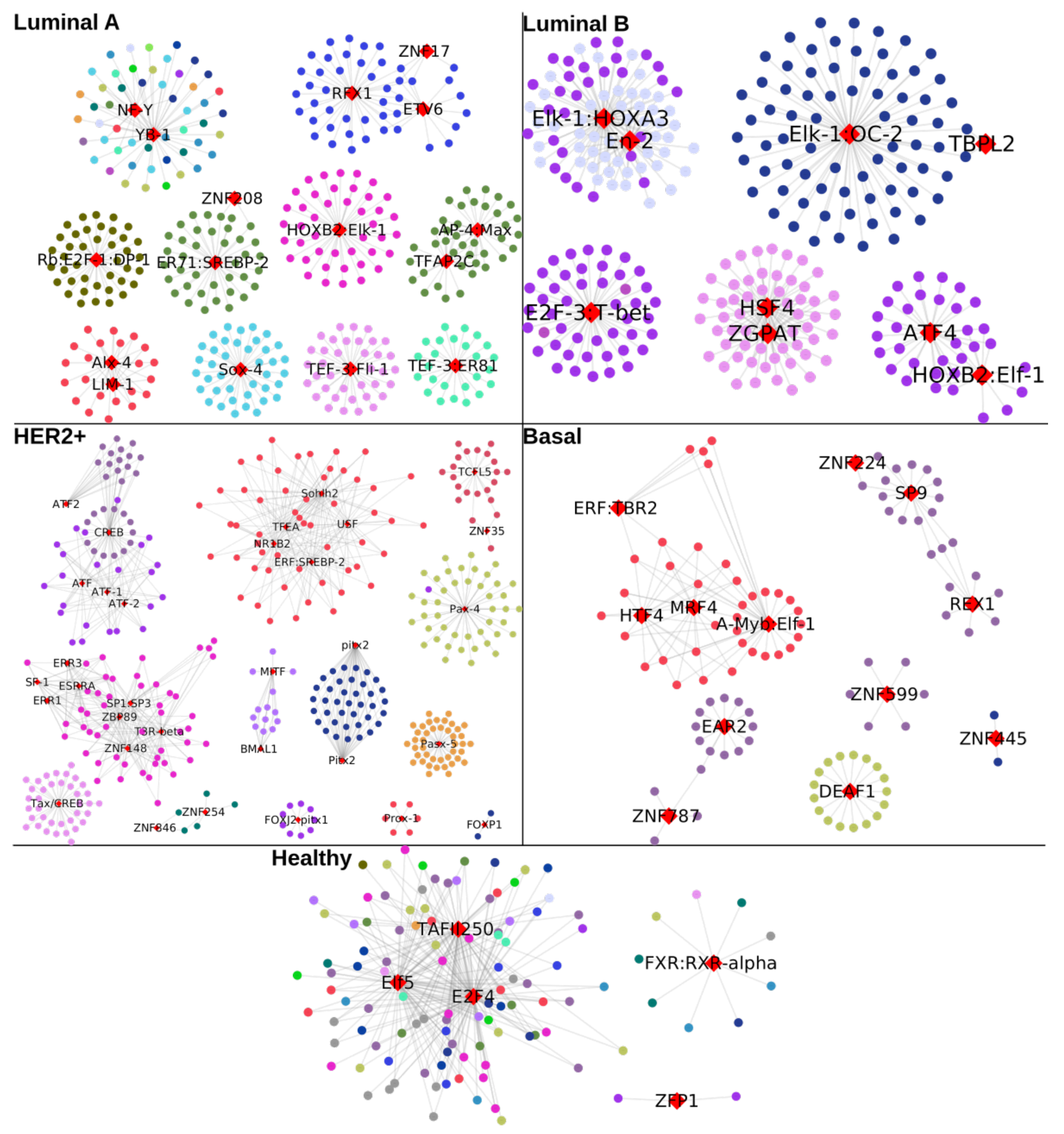
| Phenotype | Nodes | Links | Targets | TFs |
|---|---|---|---|---|
| Healthy | 572 | 6962 | 521 | 51 |
| Luminal A | 1239 | 51,439 | 1060 | 179 |
| Luminal B | 820 | 33,343 | 661 | 159 |
| HER2+ | 1181 | 46,965 | 963 | 218 |
| Basal | 903 | 29,118 | 719 | 184 |
| Phenotype | TF | # Targets | Motif |
|---|---|---|---|
| ZF5 | 214 | GSGCGCGR | |
| E2F | 188 | GGCGSG | |
| ER81 | 158 | RCCGGAARYN | |
| Elk-1 | 155 | RCCGGAAGTGN | |
| Control | E2F-3:HES-7 | 152 | NNNSGCGCSNNNNNCRCGYGNN |
| ELF4 | 145 | NCCGGAARTN | |
| Erg | 142 | ACCGGAAGTN | |
| PEA3 | 134 | NACCGGAAGTN | |
| c-Ets-1 | 132 | NNNRCCGGAWRYNNNN | |
| ERG | 129 | ACCGGAART | |
| ZF5 | 714 | GGSGCGCGS | |
| ER81 | 688 | RCCGGAARYN | |
| Elk-1 | 651 | RACCGGAAGTR | |
| Erg | 650 | NACCGGAARTN | |
| Luminal A | E2F-4 | 626 | NTTTCSCGCC |
| PEA3 | 599 | NACCGGAAGTN | |
| Elf-1 | 593 | NANGCGGAAGTN | |
| Fli-1 | 578 | NACCGGAARTN | |
| ERG | 530 | ACCGGAARTN | |
| E2F-1 | 502 | NGGGCGGGARV | |
| Elk-1 | 453 | NNCCGGAAGTN | |
| Elf-1 | 405 | NNANCCGGAAGTGS | |
| ER81 | 405 | NNCCGGAAGYG | |
| PEA3 | 397 | RCCGGAAGYN | |
| Luminal B | Erg | 347 | NACCGGAARTN |
| ELK-1 | 346 | ACCGGAAGTN | |
| Fli-1 | 344 | NACCGGAARTN | |
| ERG | 339 | ACCGGAARTN | |
| GABP-alpha | 338 | NRCCGGAAGTN | |
| Erm | 333 | NNSCGGAWGYN | |
| ZF5 | 627 | GSGCGCGR | |
| E2F-4 | 566 | SNGGGCGGGAANN | |
| E2F | 535 | GGCGSG | |
| E2F-3:HES-7 | 472 | NNNSGCGCSNNNNNCRCGYGNN | |
| HER2+ | E2F-2 | 464 | GCGCGCGCNCS |
| E2F-1 | 423 | NKTSSCGC | |
| pax-6 | 421 | NYACGCNYSANYGMNCN | |
| Sp1 | 409 | GGGGCGGGGC | |
| Elk-1 | 391 | NRSCGGAAGNN | |
| GABP-alpha | 378 | NNNRCCGGAAGTGN | |
| ZF5 | 316 | GSGCGCGR | |
| E2F-2 | 311 | GCGCGCGCNCS | |
| Elk-1 | 304 | NRSCGGAAGNN | |
| E2F | 299 | GGCGSG | |
| Basal | GABP-alpha | 285 | NNNRCCGGAAGTGN |
| E2F-3:HES-7 | 270 | NNNSGCGCSNNNNNCRCGYGNN | |
| Elf-1 | 265 | NAMCCGGAAGTN | |
| TCF-1 | 258 | ACATCGRGRCGCTGW | |
| E2F-4 | 247 | SNGGGCGGGAANN | |
| ETV7 | 238 | NCCGGAANNN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trujillo-Ortíz, R.; Espinal-Enríquez, J.; Hernández-Lemus, E. The Role of Transcription Factors in the Loss of Inter-Chromosomal Co-Expression for Breast Cancer Subtypes. Int. J. Mol. Sci. 2023, 24, 17564. https://doi.org/10.3390/ijms242417564
Trujillo-Ortíz R, Espinal-Enríquez J, Hernández-Lemus E. The Role of Transcription Factors in the Loss of Inter-Chromosomal Co-Expression for Breast Cancer Subtypes. International Journal of Molecular Sciences. 2023; 24(24):17564. https://doi.org/10.3390/ijms242417564
Chicago/Turabian StyleTrujillo-Ortíz, Rodrigo, Jesús Espinal-Enríquez, and Enrique Hernández-Lemus. 2023. "The Role of Transcription Factors in the Loss of Inter-Chromosomal Co-Expression for Breast Cancer Subtypes" International Journal of Molecular Sciences 24, no. 24: 17564. https://doi.org/10.3390/ijms242417564
APA StyleTrujillo-Ortíz, R., Espinal-Enríquez, J., & Hernández-Lemus, E. (2023). The Role of Transcription Factors in the Loss of Inter-Chromosomal Co-Expression for Breast Cancer Subtypes. International Journal of Molecular Sciences, 24(24), 17564. https://doi.org/10.3390/ijms242417564







