MiR-221-3p/222-3p Cluster Expression in Human Adipose Tissue Is Related to Obesity and Type 2 Diabetes
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
2.2. Human miR-221-3p and miR-222-3p Expression Profiles and Their Association with Obesity, T2D, and AT Depot
2.3. Human miR-221-3p and miR-222-3p Expression Profiles Were Analyzed in Relation to Obesity, T2D, and AT Depot, with Consideration of Sex-Based Differences
2.4. Mouse miR-221-3p and miR-222-3p Expression Profiles and Their Association with Obesity, T2D, and AT Depot
2.5. In Silico Analysis of miRNA-221-3p/222-3p Target Genes
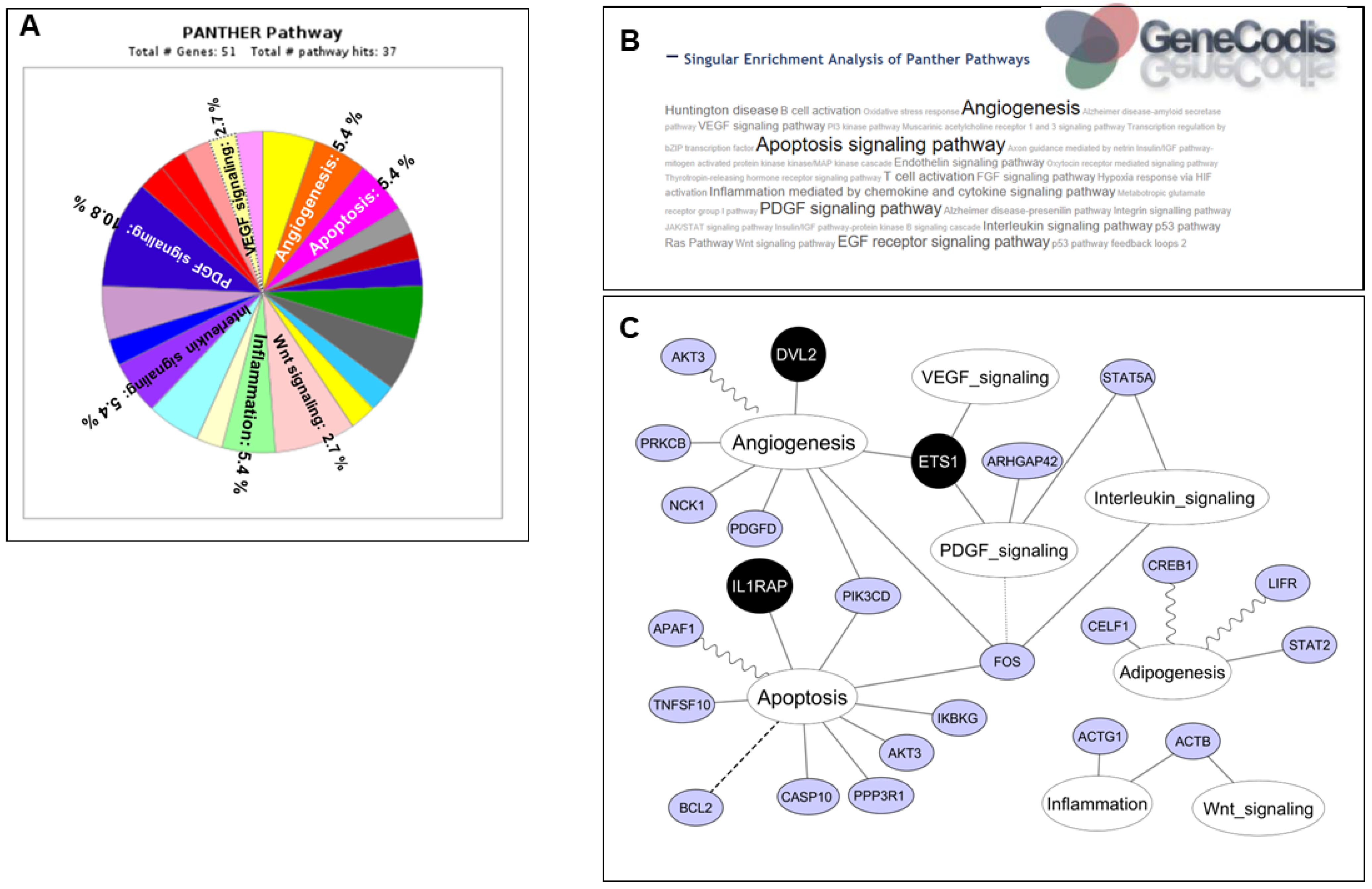
2.6. Annotation Enrichment Analysis of miR-221-3p/222-3p Target Genes
2.7. Identification of miRNA Binding Sites within Target Genes
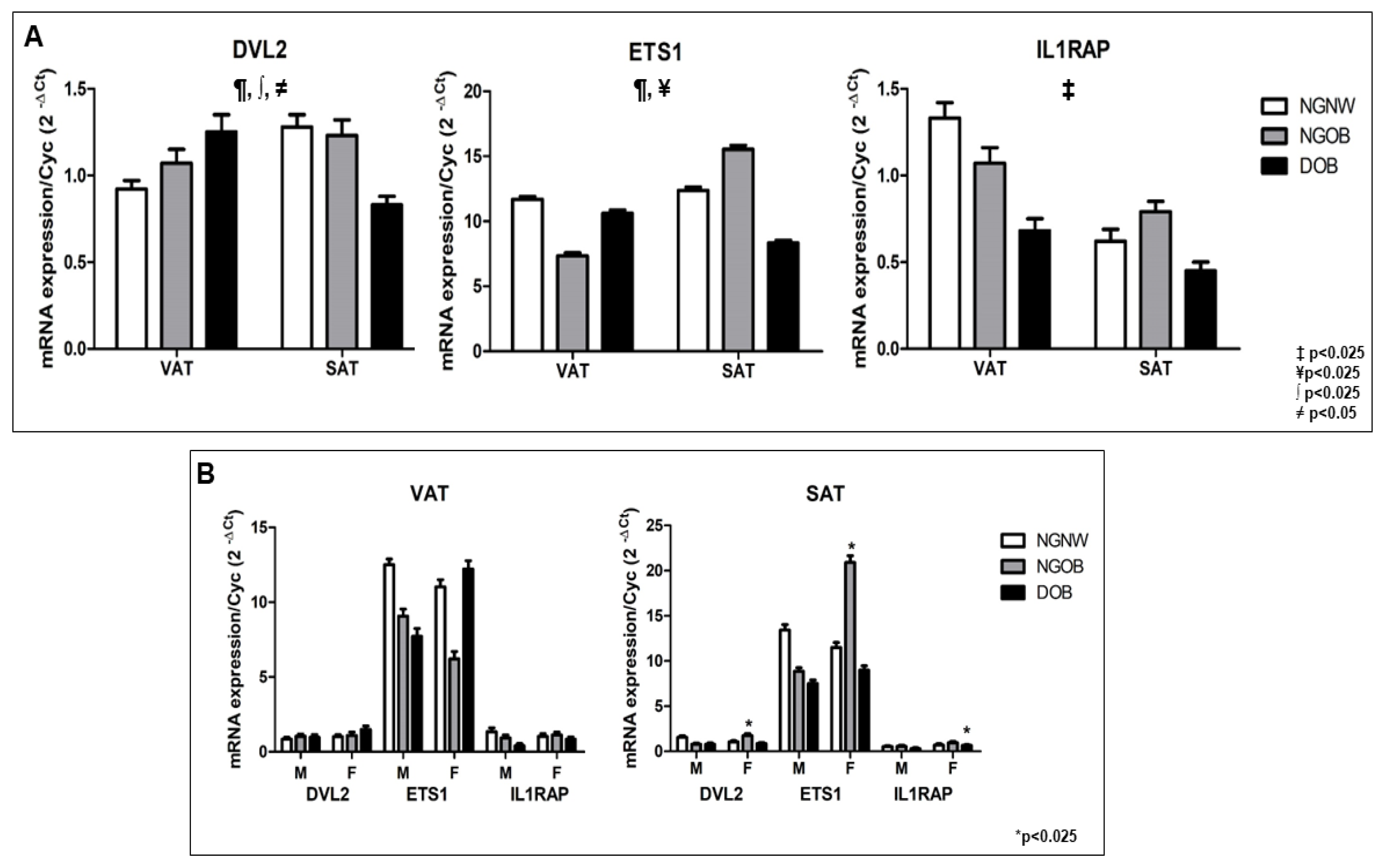
2.8. Gene Expression of ETS1, DVL2, and IL1RAP in Human VAT and SAT: Impact of Obesity, Glycemic Status, and Sex
3. Discussion
4. Materials and Methods
4.1. Patients, Study Design, and AT Collection
4.2. Laboratory Analysis
4.3. Generation of Diet-Induced Obese and Diabetic Mice
4.4. MiRNA Extraction and Real-Time Quantitative PCR (qPCR)
4.5. mRNA Isolation and qPCR
4.6. Bioinformatics Analysis
4.7. Statistical Analysis
5. Conclusions
6. Study Limitation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chooi, Y.C.; Ding, C.; Magkos, F. The epidemiology of obesity. Metabolism 2019, 92, 6–10. [Google Scholar] [CrossRef]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, M. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef]
- Ferrara, D.; Montecucco, F.; Dallegri, F.; Carbone, F. Impact of different ectopic fat depots on cardiovascular and metabolic diseases. J. Cell Physiol. 2019, 234, 21630–21641. [Google Scholar] [CrossRef]
- Barchetta, I.; Cimini, F.A.; Ciccarelli, G.; Baroni, M.G.; Cavallo, M.G. Sick fat: The good and the bad of old and new circulating markers of adipose tissue inflammation. J. Endocrinol. Investig. 2019, 42, 1257–1272. [Google Scholar] [CrossRef]
- Smith, U. Abdominal obesity: A marker of ectopic fat accumulation. J. Clin. Investig. 2015, 125, 1790–1792. [Google Scholar] [CrossRef]
- Blüher, M. Adipose tissue dysfunction in obesity. Exp. Clin. Endocrinol. Diabetes 2009, 117, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Oliva-Olivera, W.; Coín-Aragüez, L.; Lhamyani, S.; Clemente-Postigo, M.; Torres, J.A.; Bernal-López, M.R.; El Bekay, R.; Tinahones, F.J. Adipogenic Impairment of Adipose Tissue-Derived Mesenchymal Stem Cells in Subjects with Metabolic Syndrome: Possible Protective Role of FGF2. J. Clin. Endocrinol. Metab. 2017, 102, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Manna, P.; Jain, S.K. Obesity, Oxidative Stress, Adipose Tissue Dysfunction, and the Associated Health Risks: Causes and Therapeutic Strategies. Metab. Syndr. Relat. Disord. 2015, 13, 423–444. [Google Scholar] [CrossRef] [PubMed]
- Jung, U.J.; Choi, M.S. Obesity and its metabolic complications: The role of adipokines and the relationship 568 between obesity, inflammation, insulin resistance, dyslipidemia and nonalcoholic fatty liver disease. Int. J. Mol. Sci. 2014, 15, 6184–6223. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y. Angiogenesis modulates adipogenesis and obesity. J. Clin. Investig. 2007, 117, 2362–2368. [Google Scholar] [CrossRef]
- Crewe, C.; An, Y.A.; Scherer, P.E. The ominous triad of adipose tissue dysfunction: Inflammation, fibrosis, and impaired angiogenesis. J. Clin. Investig. 2017, 127, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, A.B.; Massihnia, D.; Grisafi, F.; Castiglia, M.; Toia, F.; Montesano, L.; Russo, A.; Moschella, F.; Cordova, A. Adipose tissue, angiogenesis and angio-MIR under physiological and pathological conditions. Eur. J. Cell Biol. 2019, 98, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Bråkenhielm, E.; Cao, R.; Gao, B.; Angelin, B.; Cannon, B.; Parini, P.; Cao, Y. Angiogenesis inhibitor, TNP-470, prevents diet-induced and genetic obesity in mice. Circ. Res. 2004, 94, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Neels, J.G.; Thinnes, T.; Loskutoff, D.J. Angiogenesis in an in vivo model of adipose tissue development. FASEB J. 2004, 18, 983–985. [Google Scholar] [CrossRef]
- Rupnick, M.A.; Panigrahy, D.; Zhang, C.Y.; Dallabrida, S.M.; Lowell, B.B.; Langer, R.; Folkman, M.J. Adipose tissue mass can be regulated through the vasculature. Proc. Natl. Acad. Sci. USA 2002, 99, 10730–10735. [Google Scholar] [CrossRef] [PubMed]
- Arner, P.; Kulyté, A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat. Rev. Endocrinol. 2015, 11, 276–288. [Google Scholar] [CrossRef]
- Ortega, F.J.; Moreno-Navarrete, J.M.; Pardo, G.; Monica Sabater, M.; Hummel, M.; Ferrer, A.; Rodriguez-Hermosa, J.I.; Ruiz, B.; Ricart, W.; Peral, B.; et al. miRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS ONE 2010, 5, e9022. [Google Scholar] [CrossRef] [PubMed]
- Gentile, A.M.; Lhamyani, S.; Coín-Aragüez, L.; Clemente-Postigo, M.; Oliva Olivera, W.; Romero-Zerbo, S.Y.; García-Serrano, S.; García-Escobar, E.; Zayed, H.; Doblado, E.; et al. miR-20b, miR-296, and Let-7f Expression in Human Adipose Tissue is Related to Obesity and Type 2 Diabetes. Obesity 2019, 27, 245–254. [Google Scholar] [CrossRef]
- Lhamyani, S.; Gentile, A.M.; Giráldez-Pérez, R.M.; Feijóo-Cuaresma, M.; Romero-Zerbo, S.Y.; Clemente-Postigo, M.; Zayed, H.; Olivera, W.O.; Bermúdez-Silva, F.J.; Salas, J.; et al. miR-21 mimic blocks obesity in mice: A novel therapeutic option. Mol. Ther. Nucleic Acids 2021, 26, 401–416. [Google Scholar] [CrossRef]
- Deiuliis, J.A. MicroRNAs as regulators of metabolic disease: Pathophysiologic significance and emerging role as biomarkers and therapeutics. Int. J. Obes. 2016, 40, 88–101. [Google Scholar] [CrossRef]
- Chartoumpekis, D.V.; Zaravinos, A.; Ziros, P.G.; Iskrenova, R.P.; Psyrogiannis, A.I.; Kyriazopoulou, V.E.; Habeos, I.G. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS ONE 2012, 7, e34872. [Google Scholar] [CrossRef]
- Wang, Y.T.; Tsai, P.C.; Liao, Y.C.; Hsu, C.Y.; Juo, S.H. Circulating microRNAs have a sex-specific association with metabolic syndrome. J. Biomed. Sci. 2013, 20, 72. [Google Scholar] [CrossRef]
- Song, J.; Ouyang, Y.; Che, J.; Li, X.; Zhao, Y.; Yang, K.; Zhao, X.; Chen, Y.; Fan, C.; Yuan, W. Potential Value of miR-221/222 as Diagnostic, Prognostic, and Therapeutic Biomarkers for Diseases. Front. Immunol. 2017, 8, 56. [Google Scholar] [CrossRef]
- Vivacqua, A.; De Marco, P.; Belfiore, A.; Maggiolini, M. Recent Advances on the Role of microRNAs in both Insulin Resistance and Cancer. Curr. Pharm. Des. 2017, 23, 3658–3666. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Mysore, R.; Ortega, F.J.; Latorre, J.; Ahonen, M.; Savolainen-Peltonen, H.; Fischer-Posovszky, P.; Wabitsch, M.; Olkkonen, V.M.; Fernández-Real, J.M.; Haridas, P.A.N. MicroRNA-221-3p Regulates Angiopoietin-Like 8 (ANGPTL8) Expression in Adipocytes. J. Clin. Endocrinol. Metab. 2017, 102, 4001–4012. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Garcia, J.D.; Briones-Espinoza, M.J.; Vega-Cárdenas, M.; Ruíz-Rodríguez, V.M.; Mendez-Mancilla, A.; Gómez-Otero, A.E.; Vargas Morales, J.M.; García-Hernández, M.H.; Portales-Pérez, D.P. The inflammatory state of adipose tissue is not affected by the anti-inflammatory response of the A2a-adenosine system and miR-221/PTEN. Int. J. Biochem. Cell Biol. 2018, 100, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Sobenin, I.A.; Orekhov, A.N.; Bobryshev, Y.V. Human miR-221/222 in Physiological and Atherosclerotic Vascular Remodeling. Biomed. Res. Int. 2015, 2015, 354517. [Google Scholar] [CrossRef] [PubMed]
- Kehl, T.; Backes, C.; Kern, F.; Fehlmann, T.; Ludwig, N.; Meese, E.; Lenhof, H.P.; Keller, A. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget 2017, 8, 107167–107175. [Google Scholar] [CrossRef]
- Xie, J.; Han, M.; Zhang, M.; Deng, H.; Wu, W. PP5 (PPP5C) is a phosphatase of Dvl2. Sci. Rep. 2018, 8, 2715. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, S.; Wang, X.; Ma, X.; Huang, B.; Chen, H.; Chen, D. ETS1 targets RYBP transcription to inhibit tumor cell proliferation. Biochem. Biophys. Res. Commun. 2019, 509, 810–816. [Google Scholar] [CrossRef]
- Banerjee, M.; Saxena, M. Interleukin-1 (IL-1) family of cytokines: Role in type 2 diabetes. Clin. Chim. Acta 2012, 413, 1163–1170. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, S.; Stansberg, C.; Cunningham, C. The interleukin 1 receptor family. Dev. Comp. Immunol. 2004, 28, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Li, J.; Wang, Y.; Meng, J.; Zheng, G. miR-221 promotes lens epithelial cells apoptosis through interacting with SIRT1 and E2F3. Chem. Biol. Interact. 2019, 306, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.H.; Tian, L.; Wang, Y.F.; Liu, J.S.; Guo, M.F.; Liu, W. Effects of MiR-221-Mediated Wnt/β-Catenin Signaling Pathway on Biological Activity of Childhood Acute Lymphoblastic Leukemia Cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2019, 27, 1367–1373. [Google Scholar] [PubMed]
- Xu, C.H.; Liu, Y.; Xiao, L.M.; Chen, L.K.; Zheng, S.Y.; Zeng, E.M.; Li, D.H.; Li, Y.P. Silencing microRNA-221/222 cluster suppresses glioblastoma angiogenesis by suppressor of cytokine signaling-3-dependent JAK/STAT pathway. J. Cell Physiol. 2019, 234, 22272–22284. [Google Scholar] [CrossRef]
- Wang, L.; Liu, C.; Li, C.; Xue, J.; Zhao, S.; Zhan, P.; Lin, Y.; Zhang, P.; Jiang, A.; Chen, W. Effects of microRNA-221/222 on cell proliferation and apoptosis in prostate cancer cells. Gene 2015, 572, 252–258. [Google Scholar] [CrossRef]
- Jia, Z.; Yang, Y.; Dengyan, Z.; Chunyang, Z.; Donglei, L.; Kai, W.; Song, Z. RAP1B, a DVL2 binding protein, activates Wnt/beta-catenin signaling in esophageal squamous cell carcinoma. Gene 2017, 611, 15–20. [Google Scholar] [CrossRef]
- Jang, S.; Park, J.S.; Jeong, H.S. Neural Differentiation of Human Adipose Tissue-Derived Stem Cells Involves Activation of the Wnt5a/JNK Signalling. Stem Cells Int. 2015, 2015, 178618. [Google Scholar] [CrossRef]
- Barreyro, L.; Will, B.; Bartholdy, B.; Zhou, L.; Todorova, T.I.; Stanley, R.F.; Ben-Neriah, S.; Montagna, C.; Parekh, S.; Pellagatti, A.; et al. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood 2012, 120, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
- Appari, M.; Channon, K.M.; McNeill, E. Metabolic Regulation of Adipose Tissue Macrophage Function in Obesity and Diabetes. Antioxid. Redox. Signal. 2018, 29, 297–312. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; D’ Souza, J.; Chakrabarty, S.; Satyamoorthy, K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1955–1986. [Google Scholar] [CrossRef]
- Karczewska-Kupczewska, M.; Stefanowicz, M.; Matulewicz, N.; Nikołajuk, A.; Strączkowski, M. Wnt Signaling Genes in Adipose Tissue and Skeletal Muscle of Humans with Different Degrees of Insulin Sensitivity. J. Clin. Endocrinol. Metab. 2016, 101, 3079–3087. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Postigo, M.; Roca-Rodriguez, M.; Camargo, A.; Ocaña-Wilhelmi, L.; Cardona, F.; Tinahones, F.J. Lipopolysaccharide and lipopolysaccharide-binding protein levels and their relationship to early metabolic improvement after bariatric surgery. Surg. Obes. Relat. Dis. 2015, 11, 933–939. [Google Scholar] [CrossRef] [PubMed]


| NG-NW (n = 11) | NG-OB (n = 11) | D-OB (n = 11) | |
|---|---|---|---|
| Age (years) | 41.32 ± 4.11 | 46.91 ± 3.55 | 43.91 ± 4.13 |
| Sex (Man/Woman) | 5/6 | 5/6 | 5/6 |
| BMI (kg/m2) | 22.48 ± 0.46 | 37.48 ± 1.39 * | 47.07 ± 2.52 *# |
| Glucose (mg/dL) | 95.73 ± 4.05 | 89.91 ± 3.11 | 172.86 ± 22.73 *# |
| HOMA-IR | 1.18 ± 1.56 | 2.28 ± 0.3 * | 7.90 ± 0.75 *# |
| Triglycerides (mg/dL) | 72.23 ± 6.29 | 144.45 ± 27.39 * | 166.86 ± 20.45 *# |
| Cholesterol (mg/dL) | 183.77 ± 5.15 | 202.05 ± 11.90 | 186.41 ± 14.99 |
| HDL-c (mg/dL) | 62.5 ± 3.83 | 47.82 ± 5.39 * | 41.91 ± 2.65 *# |
| LDL-c (mg/dL) | 111.36 ± 5.53 | 126.7 ± 9.54 | 115 ± 14.56 |
| Validated Genes | Non-Validated Genes | Total | |
|---|---|---|---|
| miRNA | miRTarBase | miRWalk | |
| miR-221 | 467 | 16 | 483 |
| miR-222 | 444 | 13 | 457 |
| TOTAL | 911 | 29 | 940 |
| COMMON | 53 | 14 | 67 |
| ENRICHMENT ANALYSIS | 7 | 17 | 24 |
| SELECTED | 1 | 2 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gentile, A.-M.; Lhamyani, S.; Mengual-Mesa, M.; García-Fuentes, E.; Bermúdez-Silva, F.-J.; Rojo-Martínez, G.; Clemente-Postigo, M.; Rodriguez-Cañete, A.; Olveira, G.; El Bekay, R. MiR-221-3p/222-3p Cluster Expression in Human Adipose Tissue Is Related to Obesity and Type 2 Diabetes. Int. J. Mol. Sci. 2023, 24, 17449. https://doi.org/10.3390/ijms242417449
Gentile A-M, Lhamyani S, Mengual-Mesa M, García-Fuentes E, Bermúdez-Silva F-J, Rojo-Martínez G, Clemente-Postigo M, Rodriguez-Cañete A, Olveira G, El Bekay R. MiR-221-3p/222-3p Cluster Expression in Human Adipose Tissue Is Related to Obesity and Type 2 Diabetes. International Journal of Molecular Sciences. 2023; 24(24):17449. https://doi.org/10.3390/ijms242417449
Chicago/Turabian StyleGentile, Adriana-Mariel, Said Lhamyani, María Mengual-Mesa, Eduardo García-Fuentes, Francisco-Javier Bermúdez-Silva, Gemma Rojo-Martínez, Mercedes Clemente-Postigo, Alberto Rodriguez-Cañete, Gabriel Olveira, and Rajaa El Bekay. 2023. "MiR-221-3p/222-3p Cluster Expression in Human Adipose Tissue Is Related to Obesity and Type 2 Diabetes" International Journal of Molecular Sciences 24, no. 24: 17449. https://doi.org/10.3390/ijms242417449
APA StyleGentile, A.-M., Lhamyani, S., Mengual-Mesa, M., García-Fuentes, E., Bermúdez-Silva, F.-J., Rojo-Martínez, G., Clemente-Postigo, M., Rodriguez-Cañete, A., Olveira, G., & El Bekay, R. (2023). MiR-221-3p/222-3p Cluster Expression in Human Adipose Tissue Is Related to Obesity and Type 2 Diabetes. International Journal of Molecular Sciences, 24(24), 17449. https://doi.org/10.3390/ijms242417449








