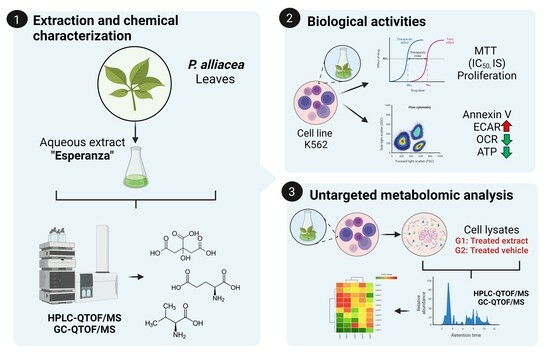
Effect of Petiveria alliacea Extracts on Metabolism of K562 Myeloid Leukemia Cells
Abstract
:1. Introduction
2. Results
2.1. The Esperanza Extract Reduces the Cell Proliferation Rate without Inducing Apoptosis
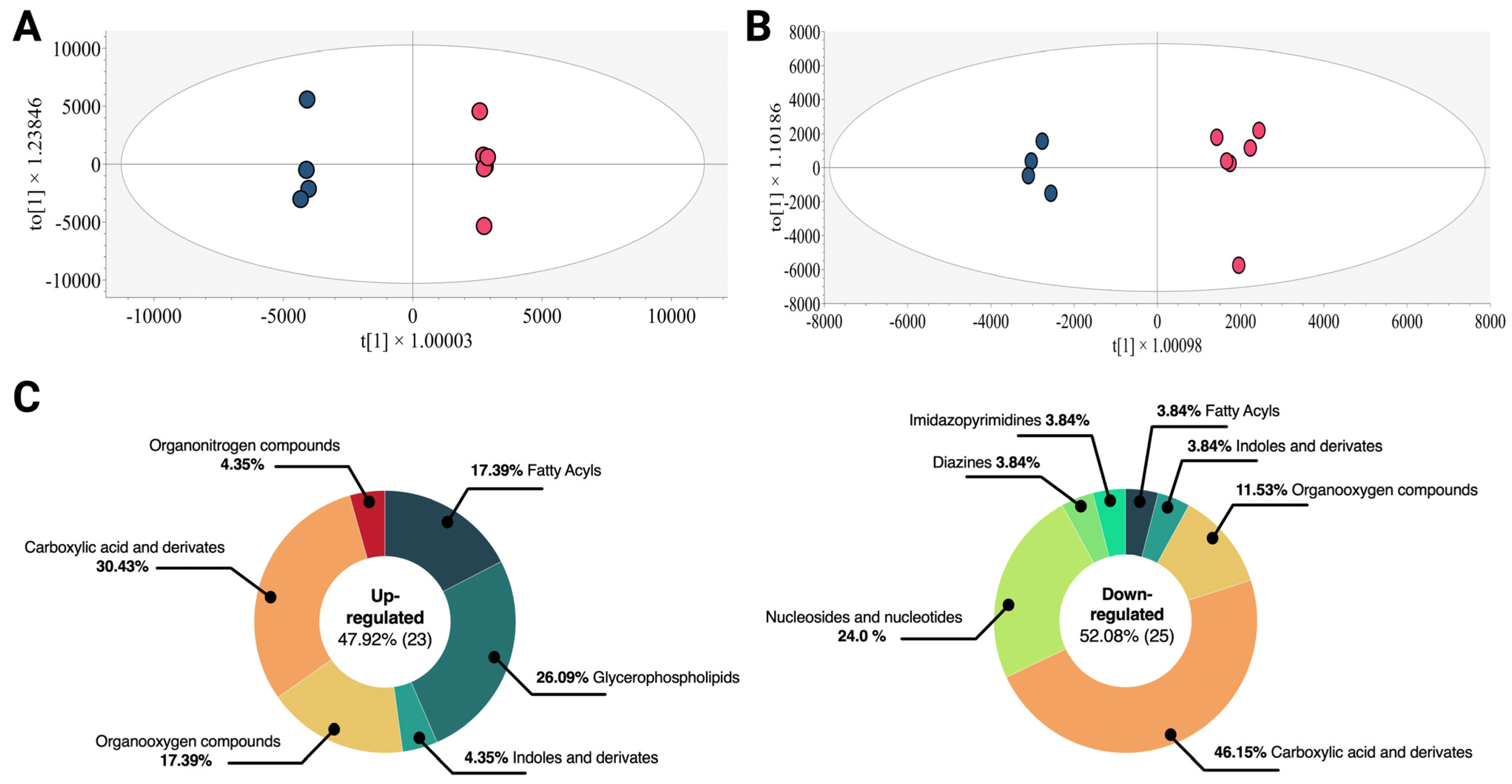
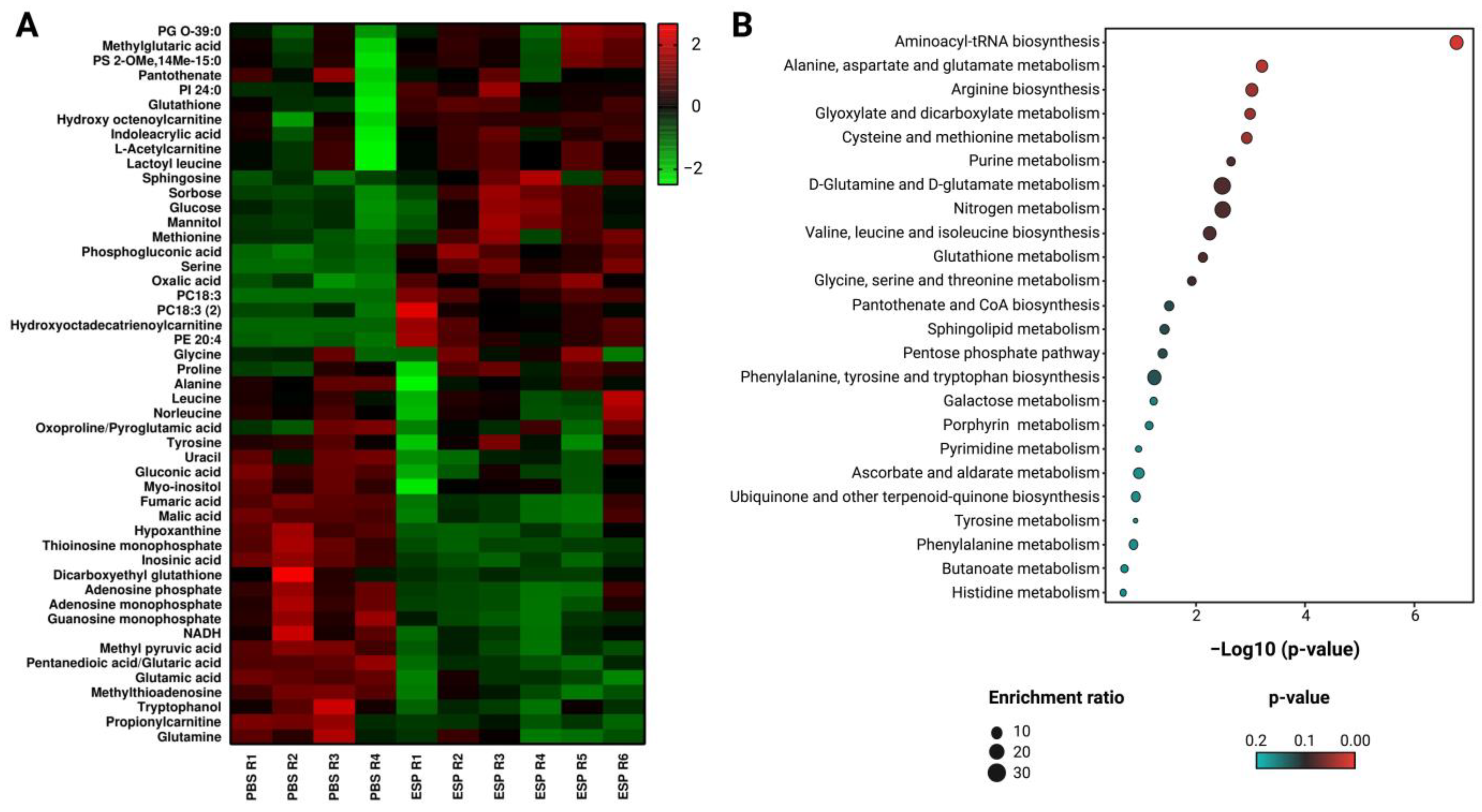
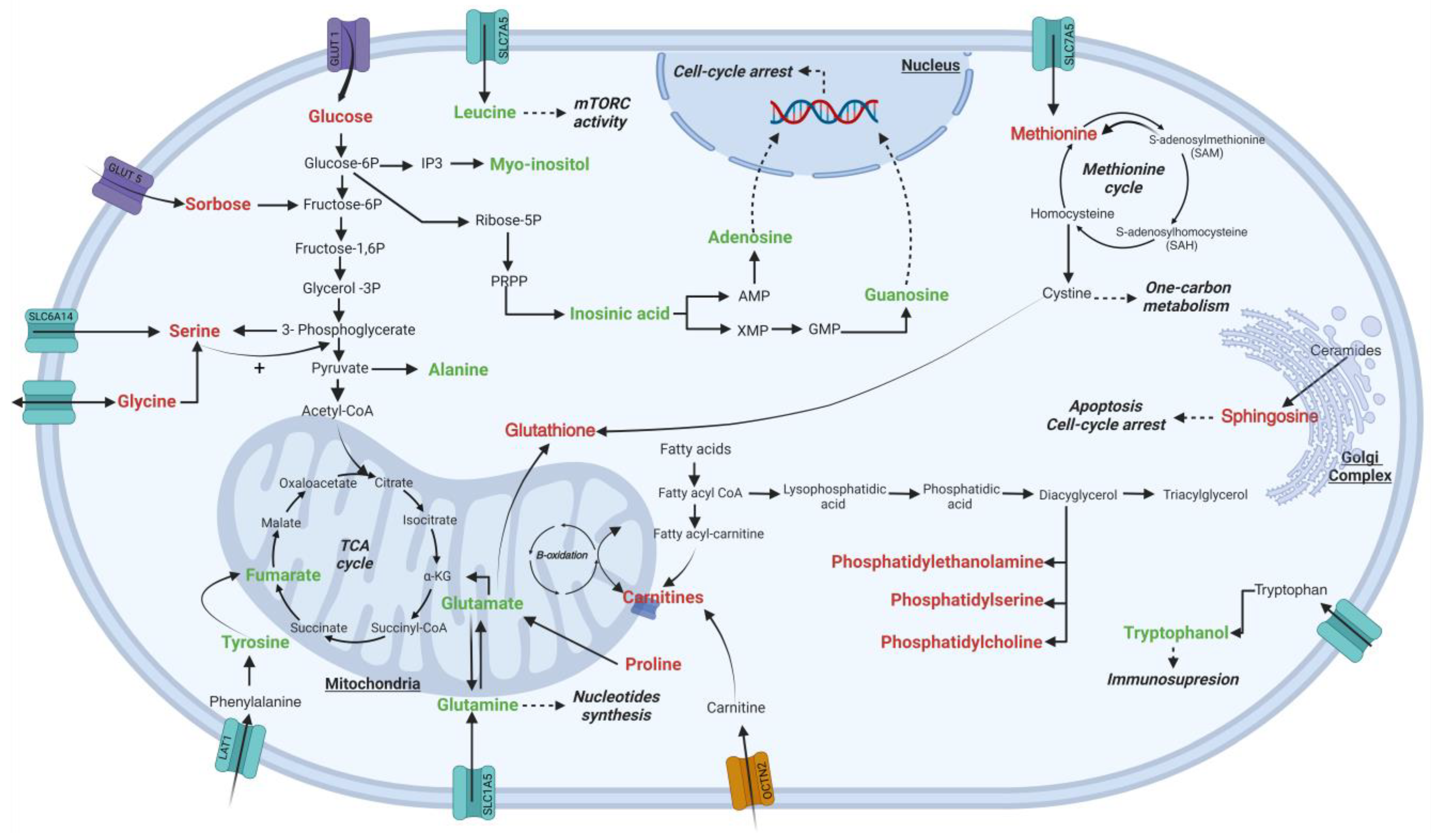
2.2. Alterations in the Endometabolome of the K562 Cell Line Treated with Esperanza Extract
3. Discussion
4. Materials and Methods
4.1. Plant Material and Extraction
4.2. In Vitro Cytotoxicity and Proliferation Assays
4.3. Cell Death Evaluation
4.4. OCR and ECAR Evaluation
4.5. ATP Determination
4.6. Untargeted Metabolomics Analysis
4.6.1. Sample Preparation
4.6.2. Untargeted Metabolomics by LC-QTOF-MS
4.6.3. Untargeted Metabolomics by GC-QTOF-MS
4.6.4. Quality Control Samples
4.7. Data Processing and Analysis
4.8. Annotation of Statistically Significant Molecular Features
4.9. Altered Metabolite Pathway Mapping
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 2017, 7, 339. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Li, W. Genomics and pharmacogenomics of pediatric acute lymphoblastic leukemia. Crit. Rev. Oncol./Hematol. 2018, 126, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Suganuma, K.; Miwa, H.; Imai, N.; Shikami, M.; Gotou, M.; Goto, M.; Mizuno, S.; Takahashi, M.; Yamamoto, H.; Hiramatsu, A. Energy metabolism of leukemia cells: Glycolysis versus oxidative phosphorylation. Leuk. Lymphoma 2010, 51, 2112–2119. [Google Scholar] [CrossRef] [PubMed]
- Emadi, A.; Jun, S.A.; Tsukamoto, T.; Fathi, A.T.; Minden, M.D.; Dang, C.V. Inhibition of glutaminase selectively suppresses the growth of primary acute myeloid leukemia cells with IDH mutations. Exp. Hematol. 2014, 42, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.A.; Kampjut, D.; Harrison, S.A.; Ralser, M. Sulfate radicals enable a non-enzymatic Krebs cycle precursor. Nat. Ecol. Evol. 2017, 1, 0083. [Google Scholar] [CrossRef] [PubMed]
- Kültz, D. Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 2005, 67, 225–257. [Google Scholar] [CrossRef] [PubMed]
- Pistollato, F.; Giampieri, F.; Battino, M. The use of plant-derived bioactive compounds to target cancer stem cells and modulate tumor microenvironment. Food Chem. Toxicol. 2015, 75, 58–70. [Google Scholar] [CrossRef]
- Goel, H.; Kumar, R.; Tanwar, P.; Upadhyay, T.K.; Khan, F.; Pandey, P.; Kang, S.; Moon, M.; Choi, J.; Choi, M. Unraveling the therapeutic potential of natural products in the prevention and treatment of leukemia. Biomed. Pharmacother. 2023, 160, 114351. [Google Scholar] [CrossRef]
- Luz, D.A.; Pinheiro, A.M.; Silva, M.L.; Monteiro, M.C.; Prediger, R.D.; Maia, C.S.F.; Fontes-Junior, E.A. Ethnobotany, phytochemistry and neuropharmacological effects of Petiveria alliacea L.(Phytolaccaceae): A review. J. Ethnopharmacol. 2016, 185, 182–201. [Google Scholar] [CrossRef]
- Di Stasi, L.C.; Costa, M.; Mendaçolli, S.L.; Kirizawa, M.; Gomes, C.; Trolin, G. Screening in mice of some medicinal plants used for analgesic purposes in the state of São Paulo. J. Ethnopharmacol. 1988, 24, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Martins, R.; Pegoraro, D.; Woisky, R.; Penna, S.; Sertié, J.A.A. The anti-inflammatory and analgesic effects of a crude extract of Petiveria alliacea L.(Phytolaccaceae). Phytomedicine 2002, 9, 245–248. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Zhu, J.; Wang, X.; Xu, X. Synthesis and anti-tumor evaluation of new trisulfide derivatives. Bioorganic Med. Chem. Lett. 2006, 16, 4826–4829. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.; Rosner, H.; Levy, H.; Barton, E. A critical review of the therapeutic potential of dibenzyl trisulphide isolated from Petiveria alliacea L (guinea hen weed, anamu). West Indian Med. J. 2007, 56, 17. [Google Scholar] [CrossRef]
- Cifuentes, M.C.; Castañeda, D.M.; Urueña, C.P.; Fiorentino, S. A fraction from Petiveria alliacea induces apoptosis via a mitochondria-dependent pathway and regulates HSP70 expression. Univ. Sci. 2009, 14, 125–134. [Google Scholar] [CrossRef]
- Urueña, C.; Cifuentes, C.; Castañeda, D.; Arango, A.; Kaur, P.; Asea, A.; Fiorentino, S. Petiveria alliacea extracts uses multiple mechanisms to inhibit growth of human and mouse tumoral cells. BMC Complement. Altern. Med. 2008, 8, 60. [Google Scholar] [CrossRef]
- Hernández, J.F.; Urueña, C.P.; Sandoval, T.A.; Cifuentes, M.C.; Formentini, L.; Cuezva, J.M.; Fiorentino, S. A cytotoxic Petiveria alliacea dry extract induces ATP depletion and decreases β-F1-ATPase expression in breast cancer cells and promotes survival in tumor-bearing mice. Rev. Bras. Farmacogn. 2017, 27, 306–314. [Google Scholar] [CrossRef]
- Hernández, J.F.; Urueña, C.P.; Cifuentes, M.C.; Sandoval, T.A.; Pombo, L.M.; Castañeda, D.; Asea, A.; Fiorentino, S. A Petiveria alliacea standardized fraction induces breast adenocarcinoma cell death by modulating glycolytic metabolism. J. Ethnopharmacol. 2014, 153, 641–649. [Google Scholar] [CrossRef]
- Sriskanthadevan, S.; Jeyaraju, D.V.; Chung, T.E.; Prabha, S.; Xu, W.; Skrtic, M.; Jhas, B.; Hurren, R.; Gronda, M.; Wang, X. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood J. Am. Soc. Hematol. 2015, 125, 2120–2130. [Google Scholar] [CrossRef]
- Panina, S.B.; Baran, N.; Brasil da Costa, F.H.; Konopleva, M.; Kirienko, N.V. A mechanism for increased sensitivity of acute myeloid leukemia to mitotoxic drugs. Cell Death Dis. 2019, 10, 617. [Google Scholar] [CrossRef]
- Murillo, N.; Lasso, P.; Urueña, C.; Pardo-Rodriguez, D.; Ballesteros-Ramírez, R.; Betancourt, G.; Rojas, L.; Cala, M.P.; Fiorentino, S. Petiveria alliacea Reduces Tumor Burden and Metastasis and Regulates the Peripheral Immune Response in a Murine Myeloid Leukemia Model. Int. J. Mol. Sci. 2023, 24, 12972. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, Å.M.; Wheelock, C.E. Trials and tribulations of ‘omics data analysis: Assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol. BioSystems 2013, 9, 2589–2596. [Google Scholar] [CrossRef] [PubMed]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, J.I. Principales referencias etnomedicas sobre el anamu (petiveria alliacea linn) y principios activos encontrados en la planta. Un acercamiento al tema. Rev. CENIC. Cienc. Biológicas 2007, 38, 27–30. [Google Scholar]
- Correa, Q.; Jaime, E.; Bernal, H.Y.; Bello, C.A.; de Cartagena, J.d.A. Especies Vegetales Promisorias de los Países del Convenio Andrés Bello; SECAB: Vijayapura, India, 1990. [Google Scholar]
- Romero-Garcia, S.; Lopez-Gonzalez, J.S.; B’ez-Viveros, J.L.; Aguilar-Cazares, D.; Prado-Garcia, H. Tumor cell metabolism: An integral view. Cancer Biol. Ther. 2011, 12, 939–948. [Google Scholar] [CrossRef]
- Chelakkot, C.; Chelakkot, V.S.; Shin, Y.; Song, K. Modulating glycolysis to improve cancer therapy. Int. J. Mol. Sci. 2023, 24, 2606. [Google Scholar] [CrossRef]
- Shiratori, R.; Furuichi, K.; Yamaguchi, M.; Miyazaki, N.; Aoki, H.; Chibana, H.; Ito, K.; Aoki, S. Glycolytic suppression dramatically changes the intracellular metabolic profile of multiple cancer cell lines in a mitochondrial metabolism-dependent manner. Sci. Rep. 2019, 9, 18699. [Google Scholar] [CrossRef]
- Meng, X.; Lu, Z.; Lv, Q.; Jiang, Y.; Zhang, L.; Wang, Z. Tumor metabolism destruction via metformin-based glycolysis inhibition and glucose oxidase-mediated glucose deprivation for enhanced cancer therapy. Acta Biomater. 2022, 145, 222–234. [Google Scholar] [CrossRef]
- Wang, H.; Chen, S.; Kang, W.; Ding, B.; Cui, S.; Zhou, L.; Zhang, N.; Luo, H.; Wang, M.; Zhang, F. High dose isoleucine stabilizes nuclear PTEN to suppress the proliferation of lung cancer. Discov. Oncol. 2023, 14, 25. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Yamamoto, K.; Sato, Y.; Inoue, S.; Morinaga, T.; Hirano, E. Combination of aspartic acid and glutamic acid inhibits tumor cell proliferation. Biomed. Res. 2016, 37, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Pike Winer, L.S.; Wu, M. Rapid analysis of glycolytic and oxidative substrate flux of cancer cells in a microplate. PLoS ONE 2014, 9, e109916. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Suzuki, M.; Saito, T.; Miyado, K. Emerging role of TCA cycle-related enzymes in human diseases. Int. J. Mol. Sci. 2021, 22, 13057. [Google Scholar] [CrossRef] [PubMed]
- Potter, V.R.; Elvehjem, C. The effect of inhibitors on succinoxidase. J. Biol. Chem. 1937, 117, 341–349. [Google Scholar] [CrossRef]
- Li, X.; Zhu, H.; Sun, W.; Yang, X.; Nie, Q.; Fang, X. Role of glutamine and its metabolite ammonia in crosstalk of cancer-associated fibroblasts and cancer cells. Cancer Cell Int. 2021, 21, 479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, Q.; Lin, Z.; Yang, P.; Dou, K.; Zhang, R. Berberine inhibits growth of liver cancer cells by suppressing glutamine uptake. OncoTargets Ther. 2019, 12, 11751–11763. [Google Scholar] [CrossRef]
- Ma, J.; Zhong, M.; Xiong, Y.; Gao, Z.; Wu, Z.; Liu, Y.; Hong, X. Emerging roles of nucleotide metabolism in cancer development: Progress and prospect. Aging (Albany NY) 2021, 13, 13349. [Google Scholar] [CrossRef]
- Rösner, H.; Williams, L.; Jung, A.; Kraus, W. Disassembly of microtubules and inhibition of neurite outgrowth, neuroblastoma cell proliferation, and MAP kinase tyrosine dephosphorylation by dibenzyl trisulphide. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2001, 1540, 166–177. [Google Scholar] [CrossRef]
- Bjørndal, B.; Alterås, E.K.; Lindquist, C.; Svardal, A.; Skorve, J.; Berge, R.K. Associations between fatty acid oxidation, hepatic mitochondrial function, and plasma acylcarnitine levels in mice. Nutr. Metab. 2018, 15, 10. [Google Scholar] [CrossRef]
- Wenzel, U.; Nickel, A.; Daniel, H. Increased carnitine-dependent fatty acid uptake into mitochondria of human colon cancer cells induces apoptosis. J. Nutr. 2005, 135, 1510–1514. [Google Scholar] [CrossRef]
- Long, Z.; Feng, G.; Zhao, N.; Wu, L.; Zhu, H. Isoferulic acid inhibits human leukemia cell growth through induction of G2/M-phase arrest and inhibition of Akt/mTOR signaling. Mol. Med. Rep. 2020, 21, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Niero, E.L.d.O.; Machado-Santelli, G.M. Cinnamic acid induces apoptotic cell death and cytoskeleton disruption in human melanoma cells. J. Exp. Clin. Cancer Res. 2013, 32, 31. [Google Scholar] [CrossRef] [PubMed]
- Samimi, S.; Ardestani, M.S.; Dorkoosh, F.A. Preparation of carbon quantum dots-quinic acid for drug delivery of gemcitabine to breast cancer cells. J. Drug Deliv. Sci. Technol. 2021, 61, 102287. [Google Scholar] [CrossRef]
- Sanchez-Martin, V.; Plaza-Calonge, M.d.C.; Soriano-Lerma, A.; Ortiz-Gonzalez, M.; Linde-Rodriguez, A.; Perez-Carrasco, V.; Ramirez-Macias, I.; Cuadros, M.; Gutierrez-Fernandez, J.; Murciano-Calles, J. Gallic acid: A natural phenolic compound exerting antitumoral activities in colorectal cancer via interaction with g-quadruplexes. Cancers 2022, 14, 2648. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Lim, S.-C.; Lee, T.-B.; Han, S.-I. Anticancer Effect of Gallic Acid on Acidity-Induced Invasion of MCF7 Breast Cancer Cells. Nutrients 2023, 15, 3596. [Google Scholar] [CrossRef]
- Ho, H.-H.; Chang, C.-S.; Ho, W.-C.; Liao, S.-Y.; Lin, W.-L.; Wang, C.-J. Gallic acid inhibits gastric cancer cells metastasis and invasive growth via increased expression of RhoB, downregulation of AKT/small GTPase signals and inhibition of NF-κB activity. Toxicol. Appl. Pharmacol. 2013, 266, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Urueña, C.; Mancipe, J.; Hernandez, J.; Castañeda, D.; Pombo, L.; Gomez, A.; Asea, A.; Fiorentino, S. Gallotannin-rich Caesalpinia spinosa fraction decreases the primary tumor and factors associated with poor prognosis in a murine breast cancer model. BMC Complement. Altern. Med. 2013, 13, 74. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Castañeda, D.M.; Pombo, L.M.; Urueña, C.P.; Hernandez, J.F.; Fiorentino, S. A gallotannin-rich fraction from Caesalpinia spinosa (Molina) Kuntze displays cytotoxic activity and raises sensitivity to doxorubicin in a leukemia cell line. BMC Complement. Altern. Med. 2012, 12, 38. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojas, L.; Pardo-Rodriguez, D.; Urueña, C.; Lasso, P.; Arévalo, C.; Cala, M.P.; Fiorentino, S. Effect of Petiveria alliacea Extracts on Metabolism of K562 Myeloid Leukemia Cells. Int. J. Mol. Sci. 2023, 24, 17418. https://doi.org/10.3390/ijms242417418
Rojas L, Pardo-Rodriguez D, Urueña C, Lasso P, Arévalo C, Cala MP, Fiorentino S. Effect of Petiveria alliacea Extracts on Metabolism of K562 Myeloid Leukemia Cells. International Journal of Molecular Sciences. 2023; 24(24):17418. https://doi.org/10.3390/ijms242417418
Chicago/Turabian StyleRojas, Laura, Daniel Pardo-Rodriguez, Claudia Urueña, Paola Lasso, Cindy Arévalo, Mónica P. Cala, and Susana Fiorentino. 2023. "Effect of Petiveria alliacea Extracts on Metabolism of K562 Myeloid Leukemia Cells" International Journal of Molecular Sciences 24, no. 24: 17418. https://doi.org/10.3390/ijms242417418
APA StyleRojas, L., Pardo-Rodriguez, D., Urueña, C., Lasso, P., Arévalo, C., Cala, M. P., & Fiorentino, S. (2023). Effect of Petiveria alliacea Extracts on Metabolism of K562 Myeloid Leukemia Cells. International Journal of Molecular Sciences, 24(24), 17418. https://doi.org/10.3390/ijms242417418