Spontaneous Mutations in Saccharomyces cerevisiae mtDNA Increase Cell-to-Cell Variation in mtDNA Amount
Abstract
:1. Introduction
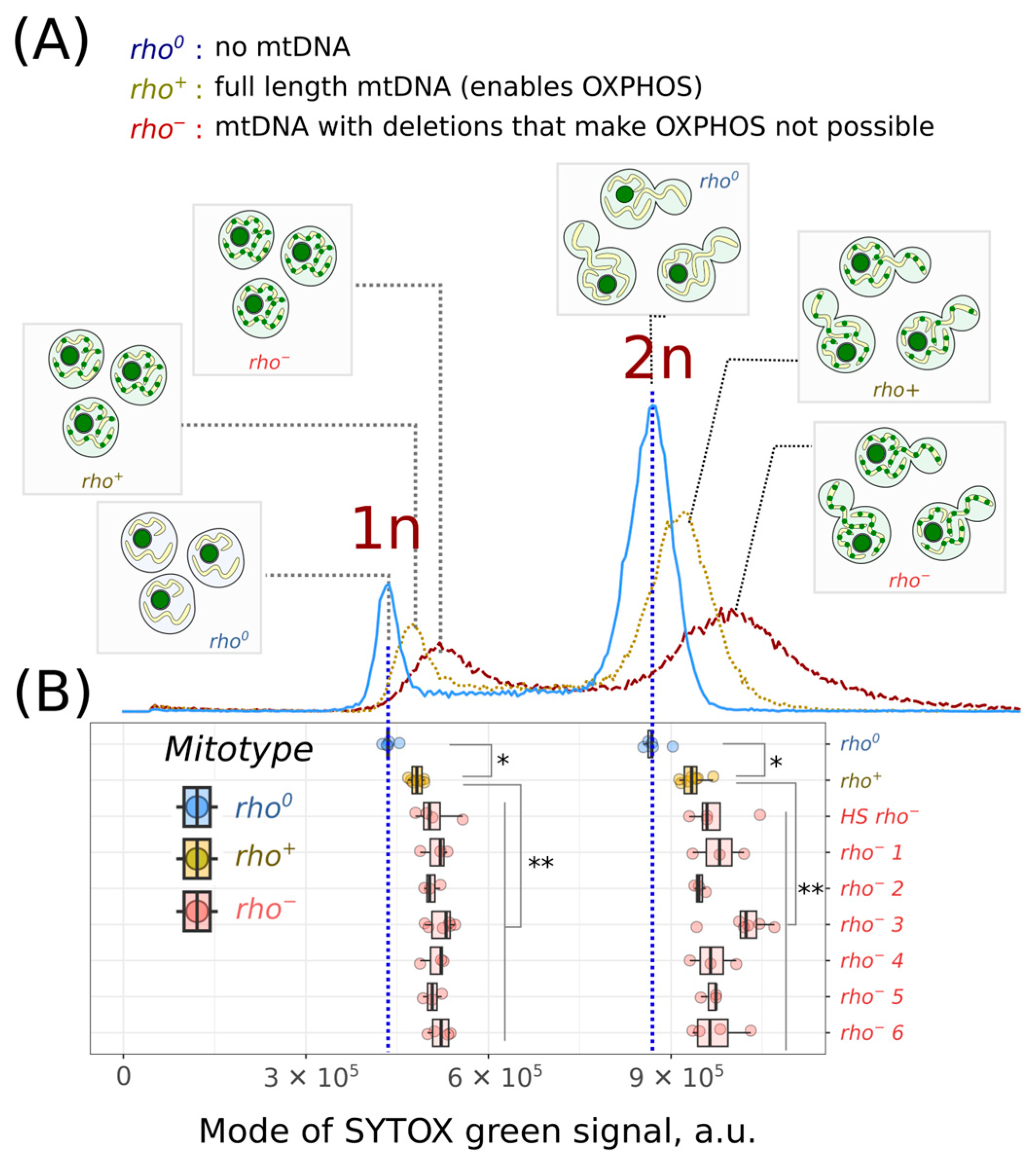
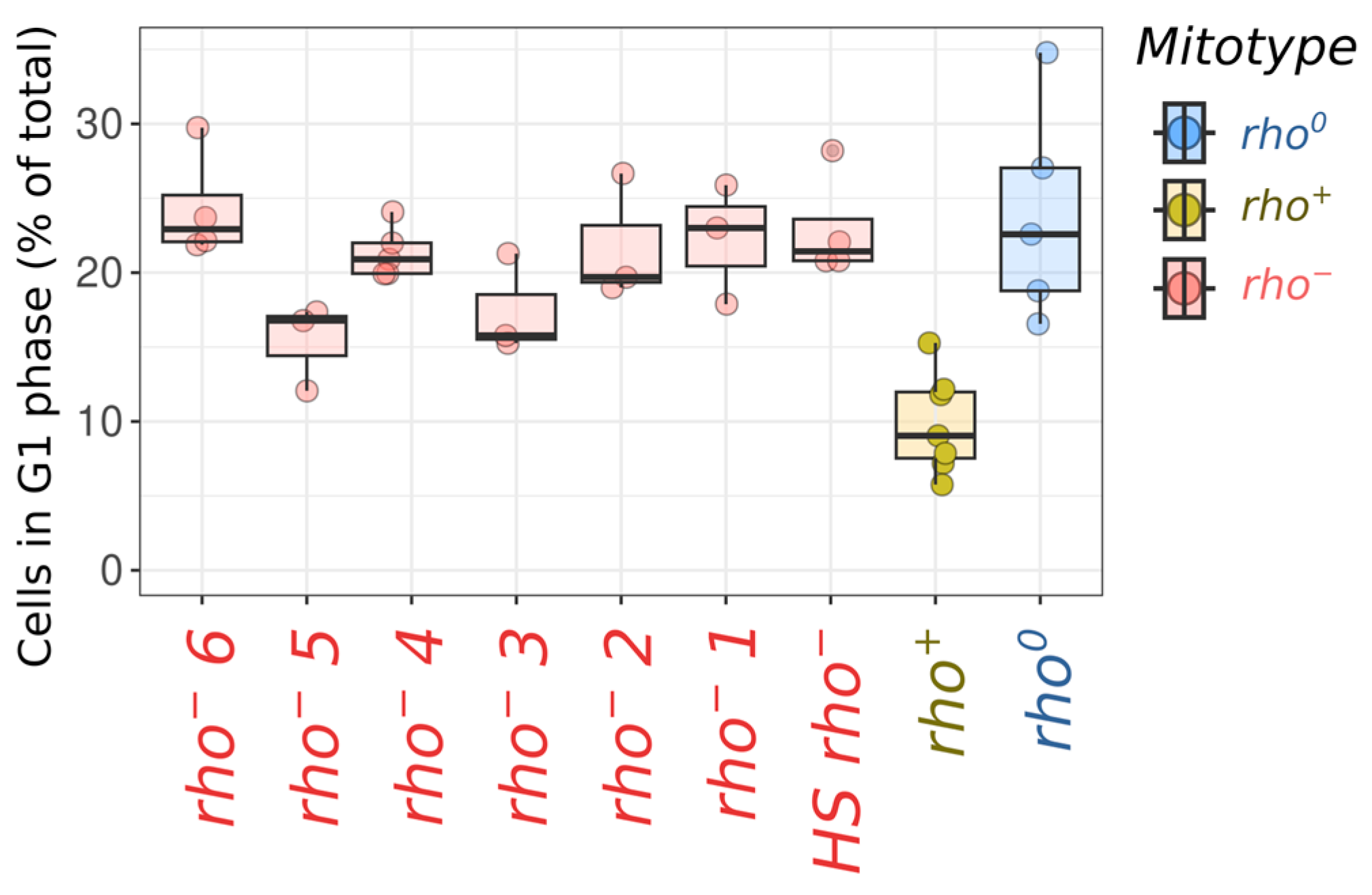
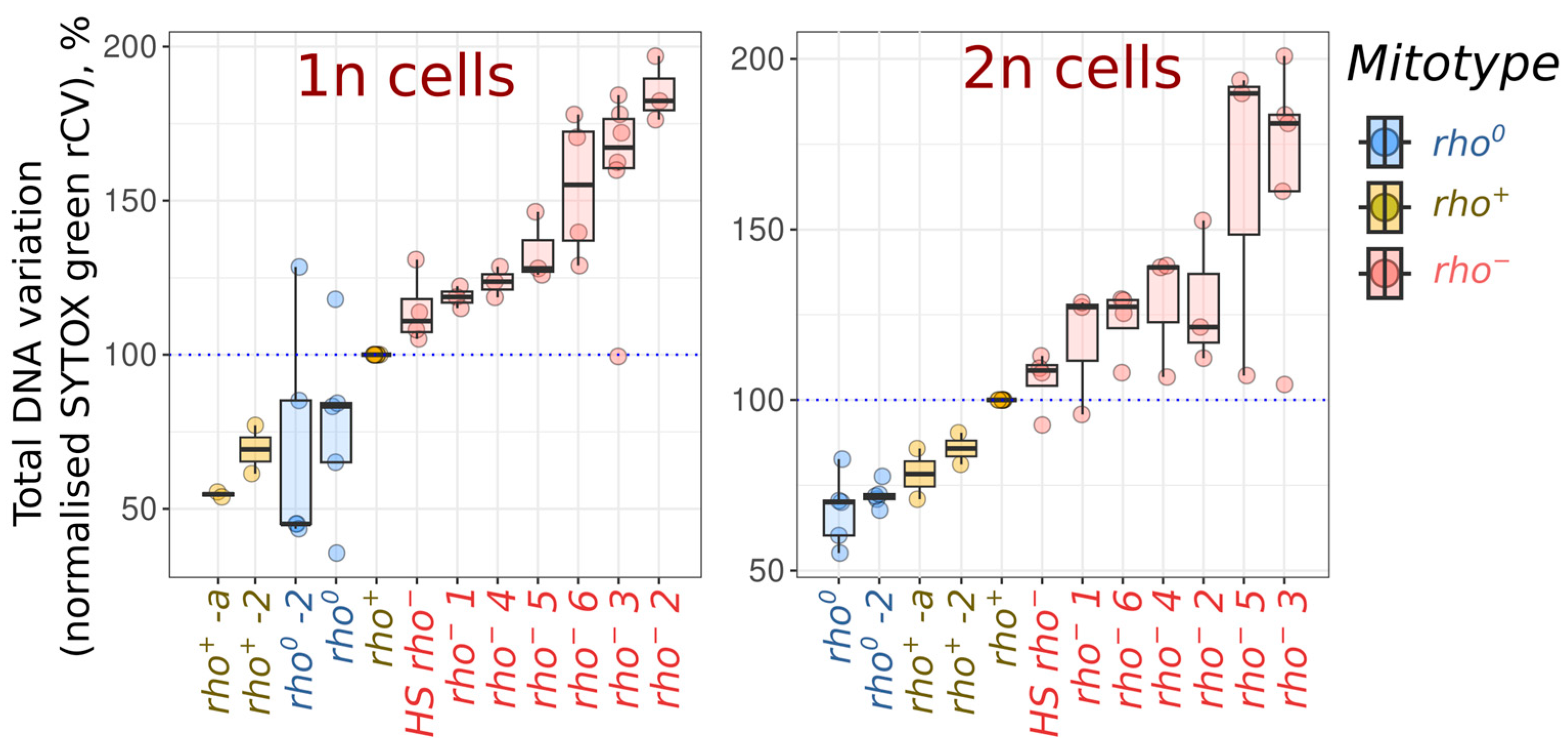
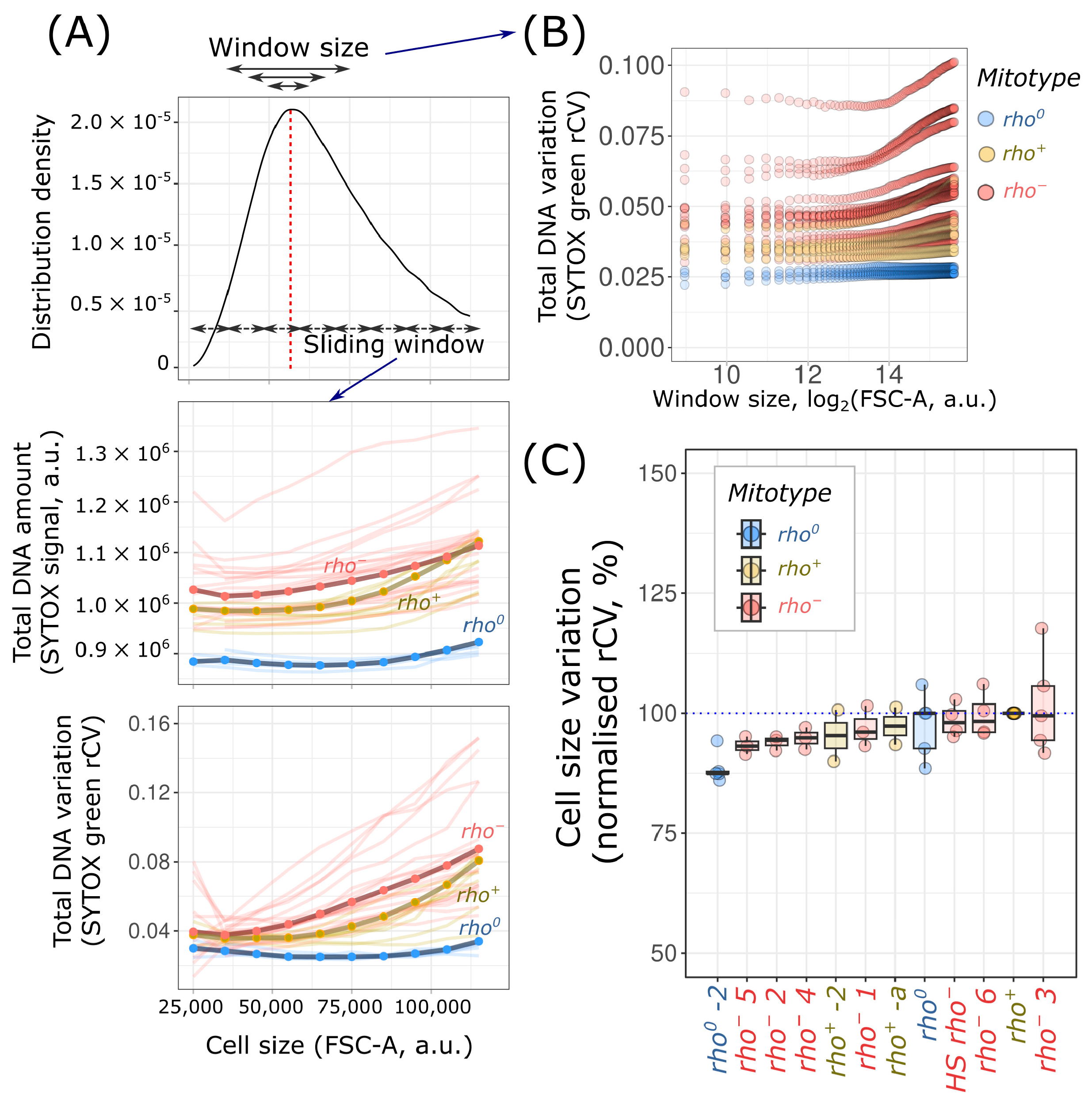
2. Results
3. Discussion
4. Materials and Methods
4.1. Yeast Strains and Growth Conditions
4.2. mtDNA Depletion
4.3. Flow Cytometry
4.4. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roger, A.J.; Muñoz-Gómez, S.A.; Kamikawa, R. The Origin and Diversification of Mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef] [PubMed]
- Taanman, J.W. The Mitochondrial Genome: Structure, Transcription, Translation and Replication. Biochim. Biophys. Acta 1999, 1410, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Newlon, C.S.; Fangman, W.L. Mitochondrial DNA Synthesis in Cell Cycle Mutants of Saccharomyces Cerevisiae. Cell 1975, 5, 423–428. [Google Scholar] [PubMed]
- Sazer, S.; Sherwood, S.W. Mitochondrial Growth and DNA Synthesis Occur in the Absence of Nuclear DNA Replication in Fission Yeast. J. Cell Sci. 1990, 97 Pt 3, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, S.; Sun, X.; Lee, J.-H.; Cho, H. Cell Cycle-Dependent Mitochondrial Biogenesis and Dynamics in Mammalian Cells. Biochem. Biophys. Res. Commun. 2007, 357, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Sato, Y.; Higashiyama, T.; Sasaki, N. Live Imaging Reveals the Dynamics and Regulation of Mitochondrial Nucleoids during the Cell Cycle in Fucci2-HeLa Cells. Sci. Rep. 2017, 7, 11257. [Google Scholar] [CrossRef]
- Chatre, L.; Ricchetti, M. Prevalent Coordination of Mitochondrial DNA Transcription and Initiation of Replication with the Cell Cycle. Nucleic Acids Res. 2013, 41, 3068–3078. [Google Scholar] [CrossRef] [PubMed]
- Glastad, R.C.; Johnston, I.G. Mitochondrial Network Structure Controls Cell-to-Cell mtDNA Variability Generated by Cell Divisions. PLoS Comput. Biol. 2023, 19, e1010953. [Google Scholar] [CrossRef]
- Summers, D.K. The Kinetics of Plasmid Loss. Trends Biotechnol. 1991, 9, 273–278. [Google Scholar] [CrossRef]
- Kumar, D.; Prajapati, H.K.; Mahilkar, A.; Ma, C.-H.; Mittal, P.; Jayaram, M.; Ghosh, S.K. The Selfish Yeast Plasmid Utilizes the Condensin Complex and Condensed Chromatin for Faithful Partitioning. PLoS Genet. 2021, 17, e1009660. [Google Scholar] [CrossRef] [PubMed]
- Schrott, S.; Osman, C. Two Mitochondrial HMG-Box Proteins, Cim1 and Abf2, Antagonistically Regulate mtDNA Copy Number in Saccharomyces cerevisiae. Nucleic Acids Res. 2023, 51, 11813–11835. [Google Scholar] [CrossRef] [PubMed]
- Puddu, F.; Herzog, M.; Selivanova, A.; Wang, S.; Zhu, J.; Klein-Lavi, S.; Gordon, M.; Meirman, R.; Millan-Zambrano, G.; Ayestaran, I.; et al. Genome Architecture and Stability in the Saccharomyces Cerevisiae Knockout Collection. Nature 2019, 573, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Göke, A.; Schrott, S.; Mizrak, A.; Belyy, V.; Osman, C.; Walter, P. Mrx6 Regulates Mitochondrial DNA Copy Number in Saccharomyces Cerevisiae by Engaging the Evolutionarily Conserved Lon Protease Pim1. Mol. Biol. Cell 2020, 31, 527–545. [Google Scholar] [CrossRef] [PubMed]
- Staneva, D.; Vasileva, B.; Podlesniy, P.; Miloshev, G.; Georgieva, M. Yeast Chromatin Mutants Reveal Altered mtDNA Copy Number and Impaired Mitochondrial Membrane Potential. J. Fungi 2023, 9, 329. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, S.; Li, J.; Gjuvsland, A.B.; Persson, K.; Demitz-Helin, E.; González Peña, C.; Yue, J.-X.; Gilchrist, C.; Ärengård, T.; Ghiaci, P.; et al. Genetically Controlled mtDNA Deletions Prevent ROS Damage by Arresting Oxidative Phosphorylation. Elife 2022, 11, e76095. [Google Scholar] [CrossRef] [PubMed]
- Hori, A.; Yoshida, M.; Shibata, T.; Ling, F. Reactive Oxygen Species Regulate DNA Copy Number in Isolated Yeast Mitochondria by Triggering Recombination-Mediated Replication. Nucleic Acids Res. 2009, 37, 749–761. [Google Scholar] [CrossRef] [PubMed]
- Moraes, C.T. What Regulates Mitochondrial DNA Copy Number in Animal Cells? Trends Genet. 2001, 17, 199–205. [Google Scholar] [CrossRef]
- Tadi, S.K.; Sebastian, R.; Dahal, S.; Babu, R.K.; Choudhary, B.; Raghavan, S.C. Microhomology-Mediated End Joining Is the Principal Mediator of Double-Strand Break Repair during Mitochondrial DNA Lesions. Mol. Biol. Cell 2016, 27, 223–235. [Google Scholar] [CrossRef]
- Rebolledo-Jaramillo, B.; Su, M.S.-W.; Stoler, N.; McElhoe, J.A.; Dickins, B.; Blankenberg, D.; Korneliussen, T.S.; Chiaromonte, F.; Nielsen, R.; Holland, M.M.; et al. Maternal Age Effect and Severe Germ-Line Bottleneck in the Inheritance of Human Mitochondrial DNA. Proc. Natl. Acad. Sci. USA 2014, 111, 15474–15479. [Google Scholar]
- Lujan, S.A.; Longley, M.J.; Humble, M.H.; Lavender, C.A.; Burkholder, A.; Blakely, E.L.; Alston, C.L.; Gorman, G.S.; Turnbull, D.M.; McFarland, R.; et al. Ultrasensitive Deletion Detection Links Mitochondrial DNA Replication, Disease, and Aging. Genome Biol. 2020, 21, 248. [Google Scholar] [CrossRef]
- Lawless, C.; Greaves, L.; Reeve, A.K.; Turnbull, D.M.; Vincent, A.E. The Rise and Rise of Mitochondrial DNA Mutations. Open Biol. 2020, 10, 200061. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, L.N.; Brem, R.B.; Kruglyak, L.; Gottschling, D.E. Polymorphisms in Multiple Genes Contribute to the Spontaneous Mitochondrial Genome Instability of Saccharomyces Cerevisiae S288C Strains. Genetics 2009, 183, 365–383. [Google Scholar] [CrossRef] [PubMed]
- de Zamaroczy, M.; Marotta, R.; Faugeron-Fonty, G.; Goursot, R.; Mangin, M.; Baldacci, G.; Bernardi, G. The Origins of Replication of the Yeast Mitochondrial Genome and the Phenomenon of Suppressivity. Nature 1981, 292, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Osman, C.; Noriega, T.R.; Okreglak, V.; Fung, J.C.; Walter, P. Integrity of the Yeast Mitochondrial Genome, but Not Its Distribution and Inheritance, Relies on Mitochondrial Fission and Fusion. Proc. Natl. Acad. Sci. USA 2015, 112, E947–E956. [Google Scholar] [CrossRef] [PubMed]
- Dzierzbicki, P.; Kaniak-Golik, A.; Malc, E.; Mieczkowski, P.; Ciesla, Z. The Generation of Oxidative Stress-Induced Rearrangements in Saccharomyces Cerevisiae mtDNA Is Dependent on the Nuc1 (EndoG/ExoG) Nuclease and Is Enhanced by Inactivation of the MRX Complex. Mutat. Res. 2012, 740, 21–33. [Google Scholar] [CrossRef]
- Fangman, W.L.; Henly, J.W.; Churchill, G.; Brewer, B.J. Stable Maintenance of a 35-Base-Pair Yeast Mitochondrial Genome. Mol. Cell. Biol. 1989, 9, 1917–1921. [Google Scholar] [PubMed]
- Foury, F.; Roganti, T.; Lecrenier, N.; Purnelle, B. The Complete Sequence of the Mitochondrial Genome of Saccharomyces Cerevisiae. FEBS Lett. 1998, 440, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.M.; Nagley, P.; Linnane, A.W. Biogenesis of Mitochondria. XLII. Genetic Analysis of the Control of Cellular Mitochondrial DNA Levels in Saccharomyces Cerevisiae. Mol. Gen. Genet. 1976, 145, 169–175. [Google Scholar] [CrossRef]
- Nunn, C.J.; Goyal, S. Contingency and Selection in Mitochondrial Genome Dynamics. Elife 2022, 11, e76557. [Google Scholar] [CrossRef]
- Karavaeva, I.E.; Golyshev, S.A.; Smirnova, E.A.; Sokolov, S.S.; Severin, F.F.; Knorre, D.A. Mitochondrial Depolarization in Yeast Zygotes Inhibits Clonal Expansion of Selfish mtDNA. J. Cell Sci. 2017, 130, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.R.S.; Ghaemmaghami, S.; Ihmels, J.; Breslow, D.K.; Noble, M.; DeRisi, J.L.; Weissman, J.S. Single-Cell Proteomic Analysis of S. Cerevisiae Reveals the Architecture of Biological Noise. Nature 2006, 441, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Peter, J.; De Chiara, M.; Friedrich, A.; Yue, J.-X.; Pflieger, D.; Bergström, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome Evolution across 1,011 Saccharomyces Cerevisiae Isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, R.; Yang, S.-Y.; Leu, J.-Y. Characterization of Chromosome Stability in Diploid, Polyploid and Hybrid Yeast Cells. PLoS ONE 2013, 8, e68094. [Google Scholar] [CrossRef] [PubMed]
- Mulla, W.; Zhu, J.; Li, R. Yeast: A Simple Model System to Study Complex Phenomena of Aneuploidy. FEMS Microbiol. Rev. 2014, 38, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.-M.; Liu, Y.-T.; Ma, C.-H.; Jayaram, M.; Sau, S. The 2 Micron Plasmid of Saccharomyces Cerevisiae: A Miniaturized Selfish Genome with Optimized Functional Competence. Plasmid 2013, 70, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.A.; Guarente, L. Extrachromosomal rDNA Circles—A Cause of Aging in Yeast. Cell 1997, 91, 1033–1042. [Google Scholar] [CrossRef]
- Knorre, D.A.; Azbarova, A.V.; Galkina, K.V.; Feniouk, B.A.; Severin, F.F. Replicative Aging as a Source of Cell Heterogeneity in Budding Yeast. Mech. Ageing Dev. 2018, 176, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Seel, A.; Padovani, F.; Mayer, M.; Finster, A.; Bureik, D.; Thoma, F.; Osman, C.; Klecker, T.; Schmoller, K.M. Regulation with Cell Size Ensures Mitochondrial DNA Homeostasis during Cell Growth. Nat. Struct. Mol. Biol. 2023, 30, 1549–1560. [Google Scholar] [CrossRef]
- Veatch, J.R.; McMurray, M.A.; Nelson, Z.W.; Gottschling, D.E. Mitochondrial Dysfunction Leads to Nuclear Genome Instability via an Iron-Sulfur Cluster Defect. Cell 2009, 137, 1247–1258. [Google Scholar] [CrossRef]
- Kelso, G.F.; Porteous, C.M.; Coulter, C.V.; Hughes, G.; Porteous, W.K.; Ledgerwood, E.C.; Smith, R.A.; Murphy, M.P. Selective Targeting of a Redox-Active Ubiquinone to Mitochondria within Cells: Antioxidant and Antiapoptotic Properties. J. Biol. Chem. 2001, 276, 4588–4596. [Google Scholar] [CrossRef]
- Joseph-Horne, T.; Hollomon, D.W.; Wood, P.M. Fungal Respiration: A Fusion of Standard and Alternative Components. Biochim. Biophys. Acta 2001, 1504, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.L.; Heyting, C.; Van der Horst, G.; Borst, P. The Organization of Repeating Units in Mitochondrial DNA from Yeast Petite Mutants. Curr. Genet. 1980, 1, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Vizeacoumar, F.J.; Bahr, S.; Li, J.; Warringer, J.; Vizeacoumar, F.S.; Min, R.; Vandersluis, B.; Bellay, J.; Devit, M.; et al. Systematic Exploration of Essential Yeast Gene Function with Temperature-Sensitive Mutants. Nat. Biotechnol. 2011, 29, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Galkina, K.V.; Zyrina, A.N.; Golyshev, S.A.; Kashko, N.D.; Markova, O.V.; Sokolov, S.S.; Severin, F.F.; Knorre, D.A. Mitochondrial Dynamics in Yeast with Repressed Adenine Nucleotide Translocator AAC2. Eur. J. Cell Biol. 2020, 99, 151071. [Google Scholar] [CrossRef] [PubMed]
- Sherman, F. Getting Started with Yeast. In Methods in Enzymology; Guthrie, C., Fink, G.R., Eds.; Academic Press: Cambridge, MA, USA, 2002; Volume 350, pp. 3–41. [Google Scholar]
- Haase, S.B. Cell Cycle Analysis of Budding Yeast Using SYTOX Green. Curr. Protoc. Cytom. 2004, 26, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Use R! Springer: New York, NY, USA; Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Hahne, F.; LeMeur, N.; Brinkman, R.R.; Ellis, B.; Haaland, P.; Sarkar, D.; Spidlen, J.; Strain, E.; Gentleman, R. flowCore: A Bioconductor Package for High Throughput Flow Cytometry. BMC Bioinform. 2009, 10, 106. [Google Scholar] [CrossRef]
- Ameijeiras-Alonso, J.; Crujeiras, R.M.; Rodriguez-Casal, A. Multimode: An R Package for Mode Assessment. J. Stat. Softw. 2021, 97, 1–32. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Potapenko, E.Y.; Kashko, N.D.; Knorre, D.A. Spontaneous Mutations in Saccharomyces cerevisiae mtDNA Increase Cell-to-Cell Variation in mtDNA Amount. Int. J. Mol. Sci. 2023, 24, 17413. https://doi.org/10.3390/ijms242417413
Potapenko EY, Kashko ND, Knorre DA. Spontaneous Mutations in Saccharomyces cerevisiae mtDNA Increase Cell-to-Cell Variation in mtDNA Amount. International Journal of Molecular Sciences. 2023; 24(24):17413. https://doi.org/10.3390/ijms242417413
Chicago/Turabian StylePotapenko, Elena Yu., Nataliia D. Kashko, and Dmitry A. Knorre. 2023. "Spontaneous Mutations in Saccharomyces cerevisiae mtDNA Increase Cell-to-Cell Variation in mtDNA Amount" International Journal of Molecular Sciences 24, no. 24: 17413. https://doi.org/10.3390/ijms242417413
APA StylePotapenko, E. Y., Kashko, N. D., & Knorre, D. A. (2023). Spontaneous Mutations in Saccharomyces cerevisiae mtDNA Increase Cell-to-Cell Variation in mtDNA Amount. International Journal of Molecular Sciences, 24(24), 17413. https://doi.org/10.3390/ijms242417413






