The Profiling of 179 miRNA Expression in Serum from Limb Girdle Muscular Dystrophy Patients and Healthy Controls
Abstract
:1. Introduction
2. Results
2.1. Clinical Aspects and Muscle Biopsy
2.2. Data Control Quality
2.3. The Expression Analysis of Serum-Circulating miRNAs
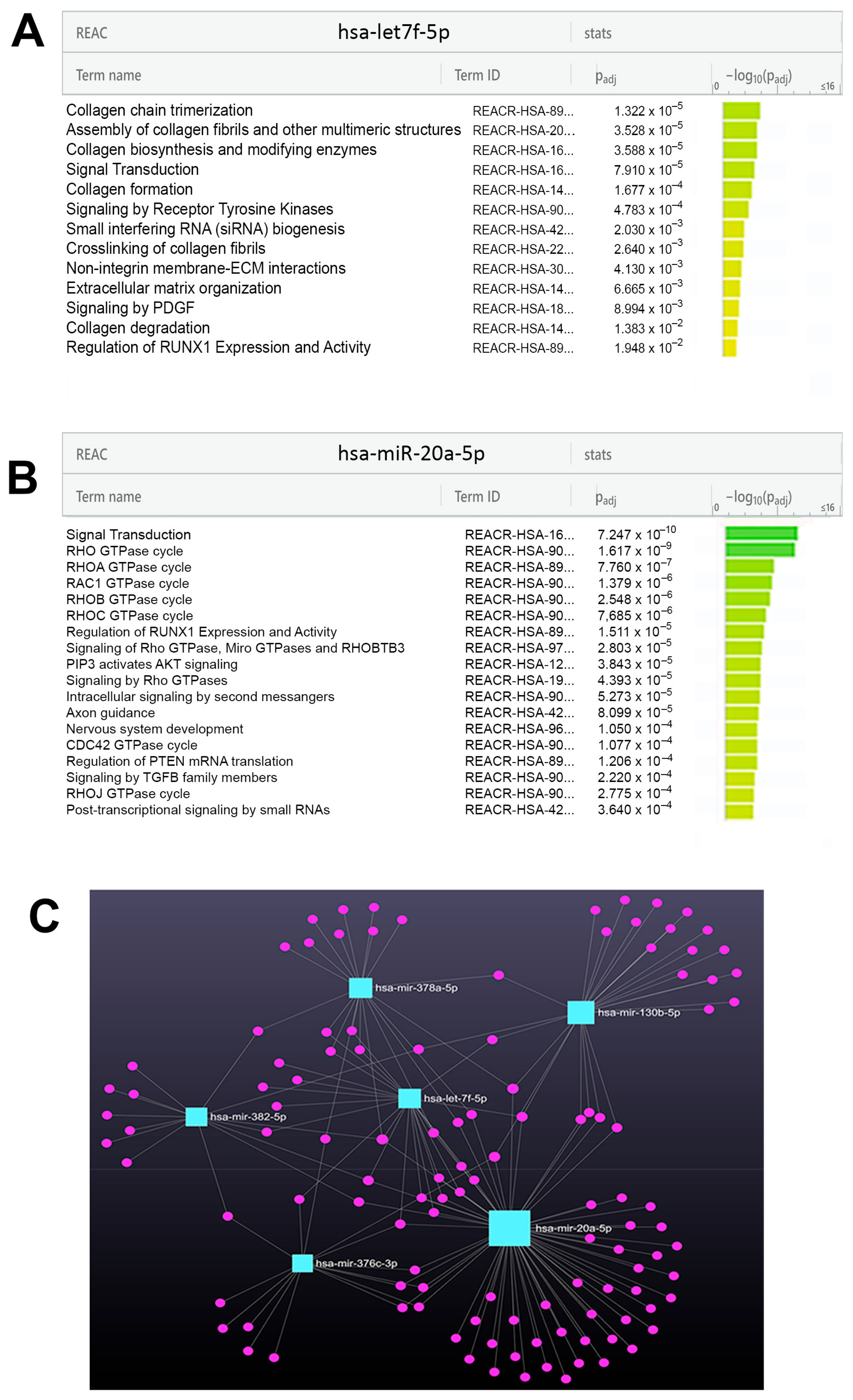
2.4. The Identification of miRNAs Differentially Expressed in Each LGMDR Pool and the Functional Enrichment of miRNA Targets
2.5. Validation and Specificity Evaluation of miRNA Signature Identification in LGMD Patients
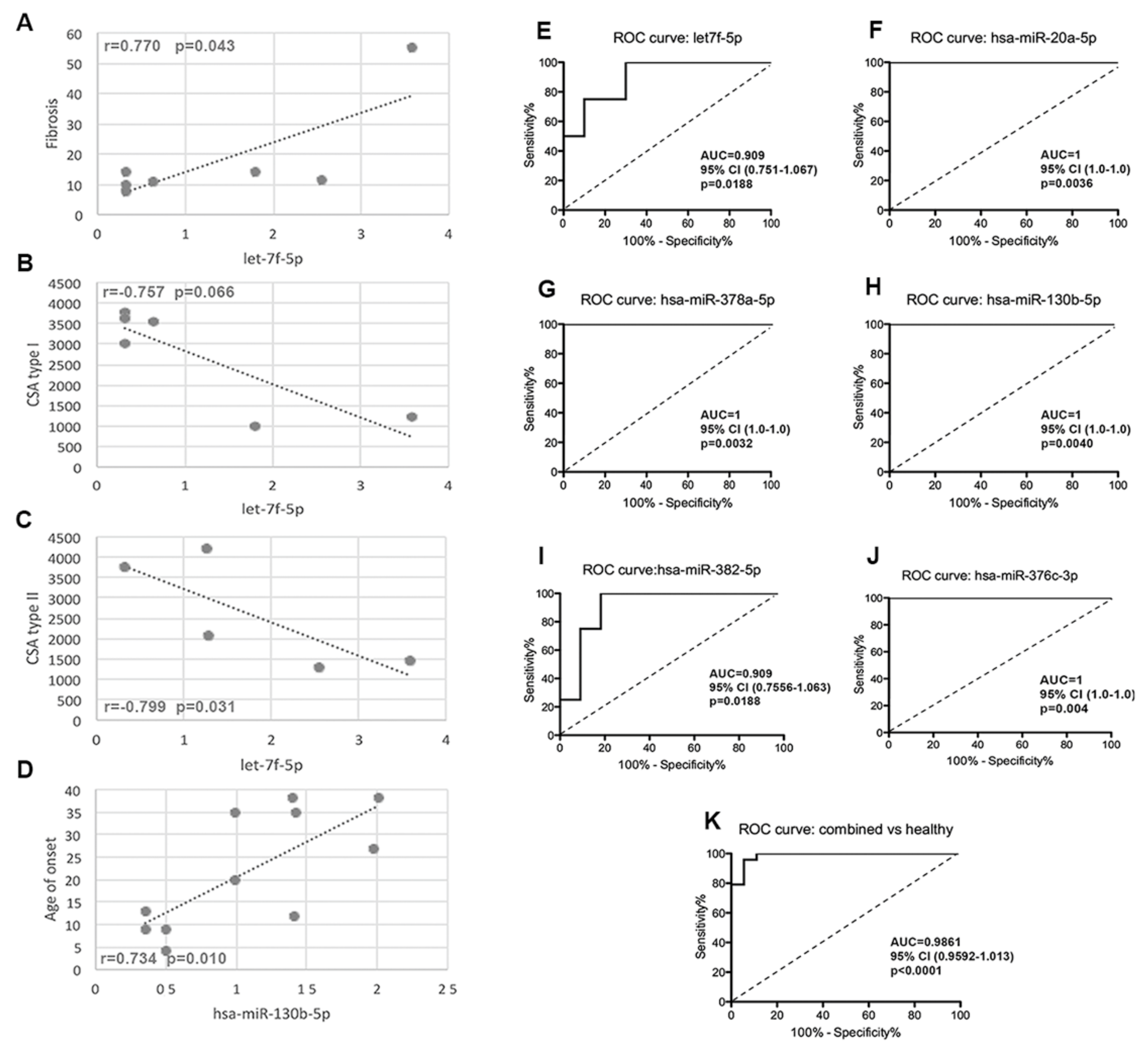
2.6. The Correlation of Circulating miRNAs with Demographic, Morphological and Functional Parameters
2.7. The Specificity of the miRNA Signature
3. Discussion
Study Limitation
4. Materials and Methods
4.1. Patients
4.2. Ethics Statement
4.3. Morphology
4.4. Serum Collection
4.5. RNA Isolation
4.6. cDNA Synthesis
4.7. RT-PCR Based miRNA Assays (Real Time PCR)
4.8. Real-Time Quantitative PCR (RT-qPCR)
4.9. Data Control Quality
4.10. Bioinformatics Tools
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Georganopoulou, D.G.; Moisiadis, V.G.; Malik, F.A.; Mohajer, A.; Dashevsky, T.M.; Wuu, S.T.; Hu, C.K. A Journey with LGMD: From Protein Abnormalities to Patient Impact. Protein J. 2021, 40, 466–488. [Google Scholar] [CrossRef] [PubMed]
- Paradas, C.; González-Quereda, L.; De Luna, N.; Gallardo, E.; García-Consuegra, I.; Gómez, H.; Cabello, A.; Illa, I.; Gallano, P. A new phenotype of dysferlinopathy with congenital onset. Neuromuscul. Disord. 2009, 19, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Barthel, B.L.; Cox, D.; Barbieri, M.; Ziemba, M.; Straub, V.; Hoffman, E.P.; Russell, A.J. Elevation of fast but not slow troponin I in the circulation of patients with Becker and Duchenne muscular dystrophy. Muscle Nerve 2021, 64, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, V.D.; van Putten, M.; Chaouch, A.; Garrood, P.; Straub, V.; Lochmüller, H.; Ginjaar, H.B.; Aartsma-Rus, A.M.; van Ommen, G.J.; den Dunnen, J.T.; et al. Serum matrix metalloproteinase-9 (MMP-9) as a biomarker for monitoring disease progression in Duchenne muscular dystrophy (DMD). Neuromuscul. Disord. 2011, 21, 569–578. [Google Scholar] [CrossRef]
- Burch, P.M.; Pogoryelova, O.; Goldstein, R.; Bennett, D.; Guglieri, M.; Straub, V.; Bushby, K.; Lochmüller, H.; Morris, C. Muscle-Derived Proteins as Serum Biomarkers for Monitoring Disease Progression in Three Forms of Muscular Dystrophy. J. Neuromuscul. Dis. 2015, 2, 241–255. [Google Scholar] [CrossRef]
- Rouillon, J.; Poupiot, J.; Zocevic, A.; Amor, F.; Léger, T.; Garcia, C.; Camadro, J.M.; Wong, B.; Pinilla, R.; Cosette, J.; et al. Serum proteomic profiling reveals fragments of MYOM3 as potential biomarkers for monitoring the outcome of therapeutic interventions in muscular dystrophies. Hum. Mol. Genet. 2015, 24, 4916–4932. [Google Scholar] [CrossRef]
- Magri, F.; Vanoli, F.; Corti, S. miRNA in spinal muscular atrophy pathogenesis and therapy. J. Cell Mol. Med. 2018, 22, 755–767. [Google Scholar] [CrossRef]
- García-Fonseca, Á.; Martin-Jimenez, C.; Barreto, G.E.; Pachón, A.F.A.; González, J. The Emerging Role of Long Non-Coding RNAs and MicroRNAs in Neurodegenerative Diseases: A Perspective of Machine Learning. Biomolecules 2021, 11, 1132. [Google Scholar] [CrossRef]
- Gentile, G.; Morello, G.; La Cognata, V.; Guarnaccia, M.; Conforti, F.L.; Cavallaro, S. Dysregulated miRNAs as Biomarkers and Therapeutical Targets in Neurodegenerative Diseases. J. Pers. Med. 2022, 12, 770. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Toden, S.; Goel, A. Non-coding RNAs as liquid biopsy biomarkers in cancer. Br. J. Cancer 2022, 126, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Langwiński, W.; Szczepankiewicz, D.; Narożna, B.; Stegmayr, J.; Wagner, D.; Alsafadi, H.; Lindstedt, S.; Stachowiak, Z.; Nowakowska, J.; Skrzypski, M.; et al. Allergic inflammation in lungs and nasal epithelium of rat model is regulated by tissue-specific miRNA expression. Mol. Immunol. 2022, 147, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Varzideh, F.; Kansakar, U.; Donkor, K.; Wilson, S.; Jankauskas, S.S.; Mone, P.; Wang, X.; Lombardi, A.; Santulli, G. Cardiac Remodeling After Myocardial Infarction: Functional Contribution of microRNAs to Inflammation and Fibrosis. Front. Cardiovasc. Med. 2022, 9, 863238. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, S.; Gibertini, S.; Curcio, M.; Savadori, P.; Pasanisi, B.; Morandi, L.; Cornelio, F.; Mantegazza, R.; Mora, M. Opposing roles of miR-21 and miR-29 in the progression of fibrosis in Duchenne muscular dystrophy. Biochim. Biophys. Acta 2015, 1852, 1451–1464. [Google Scholar] [CrossRef]
- Arrighi, N.; Moratal, C.; Savary, G.; Fassy, J.; Nottet, N.; Pons, N.; Clément, N.; Fellah, S.; Larrue, R.; Magnone, V.; et al. The FibromiR miR-214-3p Is Upregulated in Duchenne Muscular Dystrophy and Promotes Differentiation of Human Fibro-Adipogenic Muscle Progenitors. Cells 2021, 10, 1832. [Google Scholar] [CrossRef]
- Aránega, A.E.; Lozano-Velasco, E.; Rodriguez-Outeiriño, L.; Ramírez de Acuña, F.; Franco, D.; Hernández-Torres, F. MiRNAs and Muscle Regeneration: Therapeutic Targets in Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2021, 22, 4236. [Google Scholar] [CrossRef]
- Eisenberg, I.; Eran, A.; Nishino, I.; Moggio, M.; Lamperti, C.; Amato, A.A.; Lidov, H.G.; Kang, P.B.; North, K.N.; Mitrani-Rosenbaum, S.; et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. USA 2007, 104, 17016–17021. [Google Scholar] [CrossRef]
- Vignier, N.; Amor, F.; Fogel, P.; Duvallet, A.; Poupiot, J.; Charrier, S.; Arock, M.; Montus, M.; Nelson, I.; Richard, I.; et al. Distinctive serum miRNA profile in mouse models of striated muscular pathologies. PLoS ONE 2013, 8, e55281. [Google Scholar] [CrossRef]
- Zaharieva, I.T.; Calissano, M.; Scoto, M.; Preston, M.; Cirak, S.; Feng, L.; Collins, J.; Kole, R.; Guglieri, M.; Straub, V.; et al. Dystromirs as serum biomarkers for monitoring the disease severity in Duchenne muscular Dystrophy. PLoS ONE 2013, 8, e80263. [Google Scholar] [CrossRef]
- Lam, N.T.; Gartz, M.; Thomas, L.; Haberman, M.; Strande, J.L. Influence of microRNAs and exosomes in muscle health and diseases. J. Muscle Res. Cell Motil. 2020, 41, 269–284. [Google Scholar] [CrossRef]
- Gagliardi, D.; Rizzuti, M.; Brusa, R.; Ripolone, M.; Zanotti, S.; Minuti, E.; Parente, V.; Dioni, L.; Cazzaniga, S.; Bettica, P.; et al. MicroRNAs as serum biomarkers in Becker muscular dystrophy. J. Cell Mol. Med. 2022, 26, 4678–4685. [Google Scholar] [CrossRef]
- Chetta, A.; Zanini, A.; Pisi, G.; Aiello, M.; Tzani, P.; Neri, M.; Olivieri, D. Reference values for the 6-min walk test in healthy subjects 20-50 years old. Respir. Med. 2006, 100, 1573–1578. [Google Scholar] [CrossRef] [PubMed]
- Izumiya, Y.; Jinnn, M.; Kimura, Y.; Wang, Z.; Onoue, Y.; Hanatani, S.; Araki, S.; Ihn, H.; Ogawa, H. Expression of Let-7 family microRNAs in skin correlates negatively with severity of pulmonary hypertension in patients with systemic scleroderma. Int. J. Cardiol. Heart Vasc. 2015, 8, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Valle-Tenney, R.; Rebolledo, D.; Acuña, M.J.; Brandan, E. HIF-hypoxia signaling in skeletal muscle physiology and fibrosis. J. Cell Commun. Signal. 2020, 14, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Krist, B.; Florczyk, U.; Pietraszek-Gremplewicz, K.; Józkowicz, A.; Dulak, J. The Role of miR-378a in Metabolism, Angiogenesis, and Muscle Biology. Int. J. Endocrinol. 2015, 2015, 281756. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Li, H.; Zhang, B.; Li, C.; Dong, D.; Lan, X.; Huang, Y.; Bai, Y.; Lin, F.; Zhao, X.; et al. miR-378a-3p promotes differentiation and inhibits proliferation of myoblasts by targeting HDAC4 in skeletal muscle development. RNA Biol. 2016, 13, 1300–1309. [Google Scholar] [CrossRef]
- Ruckerl, D.; Jenkins, S.J.; Laqtom, N.N.; Gallagher, I.J.; Sutherland, T.E.; Duncan, S.; Buck, A.H.; Allen, J.E. Induction of IL-4Ralpha-dependent microRNAs identifies PI3K/Akt signaling as essential for IL-4-driven murine macrophage proliferation in vivo. Blood 2012, 120, 2307–2316. [Google Scholar] [CrossRef]
- Park, J.T.; Kato, M.; Lanting, L.; Castro, N.; Nam, B.Y.; Wang, M.; Kang, S.W.; Natarajan, R. Repression of let-7 by transforming growth factor-β1-induced Lin28 upregulates collagen expression in glomerular mesangial cells under diabetic conditions. Am. J. Physiol. Renal Physiol. 2014, 307, F1390–F1403. [Google Scholar] [CrossRef]
- Pandit, K.V.; Corcoran, D.; Yousef, H.; Yarlagadda, M.; Tzouvelekis, A.; Gibson, K.F.; Konishi, K.; Yousem, S.A.; Singh, M.; Handley, D.; et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2010, 182, 220–229. [Google Scholar] [CrossRef]
- Brennan, E.P.; Nolan, K.A.; Borgeson, E.; Gough, O.S.; McEvoy, C.M.; Docherty, N.G.; Higgins, D.F.; Murphy, M.; Sadlier, D.M.; Ali-Shah, S.T.; et al. Lipoxins attenuate renal fibrosis by inducing let-7c and suppressing TGFβR1. J. Am. Soc. Nephrol. 2013, 24, 627–637. [Google Scholar] [CrossRef]
- Wang, B.; Jha, J.C.; Hagiwara, S.; McClelland, A.D.; Jandeleit-Dahm, K.; Thomas, M.C.; Cooper, M.E.; Kantharidis, P. Transforming growth factor-β1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int. 2014, 85, 352–361. [Google Scholar] [CrossRef]
- Dangi-Garimella, S.; Strouch, M.J.; Grippo, P.J.; Bentrem, D.J.; Munshi, H.G. Collagen regulation of let-7 in pancreatic cancer involves TGF-β1-mediated membrane type 1-matrix metalloproteinase expression. Oncogene 2011, 30, 1002–1008. [Google Scholar] [CrossRef] [PubMed]
- Di, T.; Yang, Y.; Fu, C.; Zhang, Z.; Qin, C.; Sai, X.; Liu, J.; Hu, C.; Zheng, M.; Wu, Y.; et al. Let-7 mediated airway remodelling in chronic obstructive pulmonary disease via the regulation of IL-6. Eur. J. Clin. Investig. 2021, 51, e13425. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, J.S.; Hajira, A.; Lopez, M.A.; Boriek, A.M. Genome-wide Mechanosensitive MicroRNA (MechanomiR) Screen Uncovers Dysregulation of Their Regulatory Networks in the mdm Mouse Model of Muscular Dystrophy. J. Biol. Chem. 2015, 290, 24986–25011. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, S.; Gibertini, S.; Savadori, P.; Mantegazza, R.; Mora, M. Duchenne muscular dystrophy fibroblast nodules: A cell-based assay for screening anti-fibrotic agents. Cell Tissue Res. 2013, 352, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Qie, J.; Fu, Q.; Chen, J.; Jin, Y.; Ding, Z. Mir-20a-5p/tgfbr2 axis affects pro-inflammatory macrophages and aggravates liver fibrosis. Front. Oncol. 2020, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Selcen, D.; Stilling, G.; Engel, A.G. The earliest pathologic alterations in dysferlinopathy. Neurology 2001, 56, 1472–1481. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, M.; Sun, Y.; Li, X.; Cao, L.; Ma, X. Transforming growth factor-β receptor type 2 is required for heparin-binding protein-induced acute lung injury and vascular leakage for transforming growth factor-β/Smad/Rho signaling pathway activation. FASEB J. 2022, 36, e22580. [Google Scholar] [CrossRef]
- Guo, Q.; Zhu, X.; Wei, R.; Zhao, L.; Zhang, Z.; Yin, X.; Zhang, Y.; Chu, C.; Wang, B.; Li, X. MiR-130b-3p regulates M1 macrophage polarization via targeting IRF1. J. Cell Physiol. 2021, 236, 2008–2022. [Google Scholar] [CrossRef]
- Dahlmans, D.; Houzelle, A.; Andreux, P.; Wang, X.; Jörgensen, J.A.; Moullan, N.; Daemen, S.; Kersten, S.; Auwerx, J.; Hoeks, J. MicroRNA-382 silencing induces a mitonuclear protein imbalance and activates the mitochondrial unfolded protein response in muscle cells. J. Cell Physiol. 2019, 234, 6601–6610. [Google Scholar] [CrossRef]
- Aksu-Menges, E.; Akkaya-Ulum, Y.Z.; Dayangac-Erden, D.; Balci-Peynircioglu, B.; Yuzbasioglu, A.; Topaloglu, H.; Talim, B.; Balci-Hayta, B. The Common miRNA Signatures Associated with Mitochondrial Dysfunction in Different Muscular Dystrophies. Am. J. Pathol. 2020, 190, 2136–2145. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.J.; Kwon, E.S.; Lee, S.M.; Kim, S.K.; Min, K.W.; Lim, J.Y.; Lee, B.; Kang, J.S.; Kwak, J.Y.; Son, Y.H.; et al. A subset of microRNAs in the Dlk1-Dio3 cluster regulates age-associated muscle atrophy by targeting Atrogin-1. J. Cachexia Sarcopenia Muscle 2020, 11, 1336–1350. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.S.; Ahmad, K.; Lee, E.J.; Lee, Y.H.; Choi, I. Implications of Insulin-Like Growth Factor-1 in Skeletal Muscle and Various Diseases. Cells 2020, 9, 1773. [Google Scholar] [CrossRef] [PubMed]
- García-Giménez, J.L.; García-Trevijano, E.R.; Avilés-Alía, A.I.; Ibañez-Cabellos, J.S.; Bovea-Marco, M.; Bas, T.; Pallardó, F.V.; Viña, J.R.; Zaragozá, R. Identification of circulating miRNAs differentially expressed in patients with Limb-girdle, Duchenne or facioscapulohumeral muscular dystrophies. Orphanet J. Rare Dis. 2022, 17, 450. [Google Scholar] [CrossRef] [PubMed]
- Ripolone, M.; Velardo, D.; Mondello, S.; Zanotti, S.; Magri, F.; Minuti, E.; Cazzaniga, S.; Fortunato, F.; Ciscato, P.; Tiberio, F.; et al. Muscle histological changes in a large cohort of patients affected with Becker muscular dystrophy. Acta Neuropathol. Commun. 2022, 10, 48. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]




| Patients | LGMDR1 | LGMDR2 | LGMDR3 | LGMDR4 | Ctrl |
|---|---|---|---|---|---|
| (N = 4) | (N = 4) | (N = 4) | (N = 4) | (N = 8) | |
| Sex | 1F/3M | 3F/1M | 3F/1M | 1F/3M | 4F/4M |
| Age | 47.25 ± 15.51 | 38.25 ± 14.52 | 46.75 ± 16.25 | 19.50 ± 6.61 | 37.70 ± 0.63 |
| Age at onset (yrs) | 30.75 ± 7.88 | 20.75 ± 12.33 | 12.50 ± 5.44 | 7.25 ± 4.03 | NA |
| Disease duration (yrs) | 16.5 ± 10.37 | 17.25 ± 5.85 | 32.50 ± 17.74 | 9.0 ± 8.36 | NA |
| Morphology | |||||
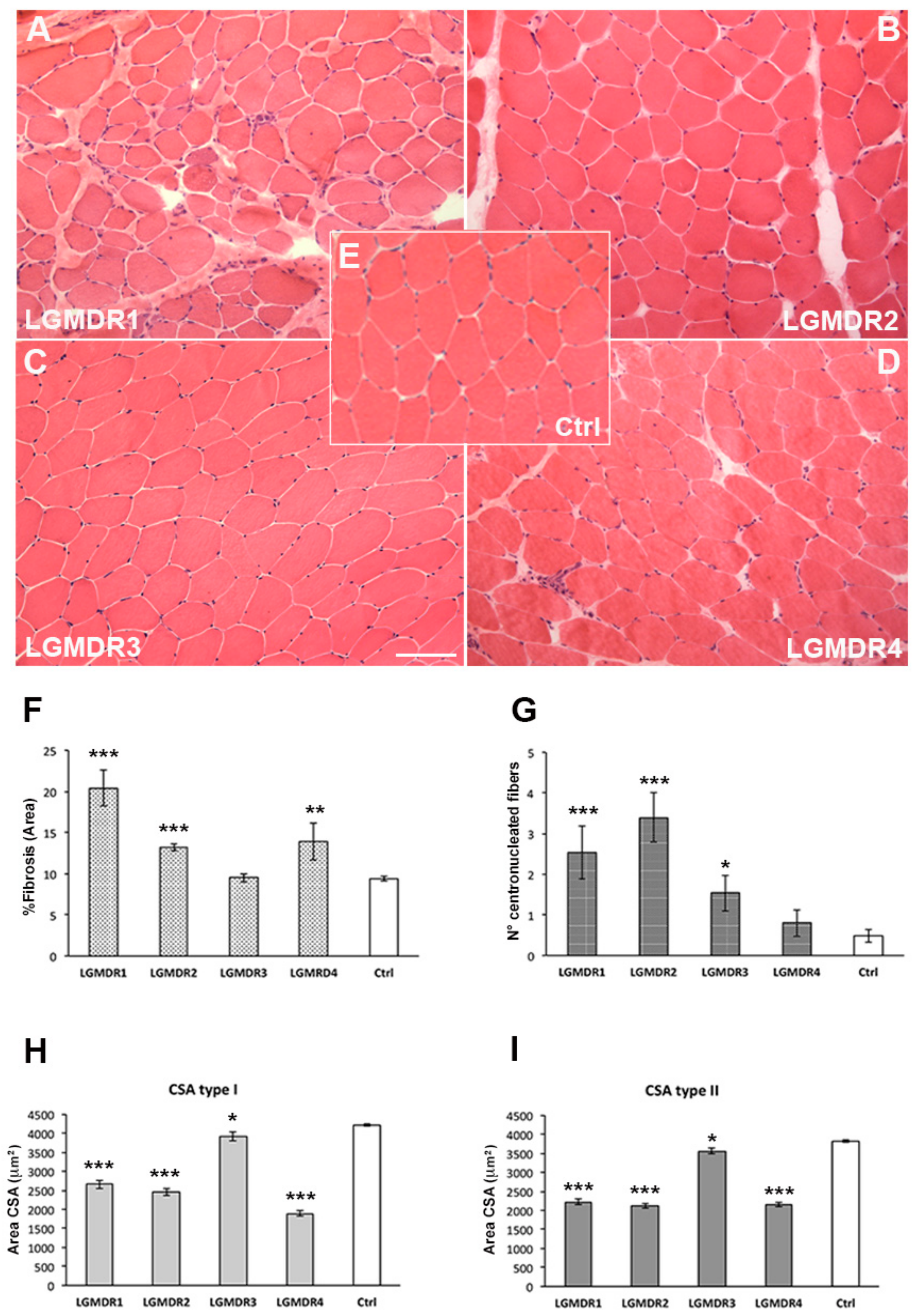
| Fibrosis (% area) | 20.44 ± 2.23 p < 0.0001 | 13.24 ± 0.45 p < 0.0001 | 9.49 ± 0.48 p = 0.8619 | 13.90 ± 2.22 p = 0.0002 | 9.39 ± 0.29 |
| Centronucleated Fiber | 2.53 ± 0.66 p < 0.0001 | 3.40 ± 0.61 p < 0.0001 | 1.53 ± 0.43 p = 0.0081 | 0.80 ± 1.03 p = 0.320 | 0.48 ± 0.16 |
| CSA type I (mm2) | 2657 ± 116 p < 0.0001 | 2451 ± 89 p < 0.0001 | 3920 ± 114 p = 0.0145 | 1893 ± 67 p < 0.0001 | 4215 ± 38 |
| CSA type II (mm2) | 2227 ± 72 p < 0.0001 | 2118 ± 69 p < 0.0001 | 3563 ± 80 p = 0.0019 | 2158 ± 58 p < 0.0001 | 3819 ± 36 |
| Clinical data | |||||
| CK (IU/L) | 1813 ± 550 | 931 ± 185 | 1996 ± 1457 | 6151 ± 1958 | 60-190 |
| Loss of ambulation | 1/4 | 1/4 | 1/4 | 2/4 | NA |
| Functional tests | Normal values | ||||
| MFM Dimension 1 (score) | 14.75 ± 15.79 | 23.25 ± 9.84 | 21.75 ± 16.45 | 17.25 ± 20.25 | 39 |
| MFM Dimension 2 (score) | 30.00 ± 5.16 | 30.00 ± 14.69 | 32.00 ± 6.73 | 19.00 ± 19.69 | 36 |
| MFM Dimension 3 (score) | 18.75 ± 1.79 | 18.25 ± 9.33 | 20.00 ± 1.15 | 13.00 ± 8.90 | 21 |
| MFM tot (score) | 63.50 ± 21.79 | 71.5 ± 30.50 | 73.75 ± 23.76 | 49.25 ± 48.50 | 96 |
| Six Minute Walking test (6-MWT) (meter) | 284.3 ± 141.8 (n = 3) | 381.0 ± 152.0 (n = 3) | 461.2 ± 168.8 (n = 3) | 530.5 ± 67.1 (n = 2) | 615.5 ± 31.8 (n = 102) [22] |
| NSAD | 21.00 ± 22.68 | 32.25 ± 15.41 | 31.00 ± 22.13 | 23.00 ± 27.00 | 54 |
| PUL | 27.25 ± 10.14 | 35.00 ± 15.15 | 34.00 ± 10.09 | 22.50 ± 20.85 | 42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magri, F.; Napoli, L.; Ripolone, M.; Ciscato, P.; Moggio, M.; Corti, S.; Comi, G.P.; Sciacco, M.; Zanotti, S. The Profiling of 179 miRNA Expression in Serum from Limb Girdle Muscular Dystrophy Patients and Healthy Controls. Int. J. Mol. Sci. 2023, 24, 17402. https://doi.org/10.3390/ijms242417402
Magri F, Napoli L, Ripolone M, Ciscato P, Moggio M, Corti S, Comi GP, Sciacco M, Zanotti S. The Profiling of 179 miRNA Expression in Serum from Limb Girdle Muscular Dystrophy Patients and Healthy Controls. International Journal of Molecular Sciences. 2023; 24(24):17402. https://doi.org/10.3390/ijms242417402
Chicago/Turabian StyleMagri, Francesca, Laura Napoli, Michela Ripolone, Patrizia Ciscato, Maurizio Moggio, Stefania Corti, Giacomo Pietro Comi, Monica Sciacco, and Simona Zanotti. 2023. "The Profiling of 179 miRNA Expression in Serum from Limb Girdle Muscular Dystrophy Patients and Healthy Controls" International Journal of Molecular Sciences 24, no. 24: 17402. https://doi.org/10.3390/ijms242417402
APA StyleMagri, F., Napoli, L., Ripolone, M., Ciscato, P., Moggio, M., Corti, S., Comi, G. P., Sciacco, M., & Zanotti, S. (2023). The Profiling of 179 miRNA Expression in Serum from Limb Girdle Muscular Dystrophy Patients and Healthy Controls. International Journal of Molecular Sciences, 24(24), 17402. https://doi.org/10.3390/ijms242417402









