The Effects of Larval Cryopreservation on the Epigenetics of the Pacific Oyster Crassostrea gigas
Abstract
:1. Introduction
2. Results
2.1. Comparison of Larval Performance in Control and Treatment Groups across Developmental Stages in C. gigas
2.2. Comparison of DNA Methylation and Gene Expressions of Epigenetic Regulators in Control and Treatment Groups across Developmental Stages in C. gigas
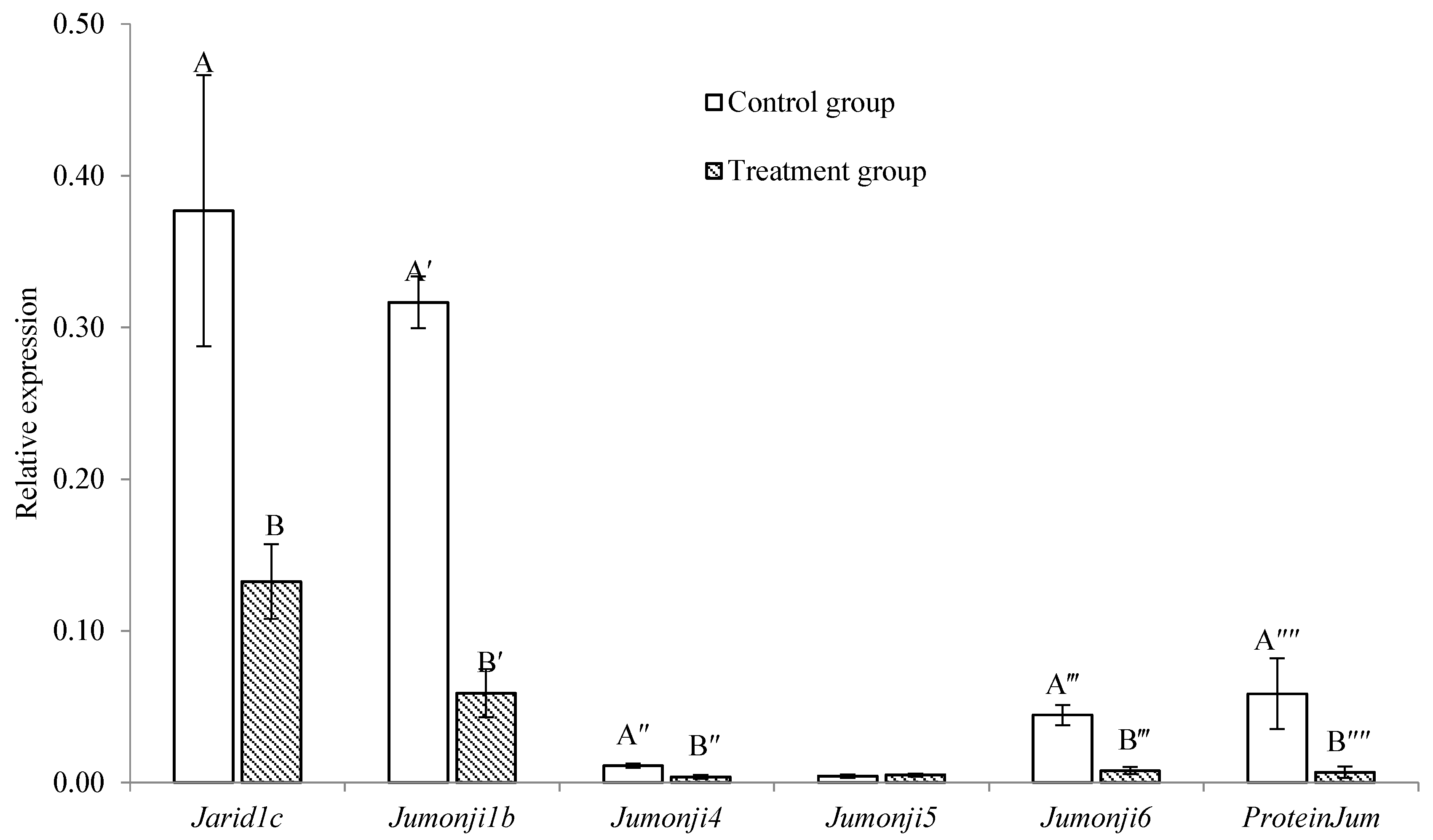
2.3. Comparison of the Expression of Jumonji Orthologs between Control and Treatment Groups in Trochophore Larvae
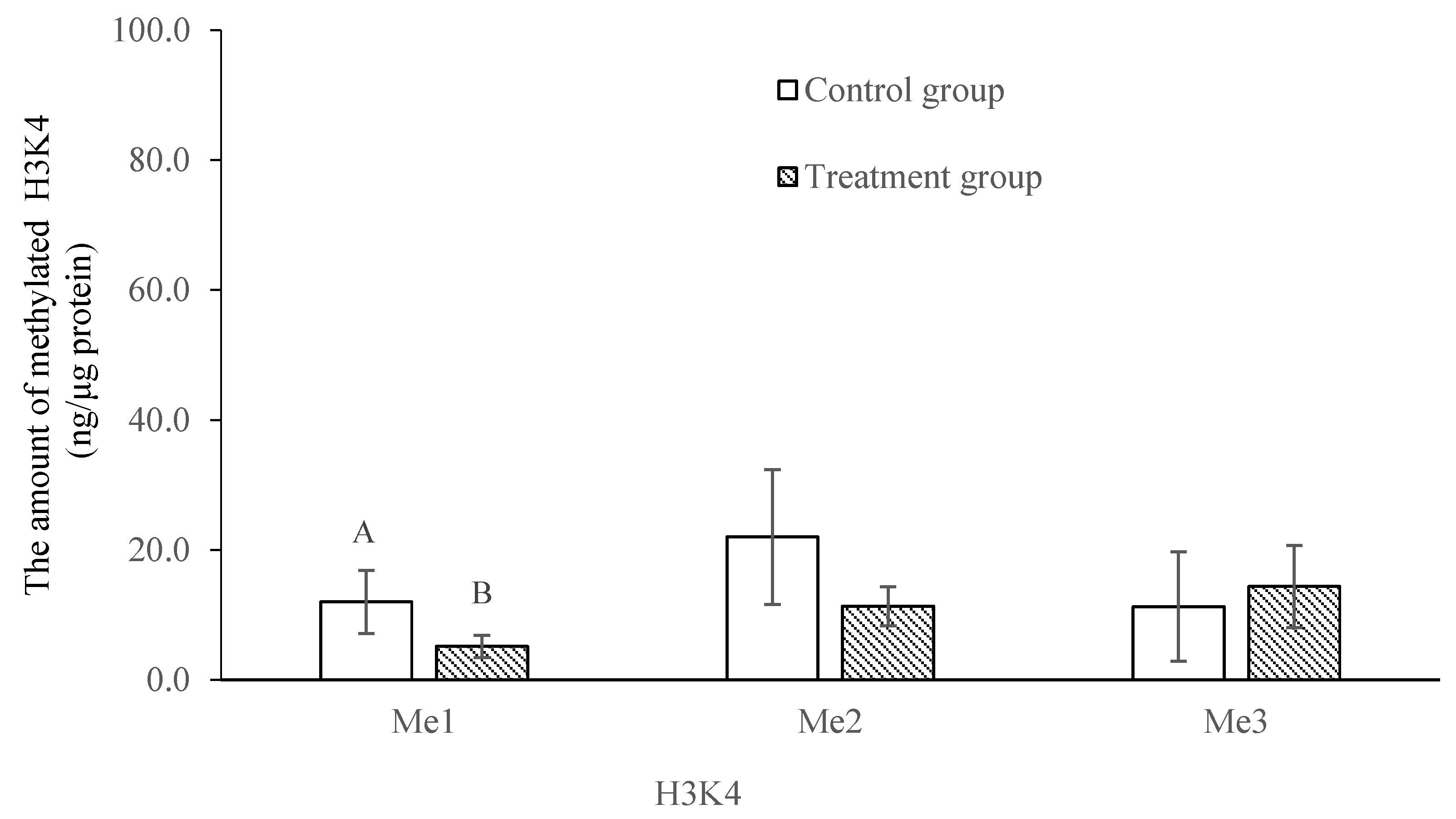
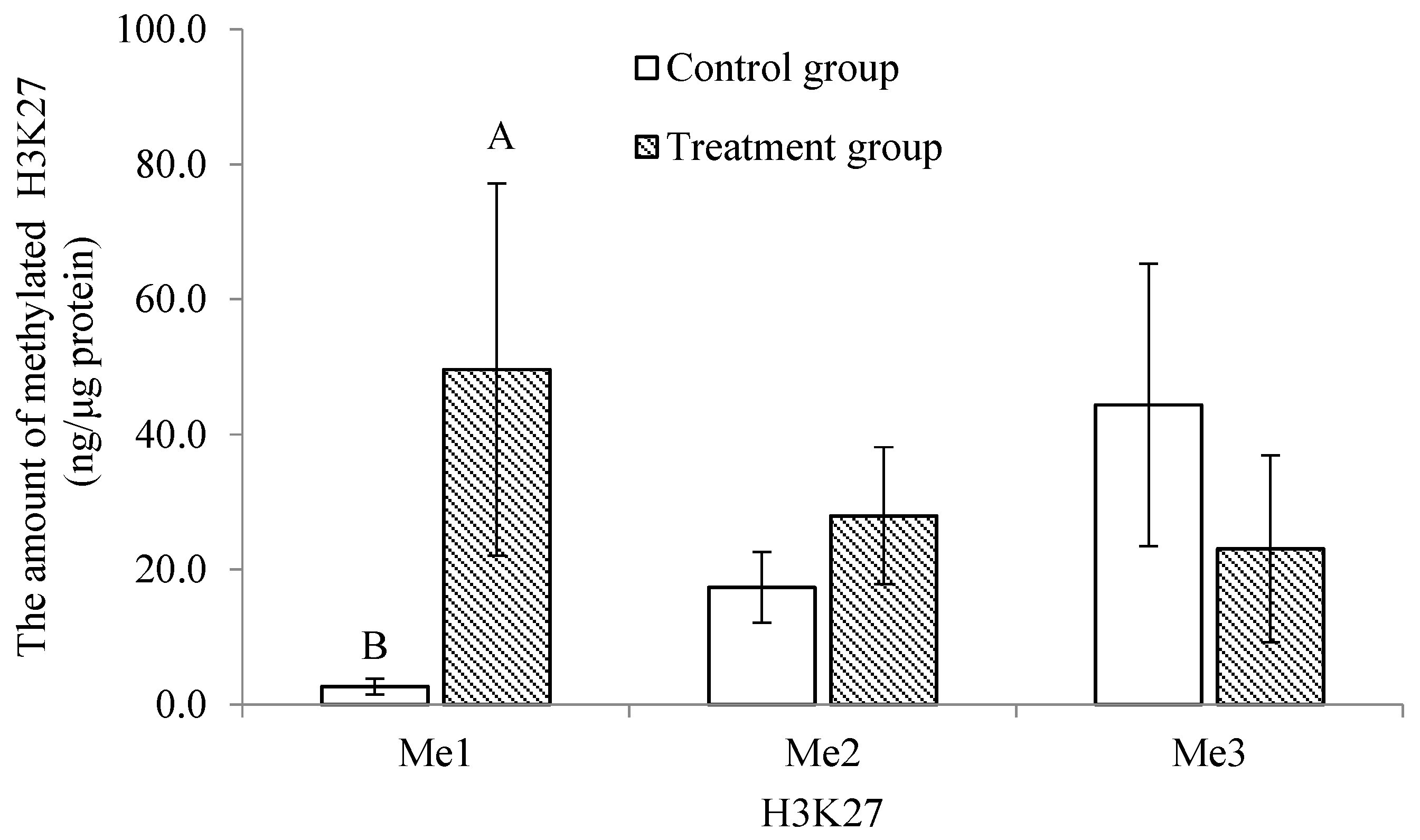
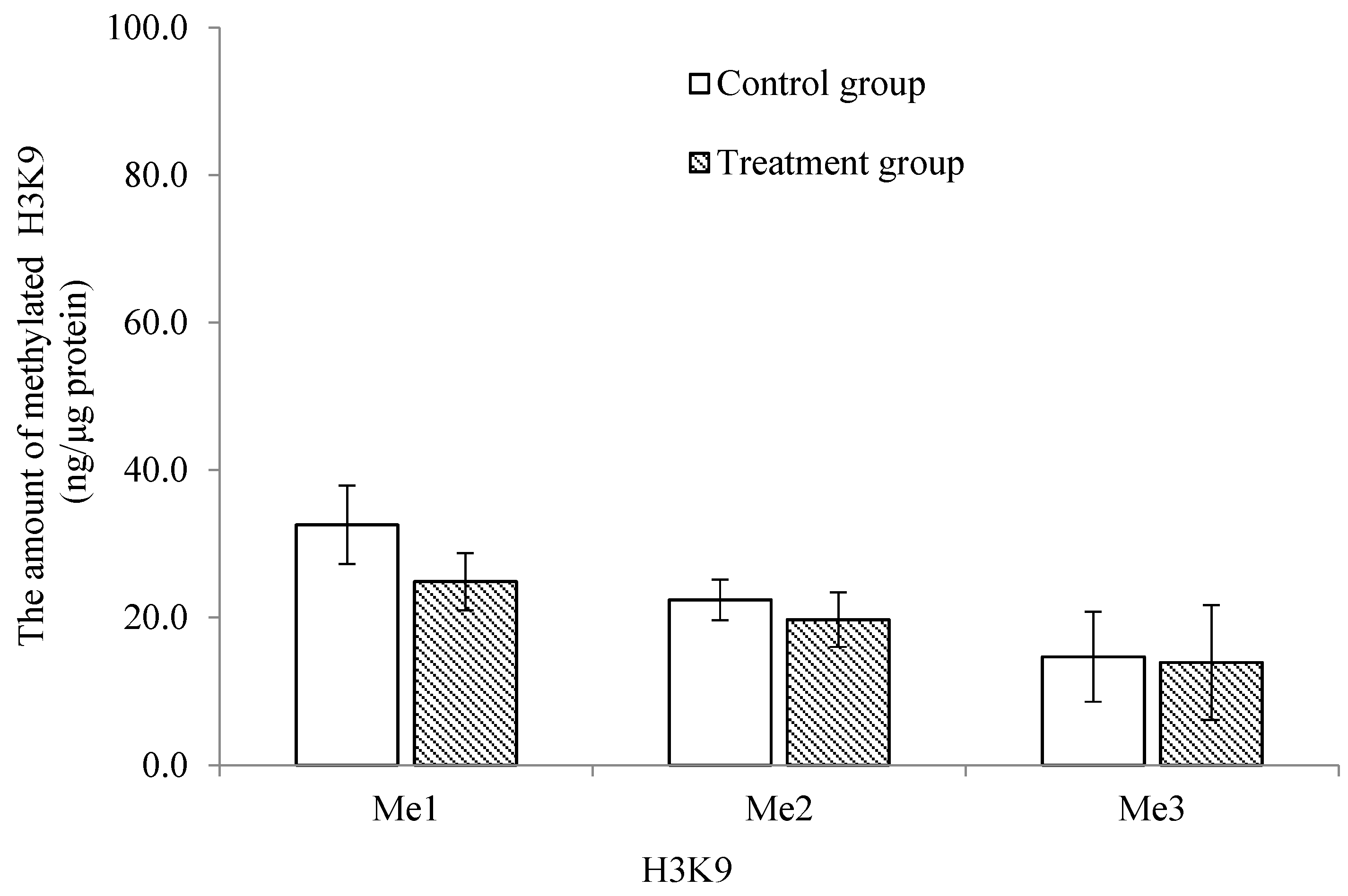
2.4. Comparison of the Mono-, Di- and Tri-Methylation Profiles between Control and Treatment Groups of Histone 3 at Lysines 4, 9 and 27 in Trochophore Larvae
3. Discussion
4. Materials and Methods
4.1. Larvae Preparation
4.2. Global DNA Methylation Quantification
4.3. Histone Extraction and Quantification
4.4. Histone Modification Quantification
4.5. RNA Extraction and cDNA Synthesis
4.6. Quantification of mRNA
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Botta, R.; Asche, F.; Borsum, J.S.; Camp, E.V. A review of global oyster aquaculture production and consumption. Mar. Policy 2020, 117, 103952. [Google Scholar] [CrossRef]
- Nascimento-Schulze, J.C.; Bean, T.P.; Houston, R.D.; Santos, E.M.; Sanders, M.B.; Lewis, C.; Ellis, R.P. Optimizing hatchery practices for genetic improvement of marine bivalves. Rev. Aquacult. 2021, 13, 2289–2304. [Google Scholar] [CrossRef]
- Hollenbeck, C.M.; Johnston, I.A. Genomics tools and selective breeding in molluscs. Front. Genet. 2018, 9, 253. [Google Scholar] [CrossRef] [PubMed]
- Ugalde, S.C.; Preston, J.; Ogier, E.; Crawford, C. Analysis of farm management strategies following herpesvirus (OsHV-1) disease outbreaks in Pacific oysters in Tasmania, Australia. Aquaculture 2018, 495, 179–186. [Google Scholar] [CrossRef]
- Azéma, P.; Lamy, J.; Boudry, P.; Renault, T.; Travers, M.; Dégremont, L. Genetic parameters of resistance to Vibrio aestuarianus, and OsHV-1 infections in the Pacific oyster, Crassostrea gigas, at three different life stages. Genet. Sel. Evol. 2017, 49, 23. [Google Scholar] [CrossRef]
- Delisle, L.; Laroche, O.; Hilton, Z.; Burguin, J.; Rolton, A.; Berry, J.; Pochon, X.; Boudry, P.; Vignier, J. Understanding the dynamic of POMS infection and the role of microbiota composition in the survival of Pacific oysters, Crassostrea gigas. Microbiol. Spectr. 2022, 10, e01959-22. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, J.P.; Hicks, M.B.R.; Hard, J.J.; Nichols, K.M.; Langdon, C.J.; Divilov, K.; Schoolfield, B.; Arkoosh, M.R. Heritability estimates of disease resistance to Vibrio coralliiyticus in Pacific oyster (Crassostrea gigas) larvae from a selective broodstock program. Aquaculture 2022, 560, 738492. [Google Scholar] [CrossRef]
- Martínez-Páramo, S.; Horváth, Á.; Labbé, C.; Zhang, T.; Robles, V.; Herráez, P.; Suquet, M.; Adams, S.; Viveiros, A.; Tiersch, T.R.; et al. Cryobanking of aquatic species. Aquaculture 2017, 472, 156–177. [Google Scholar] [CrossRef]
- Yang, H.; Huo, Y. Review of molluscan larval cryopreservation and application to germplasm cryobanking and commercial seed production. Aquaculture 2022, 547, 737491. [Google Scholar] [CrossRef]
- Labbé, C.; Haffray, P.; Mingant, C.; Quittet, B.; Diss, B.; Tervit, H.R.; Adams, S.L.; Rimond, F.; Suquet, M. Cryopreservation of Pacific oyster (Crassostrea gigas) larvae: Revisiting the practical limitations and scaling up the procedure for application to hatchery. Aquaculture 2018, 488, 227–234. [Google Scholar] [CrossRef]
- Liu, Y.; Gluis, M.; Miller-Ezzy, P.; Han, J.; Qin, J.; Zhan, X.; Li, X. Developmental of a programmable freezing technique on larval cryopreservation in the Pacific oyster Crassostrea gigas. Aquaculture 2020, 523, 735199. [Google Scholar] [CrossRef]
- Suneja, S.; Alfaro, A.C.; Rusk, A.B.; Morrish, J.R.; Tervit, H.R.; McGowan, L.T.; Adams, S.L. Multi-technique approach to characterise the effects of cryopreservation on larval development of the Pacific oyster (Crassostrea gigas). N. Z. J. Mar. Freshwater Res. 2014, 48, 335–349. [Google Scholar] [CrossRef]
- Rusk, A.B.; Alfaro, A.C.; Young, T.; Watts, E.; Adams, S.L. Development stage of cryopreserved mussel (Perna canaliculus) larvae influences post-thaw impact on shell formation, organogenesis, neurogenesis, feeding ability and survival. Cryobiology 2020, 93, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhan, X.; Catalano, S.R.; Qin, J.; Han, J.; Li, X. Investigation on redox status and gene expreesion related to larval cryopreservation in the Pacific oyster Crassostrea gigas. Fish Sci. 2022, 88, 377–386. [Google Scholar] [CrossRef]
- Barberet, J.; Barry, F.; Choux, C.; Guilleman, M.; Karoui, S.; Simonot, R.; Bruno, C.; Fauque, P. What impact does oocyte vitrification have on epigenetics and gene expression? Clin. Epigenet. 2020, 12, 121. [Google Scholar] [CrossRef] [PubMed]
- Salehi, M.; Mahdavi, A.H.; Sharafi, M.; Shahverdi, A. Cryopreservation of rooster semen: Evidence for the epigenetic modifications of thawed sperm. Theriogenology 2020, 142, 15–25. [Google Scholar] [CrossRef]
- Maldonado, M.B.C.; Penteado, J.C.T.; Faccio, B.M.C.; Lopes, F.L.; Arnold, D.R. Changes in tri-methylation profile of lysines 4 and 27 of histone H3 in bovine blastocysts after cryopreservation. Cryobiology 2015, 71, 481–485. [Google Scholar] [CrossRef]
- Gavery, M.R.; Roberts, S.B. Epigenetic considerations in aquaculture. PeerJ 2017, 5, e4147. [Google Scholar] [CrossRef]
- Jambhekar, A.; Dhall, A.; Shi, Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 2019, 20, 625–641. [Google Scholar] [CrossRef]
- Fellous, A.; Favrel, P.; Riviere, G. Temperature influences histone methylation and mRNA expression of the Jmj-C histone-demethylase orthologues during the early development of the oyster Crassostrea gigas. Mar. Genom. 2015, 19, 23–30. [Google Scholar] [CrossRef]
- Chatterjee, A.; Saha, D.; Niemann, H.; Gryshkov, O.; Glasmacher, B.; Hofmann, N. Effects of cryopreservation on the epigenetic profile of cells. Cryobiology 2017, 74, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yan, J.; Qiao, J.; Zhao, P.; Liu, P. Effects of oocyte vitrification on histone modifications. Reprod. Fertil. Dev. 2010, 22, 920–925. [Google Scholar] [CrossRef] [PubMed]
- Moulavi, F.; Saadeldin, I.M.; Swelum, A.A.; Tasighi, F.; Hosseini-Fahraji, H.; Hosseini, S.M. Oocyte vitrification induces loss of DNA methylation and histone acetylation in the resulting embryos derived using ICSI in dromedary camel. Zygote 2021, 29, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Riviere, G.; Wu, G.; Fellous, A.; Goux, D.; Sourdaine, P.; Favrel, P. DNA methylation is crucial for the early development in the oyster C. gigas. Mar. Biotechnol. 2013, 15, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Fellous, A.; Le Franc, L.; Jouaux, A.; Goux, D.; Favrel, P.; Rivière, G. Histone methylation participates in gene expression control during the early development of the Pacific oyster Crassostrea gigas. Gene 2019, 10, 695. [Google Scholar] [CrossRef]
- Gavery, M.R.; Roberts, S.B. DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (Crassostrea gigas). BMC Genom. 2010, 11, 483. [Google Scholar] [CrossRef]
- Riviere, G.; He, Y.; Tecchio, S.; Crowell, E.; Gras, M.; Sourdaine, P.; Guo, X.; Favrel, P. Dynamics of DNA methylomes underlie oyster development. PLoS Genet. 2017, 13, e1006807. [Google Scholar] [CrossRef]
- Fellous, A.; Favrel, P.; Guo, X.; Riviere, G. The Jumonji gene family in Crassostrea gigas suggests evolutionary conservation of Jmj-C histone demethylasese orthologues in the oyster gametogenesis and development. Gene 2014, 538, 164–175. [Google Scholar] [CrossRef]
- Rivière, G. Epigenetic features in the oyster Crassostrea gigas suggestive of functionally relevant promoter DNA methylation in invertebrates. Front. Physiol. 2014, 5, 129. [Google Scholar]
- Boyes, J.; Bird, A. Repression of genes by DNA methylation depends on CpG density and promoter strength: Evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992, 11, 327–333. [Google Scholar] [CrossRef]
- Suzuki, M.M.; Kerr, A.R.W.; Sousa, D.; Bird, A. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 2007, 17, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Tweedie, S.; Charlton, J.; Clark, V.; Bird, A. Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol. Cell. Biol. 1997, 17, 1469–1475. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Cheung, K.; Dang, X.; Roberts, S.B.; Wang, X.; Thiyagarajan, V. DNA methylation changes in response to ocean acidification at the time of larval metamorphosis in the edible oyster, Crassostrea hongkongensis. Mar. Environ. Res. 2021, 163, 105214. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, L.; Deng, T.; Zou, P.; Wang, Y.; Quan, F.; Zhang, Y. Effects of oocyte vitrification on epigenetic status in early bovine embryos. Theriogenology 2016, 86, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Fu, X.; Li, J.; Yuan, D.; Zhu, S. DNA methylation pattern in mouse oocytes and their in vitro fertilized early embryos: Effect of oocyte vitrification. Zygote 2014, 22, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.R.; Yarychkivska, O.; Boulard, M.; Bestor, T.H. DNA methylation and DNA methyltransferases. Epigenetics Chromatin 2017, 10, 23. [Google Scholar] [CrossRef]
- Brero, A.; Easwaran, H.P.; Nowak, D.; Grunewald, I.; Cremer, T.; Leonhardt, H.; Cardoso, M.C. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 2005, 169, 733–743. [Google Scholar] [CrossRef]
- Cross, I.; Portela-Bens, S.; García-Angulo, A.; Merlo, M.A.; Rodríguez, M.E.; Liehr, T.; Rebordinos, L. A preliminary integrated genetic map distinguishes every chromosome pair and locates essential genes related to abiotic adaptation of Crassostrea angulate gigas. BMC Genet. 2018, 19, 104. [Google Scholar] [CrossRef]
- Han, G.; Li, J.; Wang, Y.; Li, X.; Mao, H.; Liu, Y.; Chen, C.D. The hydroxylation activity of Jmjd6 is required for its homo-olimomerization. J. Cell. Biochem. 2012, 113, 1663–1670. [Google Scholar] [CrossRef]
- Zheng, Y.; Hsu, F.; Xu, W.; Xie, X.; Ren, X.; Gao, X.; Ni, J.; Ji, Y. A developmental genetic analysis of the lysine demethylase KDM2 mutations in Drosophila melanogaster. Mech. Dev. 2014, 133, 36–53. [Google Scholar] [CrossRef]
- Takeuchi, T.; Watanabe, Y.; Takano-Shimizu, T.; Kondo, S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev. Dyn. 2006, 255, 2449–2459. [Google Scholar] [CrossRef] [PubMed]
- Euskirchen, G.; Auerbach, R.K.; Snyder, M. SWI/SNF chromatin-remodeling factors: Multiscale analyses and diverse functions. J. Biol. Chem. 2012, 287, 30897–30905. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.W.M.; Orkin, S.H. The SWI/SNF complex-chromatin and cancer. Nat. Rev. Cancer. 2004, 4, 133–142. [Google Scholar] [CrossRef]
- Turberfield, A.H.; Kondo, T.; Nakayama, M.; Koseki, Y.; King, H.W.; Koseki, H.; Klose, R.J. KDM2 proteins constrain transcription from CpG island gene promoters independently of their histone demethylase activity. Nucleic Acids Res. 2019, 47, 9005–9023. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; Calcar, S.V.; Qu, C.; Ching, K.A.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007, 39, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef]
- Bernstein, B.; Kamal, M.; Lindblad-Toh, K.; Bekiranov, S.; Bailery, D.K.; Huebert, D.J.; McMahon, S.; Karlsson, E.K.; Kulbokas, E.J.; Gingeras, T.R.; et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 2005, 120, 169–181. [Google Scholar] [CrossRef]
- Spinaci, M.; Vallorani, C.; Bucci, D.; Tamanini, C.; Porcu, E.; Galeati, G. Vitrification of pig oocytes induces changes in histone H4 acetylation and histone H3 lysine 9 methylation (H3K9). Vet. Res. Commun. 2012, 36, 165–171. [Google Scholar] [CrossRef]
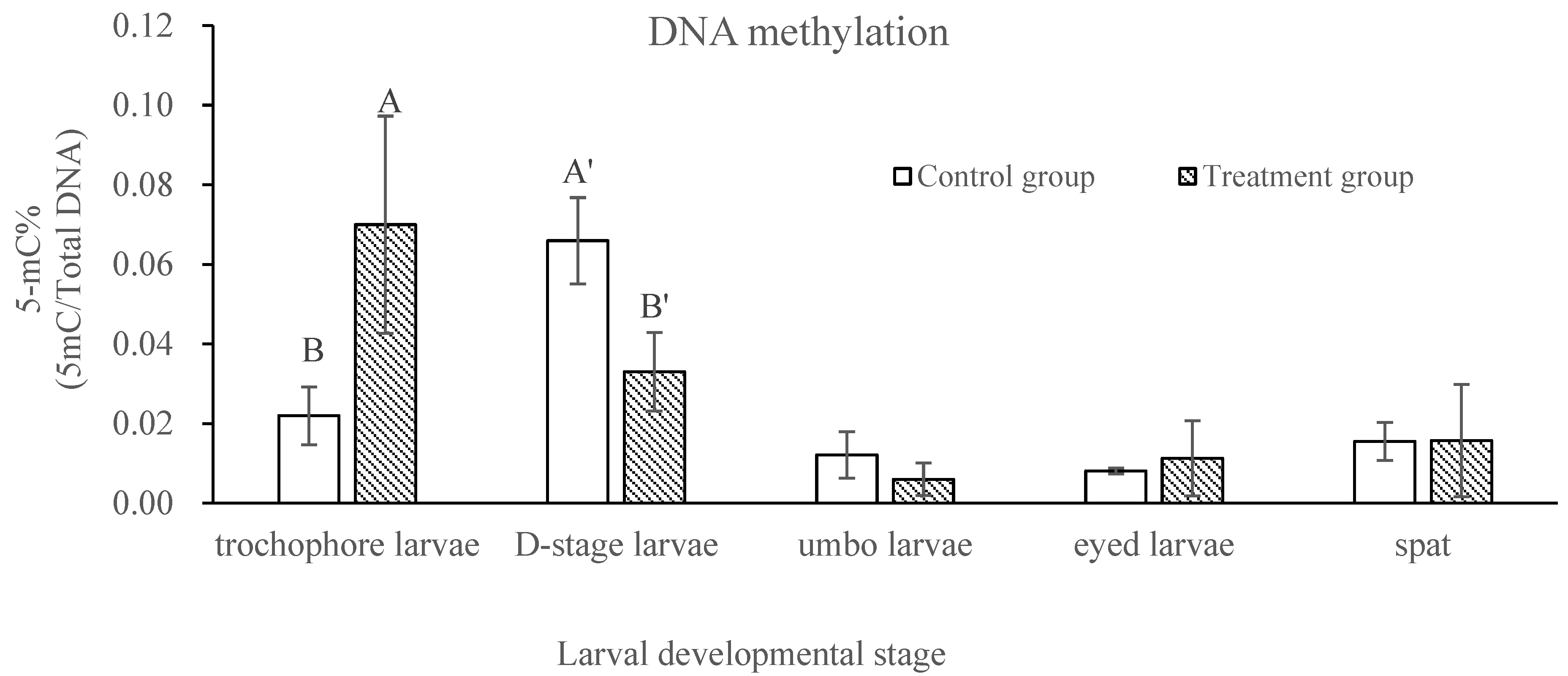
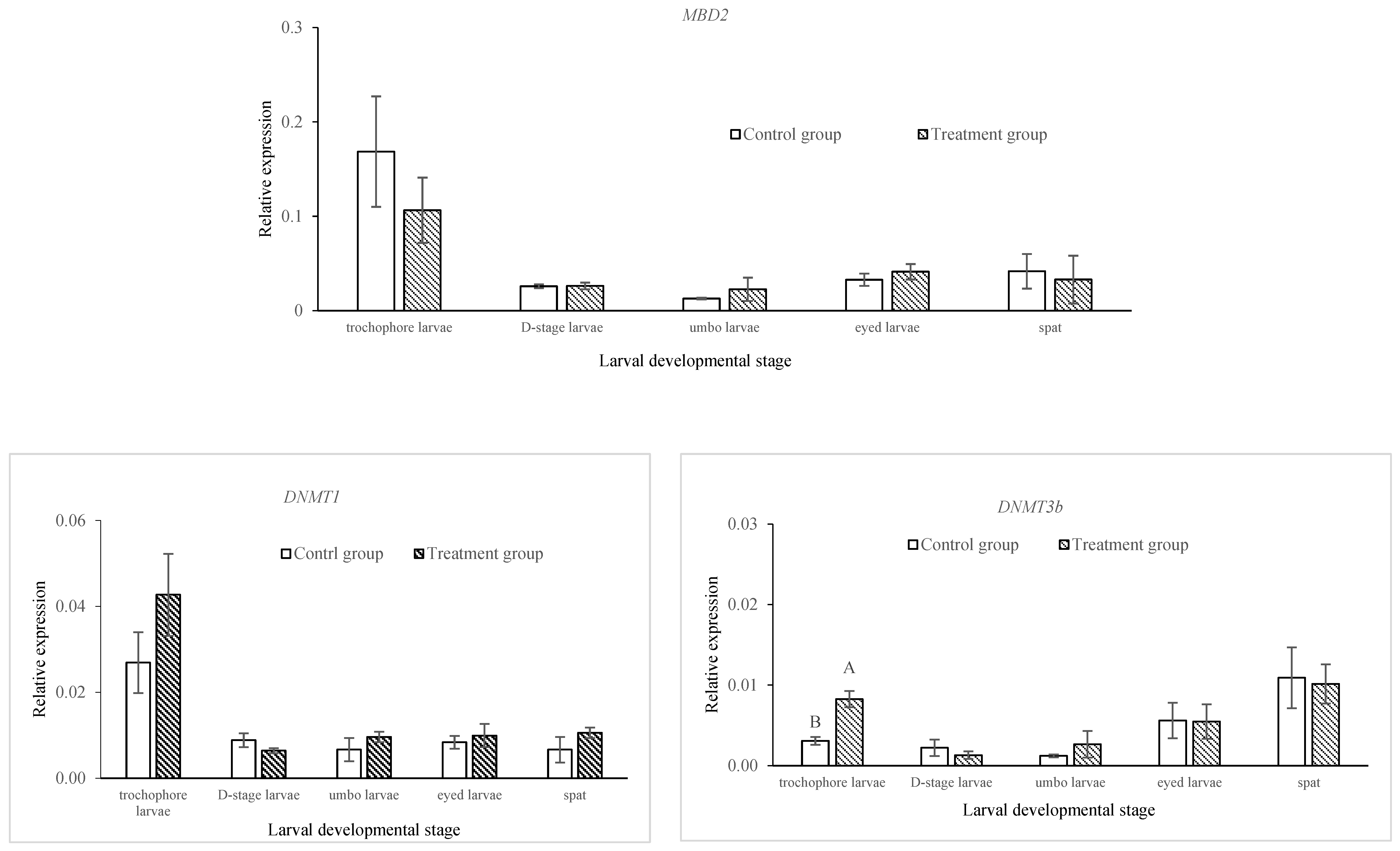
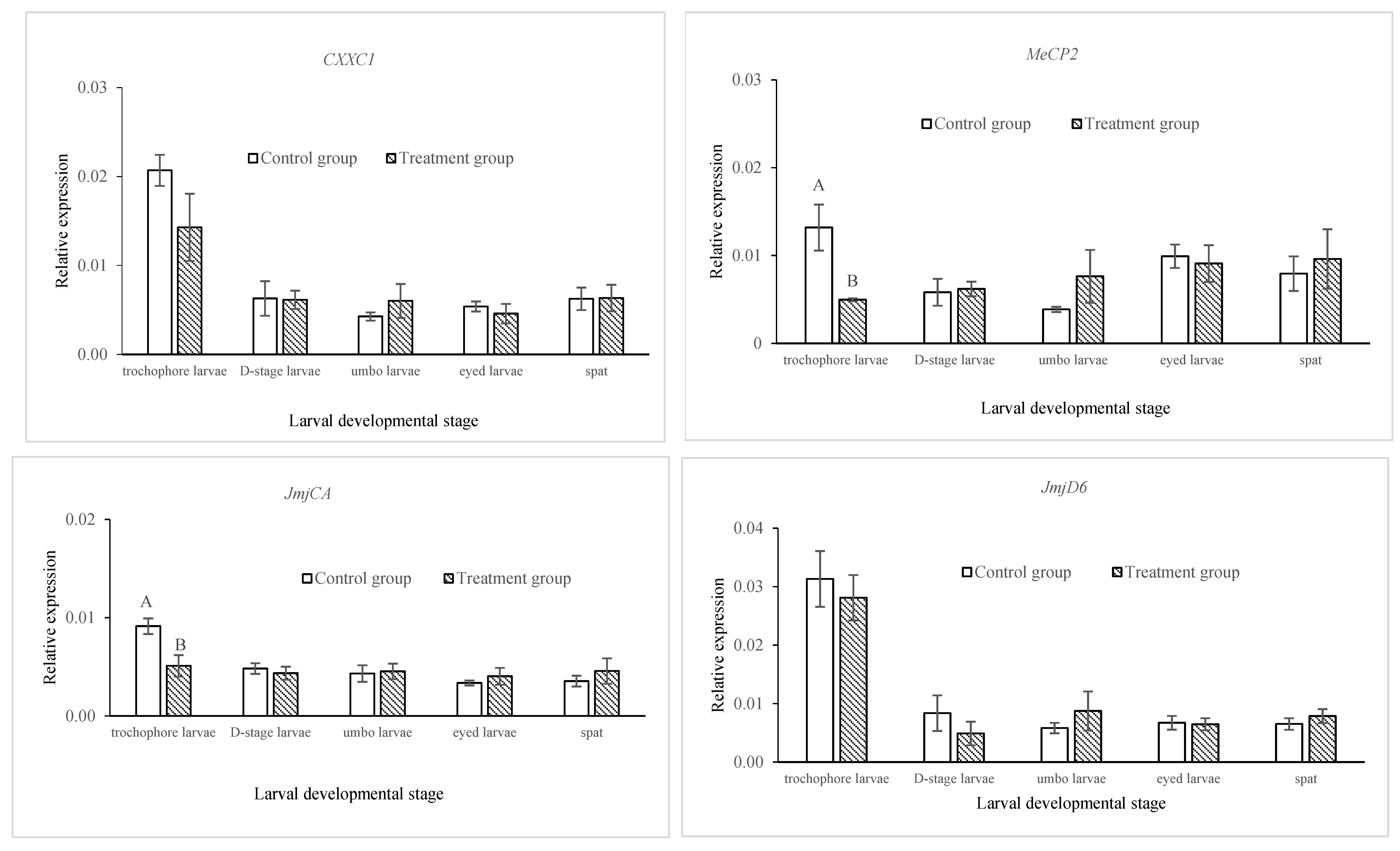
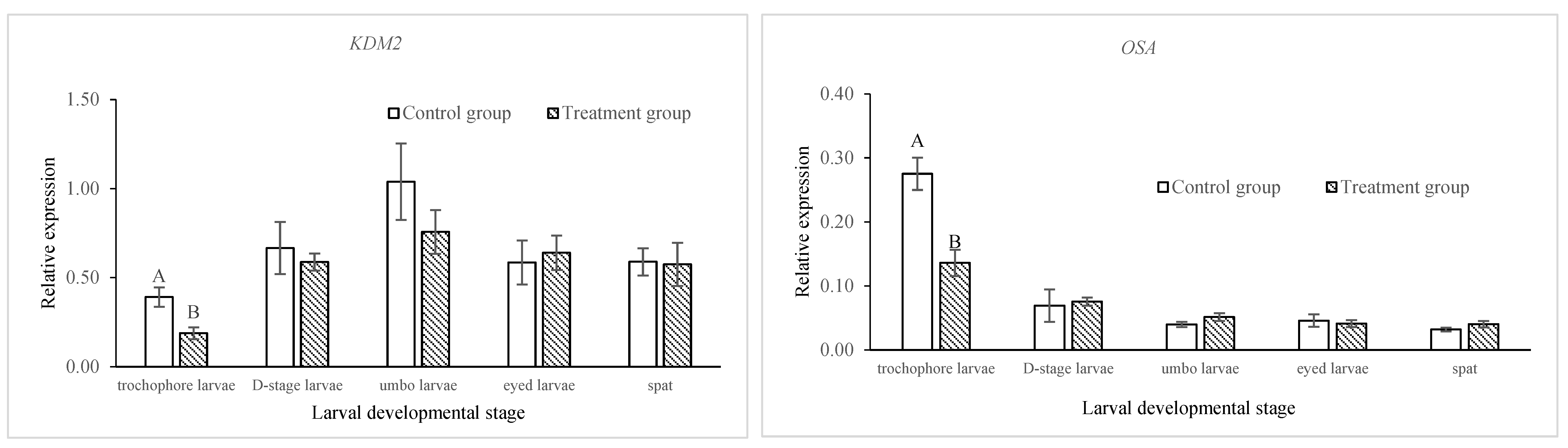
| Days Post-Fertilization | Survival Rate (%) | Relative Survival Rate (%) | Shell Length (µm) | |||
|---|---|---|---|---|---|---|
| Control Group | Treatment Group | Control Group | Treatment Group | Control Group | Treatment Group | |
| Day 1 (D-stage larvae) | 81.7 ± 5.8 | 61.3 ± 3.1 * | 81.8 ± 3.5 | 80.2 ± 2.4 | ||
| Day 8 (umbo larvae) | 51.3 ± 7.2 | 20.3 ± 4.5 * | 62.6 ± 4.6 | 33.3 ± 7.7 * | 105.8 ± 3.2 | 103.8 ± 4.2 |
| Day 22 (eyed larvae) | 31.7 ± 9.3 | 7.7 ± 3.8 * | 63.8 ± 25.5 | 38.4 ± 19.0 | 277.0 ± 12.3 | 272.0 ± 7.8 |
| Day 27 (spat) | 15.7 ± 6.0 | 4.7 ± 1.2 * | 49.4 ± 10.9 | 65.6 ± 15.0 | 465.9 ± 30.0 | 455.7 ± 40.9 |
| Gene | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|
| DNA methylation machinery | ||
| DNMT1 | TTGGCAACATTCTGGACAAA | CGGTCTTCCATTCCAGTGAC |
| DNMT3b | TCTCTCAAGCAGGGGAGAAA | TGCTCTGGAAACCCAAAGAC |
| MBD2 | TGACTTCCGCAGTGGTAGAA | ACTGTCGTGCCTCATTCCTC |
| MeCP2 | ATGCAACCCTCAACCCAATA | GCCAAACTCATCGCCTGTAT |
| CXXC1 | CGGCAAGATGCACAGTAGAA | CGGTTCATGATTGGTTGTGA |
| Histone modifier | ||
| OSA | AACGAGATTGAGGGATGCTG | CGAGTTTGCTCTCGTTCTCC |
| JmjCA | TTCCGAATAGCATCCAAAGG | CCGGATCAAATAGCACCACT |
| JmjD6 | CAGTTTGCTGGGGAGAGAAG | TGGTTCCTAGAGGGTCGATG |
| KDM2 | TGTGTGGGAGAGTCTGGTGA | ATGCCAAAGGACCTGACAGT |
| Jumonji orthologue | ||
| Jarid 1c | TCGCAGTGGATGTGGATAAA | ACCTAGCAAGCTGGTCCAAA |
| Jumonji 1b | CCCAGAACACCTGAACCACT | CTGTCCCAGCACTCACTGAA |
| Jumonji 4 | CAGCACAACCGAAGGAAGAT | AGCCGCAAGGAGTCTCATAA |
| Jumonji 5 | CCTGGACAAATACCAGAAGGAG | ATCTAGACCGTCGTTGTGTAGGAC |
| Jumonji 6 | GGTTGGAGGTCAGCTTTCAG | CTGGGCAGTTCATCCATTCT |
| Protein Jumonji | CCGAGAGCCTAATGACGAAG | GGCACAATGACCTTGACCTT |
| Reference gene | ||
| EFα | ACCACCCTGGTGAGATCAAG | ACGACGATCGCATTTCTCTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Bao, L.; Catalano, S.R.; Zhu, X.; Li, X. The Effects of Larval Cryopreservation on the Epigenetics of the Pacific Oyster Crassostrea gigas. Int. J. Mol. Sci. 2023, 24, 17262. https://doi.org/10.3390/ijms242417262
Liu Y, Bao L, Catalano SR, Zhu X, Li X. The Effects of Larval Cryopreservation on the Epigenetics of the Pacific Oyster Crassostrea gigas. International Journal of Molecular Sciences. 2023; 24(24):17262. https://doi.org/10.3390/ijms242417262
Chicago/Turabian StyleLiu, Yibing, Lisui Bao, Sarah R. Catalano, Xiaochen Zhu, and Xiaoxu Li. 2023. "The Effects of Larval Cryopreservation on the Epigenetics of the Pacific Oyster Crassostrea gigas" International Journal of Molecular Sciences 24, no. 24: 17262. https://doi.org/10.3390/ijms242417262
APA StyleLiu, Y., Bao, L., Catalano, S. R., Zhu, X., & Li, X. (2023). The Effects of Larval Cryopreservation on the Epigenetics of the Pacific Oyster Crassostrea gigas. International Journal of Molecular Sciences, 24(24), 17262. https://doi.org/10.3390/ijms242417262







