Knockouts of TNFRSF1A and TNFRSF1B Genes in K562 Cell Line Lead to Diverse Long-Lasting Responses to TNF-α
Abstract
:1. Introduction
2. Results
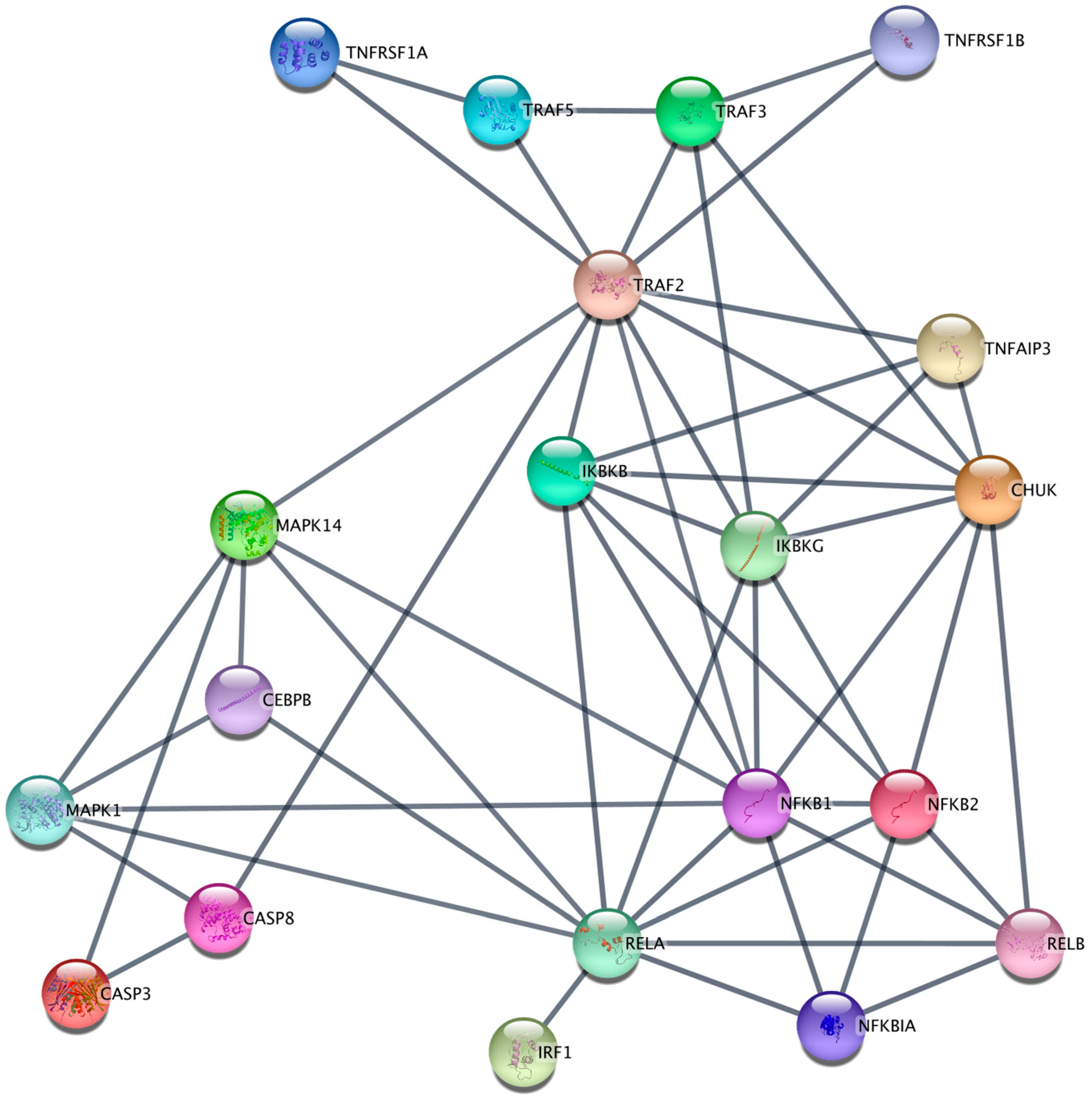
2.1. Transcriptomic Analysis of TNFR1 and TNFR2 Signaling Pathways in the K562 Cell Line
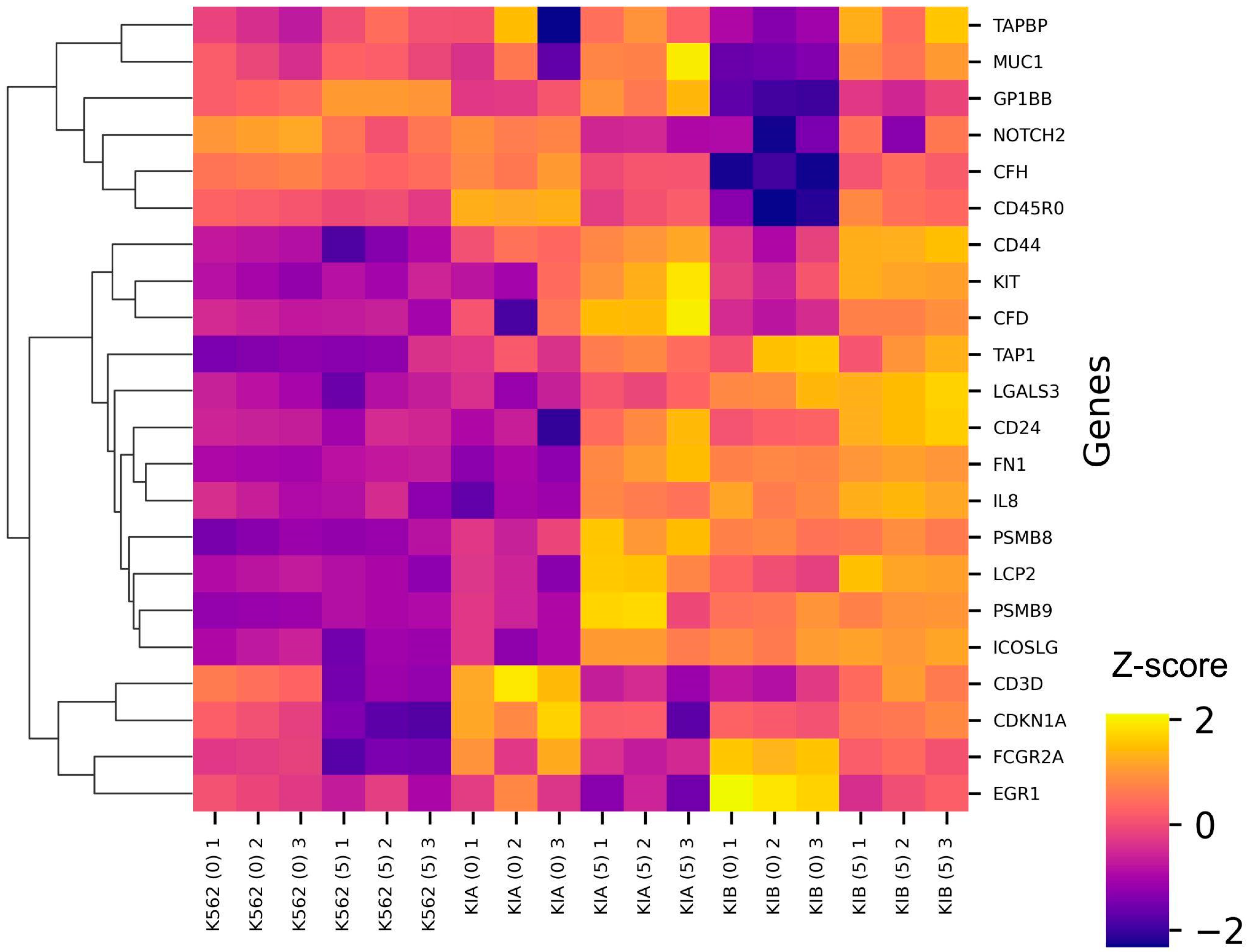
2.2. Transcriptomic Analysis of K562 with TNFR1 and TNFR2 Knockouts and TNF-α Addition
3. Discussion
4. Materials and Methods
4.1. Cell Culture
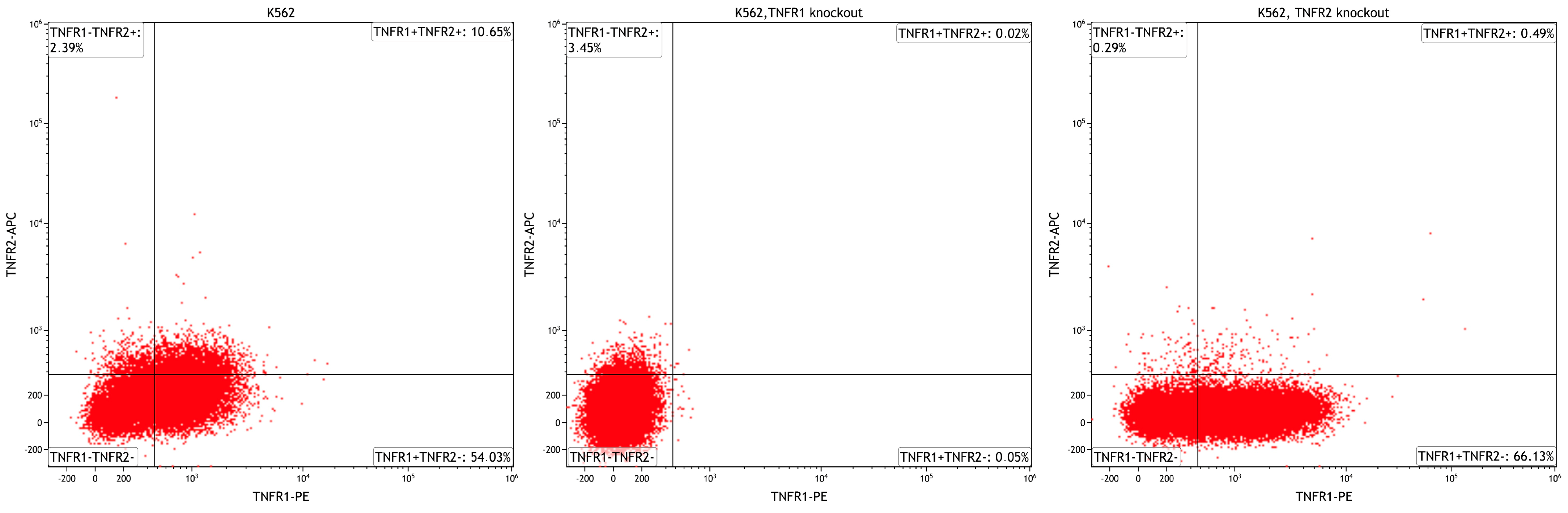
4.2. TNFRSF1A and TNFRSF1B Gene Knockouts
4.3. TNF-α Co-Culturing
4.4. Total RNA Extraction
4.5. NanoString RNA Profiling
4.6. TNFR1 and TNFR2 Signaling Gene Network Reconstruction for K562
4.7. Differential Gene Expression Testing
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salomon, B.L. Insights into the biology and therapeutic implications of TNF and regulatory T cells. Nat. Rev. Rheumatol. 2021, 17, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Siegmund, D. TNFR1 and TNFR2 in the Control of the Life and Death Balance of Macrophages. Front. Cell Dev. Biol. 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, J.; Brand, D.D.; Zheng, S.G. Role of TNF–TNF receptor 2 signal in regulatory T cells and its therapeutic implications. Front. Immunol. 2018, 9, 784. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Kontermann, R.E.; Pfizenmaier, K. Selective targeting of TNF receptors as a novel therapeutic approach. Front. Cell Dev. Biol. 2020, 8, 401. [Google Scholar] [CrossRef] [PubMed]
- Leone, G.M.; Mangano, K.; Petralia, M.C.; Nicoletti, F.; Fagone, P. Past, Present and (Foreseeable) Future of Biological Anti-TNF Alpha Therapy. J. Clin. Med. 2023, 12, 1630. [Google Scholar] [CrossRef]
- Medler, J.; Kucka, K.; Wajant, H. Tumor necrosis factor receptor 2 (TNFR2): An emerging target in cancer therapy. Cancers 2022, 14, 2603. [Google Scholar] [CrossRef]
- Sennikov, S.V.; Alshevskaya, A.A.; Zhukova, J.; Belomestnova, I.; Karaulov, A.V.; Lopatnikova, J.A. Expression density of receptors as a potent regulator of cell function and property in health and pathology. Int. Arch. Allergy Immunol. 2019, 178, 182–191. [Google Scholar] [CrossRef]
- Alshevskaya, A.; Koneva, O.; Belomestnova, I.; Lopatnikova, J.; Evsegneeva, I.; Zhukova, J.; Kireev, F.; Karaulov, A.; Sennikov, S. Ligand-regulated expression of TNF receptors 1 and 2 determines receptor-mediated functional responses. Int. Arch. Allergy Immunol. 2021, 182, 1077–1088. [Google Scholar] [CrossRef]
- Sander, C.A.; Rush, E.A.; Shi, J.; Arantes, L.M.; Tesi, R.J.; Ross, M.A.; Calderon, M.J.; Watkins, S.C.; Kirkwood, J.M.; Ferris, R.L.; et al. Co-expression of TNF receptors 1 and 2 on melanomas facilitates soluble TNF-induced resistance to MAPK pathway inhibitors. J. Transl. Med. 2022, 20, 331. [Google Scholar] [CrossRef]
- Gough, P.; Myles, I.A. Tumor necrosis factor receptors: Pleiotropic signaling complexes and their differential effects. Front. Immunol. 2020, 11, 585880. [Google Scholar] [CrossRef]
- Jang, D.I.; Lee, A.H.; Shin, H.Y.; Song, H.R.; Park, J.H.; Kang, T.B.; Lee, S.R.; Yang, S.H. The role of tumor necrosis factor alpha (TNF-α) in autoimmune disease and current TNF-α inhibitors in therapeutics. Int. J. Mol. Sci. 2021, 22, 2719. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.H.; Huber, E.C.; Sachs, J.N. Conformational states of TNFR1 as a molecular switch for receptor function. Protein Sci. 2020, 29, 1401–1415. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xie, C.; Chen, Y.; Wang, J.; Chen, X.; Lu, Z.; June, R.R.; Zheng, S.G. Differential roles of TNFα-TNFR1 and TNFα-TNFR2 in the differentiation and function of CD4+ Foxp3+ induced Treg cells in vitro and in vivo periphery in autoimmune diseases. Cell Death Dis. 2019, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, T.; Perkins, N.D.; Wilson, C.L. NFKB1: A suppressor of inflammation, ageing and cancer. FEBS J. 2016, 283, 1812–1822. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.; Mukherjee, T.; Chatterjee, B.; Vijayaragavan, B.; Banoth, B.; Basak, S. Non-canonical NFκB mutations reinforce pro-survival TNF response in multiple myeloma through an autoregulatory RelB: p50 NFκB pathway. Oncogene 2017, 36, 1417–1429. [Google Scholar] [CrossRef]
- Kunsch, C.; Rosen, C.A. NF-kappa B subunit-specific regulation of the interleukin-8 promoter. Mol. Cell. Biol. 1993, 13, 6137–6146. [Google Scholar] [CrossRef] [PubMed]
- Hopkins-Donaldson, S.; Bodmer, J.L.; Bourloud, K.B.; Brognara, C.B.; Tschopp, J.; Gross, N. Loss of caspase-8 expression in highly malignant human neuroblastoma cells correlates with resistance to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 2000, 60, 4315–4319. [Google Scholar]
- Osawa, Y.; Nagaki, M.; Banno, Y.; Brenner, D.A.; Asano, T.; Nozawa, Y.; Moriwaki, H.; Nakashima, S. Tumor necrosis factor alpha-induced interleukin-8 production via NF-κB and phosphatidylinositol 3-kinase/Akt pathways inhibits cell apoptosis in human hepatocytes. Infect. Immun. 2002, 70, 6294–6301. [Google Scholar] [CrossRef]
- Gil, A.; Aguilera, C.M.; Gil-Campos, M.; Canete, R. Altered signalling and gene expression associated with the immune system and the inflammatory response in obesity. Br. J. Nutr. 2007, 98, S121–S126. [Google Scholar] [CrossRef]
- Zhang, Y.; Harada, A.; Bluethmann, H.; Wang, J.B.; Nakao, S.; Mukaida, N.; Matsushima, K. Tumor necrosis factor (TNF) is a physiologic regulator of hematopoietic progenitor cells: Increase of early hematopoietic progenitor cells in TNF receptor p55-deficient mice in vivo and potent inhibition of progenitor cell proliferation by TNF alpha in vitro. Blood 1995, 86, 2930–2937. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network analysis and visualization of proteomics data. J. Proteome Res. 2018, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Liu, X.; Peltz, G. GSEApy: A comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics 2023, 39, btac757. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perik-Zavodskaia, O.; Alrhmoun, S.; Perik-Zavodskii, R.; Zhukova, J.; Lopatnikova, J.; Volynets, M.; Alshevskaya, A.; Sennikov, S. Knockouts of TNFRSF1A and TNFRSF1B Genes in K562 Cell Line Lead to Diverse Long-Lasting Responses to TNF-α. Int. J. Mol. Sci. 2023, 24, 17169. https://doi.org/10.3390/ijms242417169
Perik-Zavodskaia O, Alrhmoun S, Perik-Zavodskii R, Zhukova J, Lopatnikova J, Volynets M, Alshevskaya A, Sennikov S. Knockouts of TNFRSF1A and TNFRSF1B Genes in K562 Cell Line Lead to Diverse Long-Lasting Responses to TNF-α. International Journal of Molecular Sciences. 2023; 24(24):17169. https://doi.org/10.3390/ijms242417169
Chicago/Turabian StylePerik-Zavodskaia, Olga, Saleh Alrhmoun, Roman Perik-Zavodskii, Julia Zhukova, Julia Lopatnikova, Marina Volynets, Alina Alshevskaya, and Sergey Sennikov. 2023. "Knockouts of TNFRSF1A and TNFRSF1B Genes in K562 Cell Line Lead to Diverse Long-Lasting Responses to TNF-α" International Journal of Molecular Sciences 24, no. 24: 17169. https://doi.org/10.3390/ijms242417169
APA StylePerik-Zavodskaia, O., Alrhmoun, S., Perik-Zavodskii, R., Zhukova, J., Lopatnikova, J., Volynets, M., Alshevskaya, A., & Sennikov, S. (2023). Knockouts of TNFRSF1A and TNFRSF1B Genes in K562 Cell Line Lead to Diverse Long-Lasting Responses to TNF-α. International Journal of Molecular Sciences, 24(24), 17169. https://doi.org/10.3390/ijms242417169





