Advances in Detection Techniques for the H5N1 Avian Influenza Virus
Abstract
1. Introduction
2. Virus Isolation and Identification
3. Serological Detection Technology
3.1. HA and HI Test
3.2. Enzyme-Linked Immunosorbent Assays (ELISA)
4. Immunological Detection Technology
4.1. Collagen Gold Immunochromatography Technology
4.2. Fluorescence Immunodetection Technique
5. Molecular Biological Detection Technology
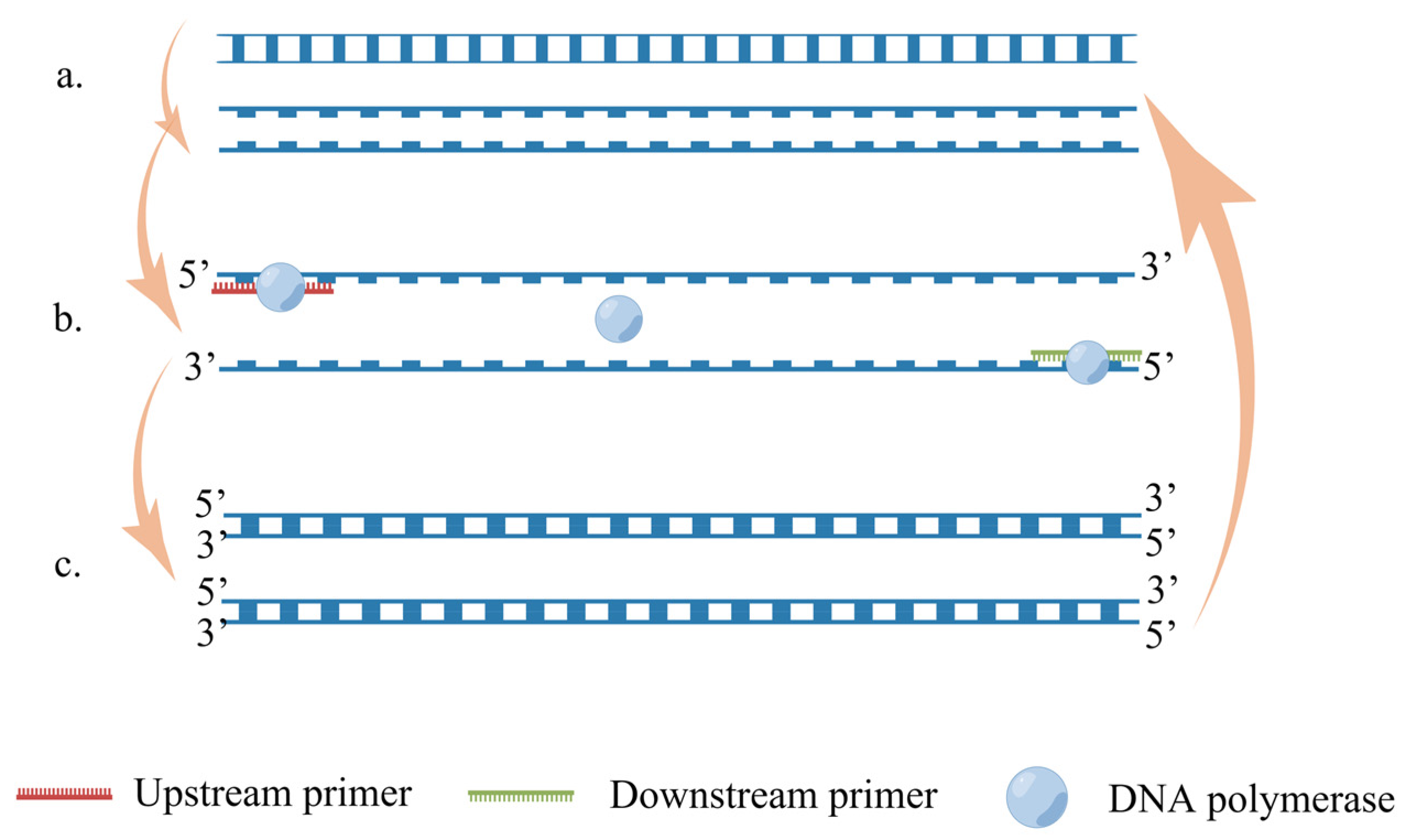
5.1. Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
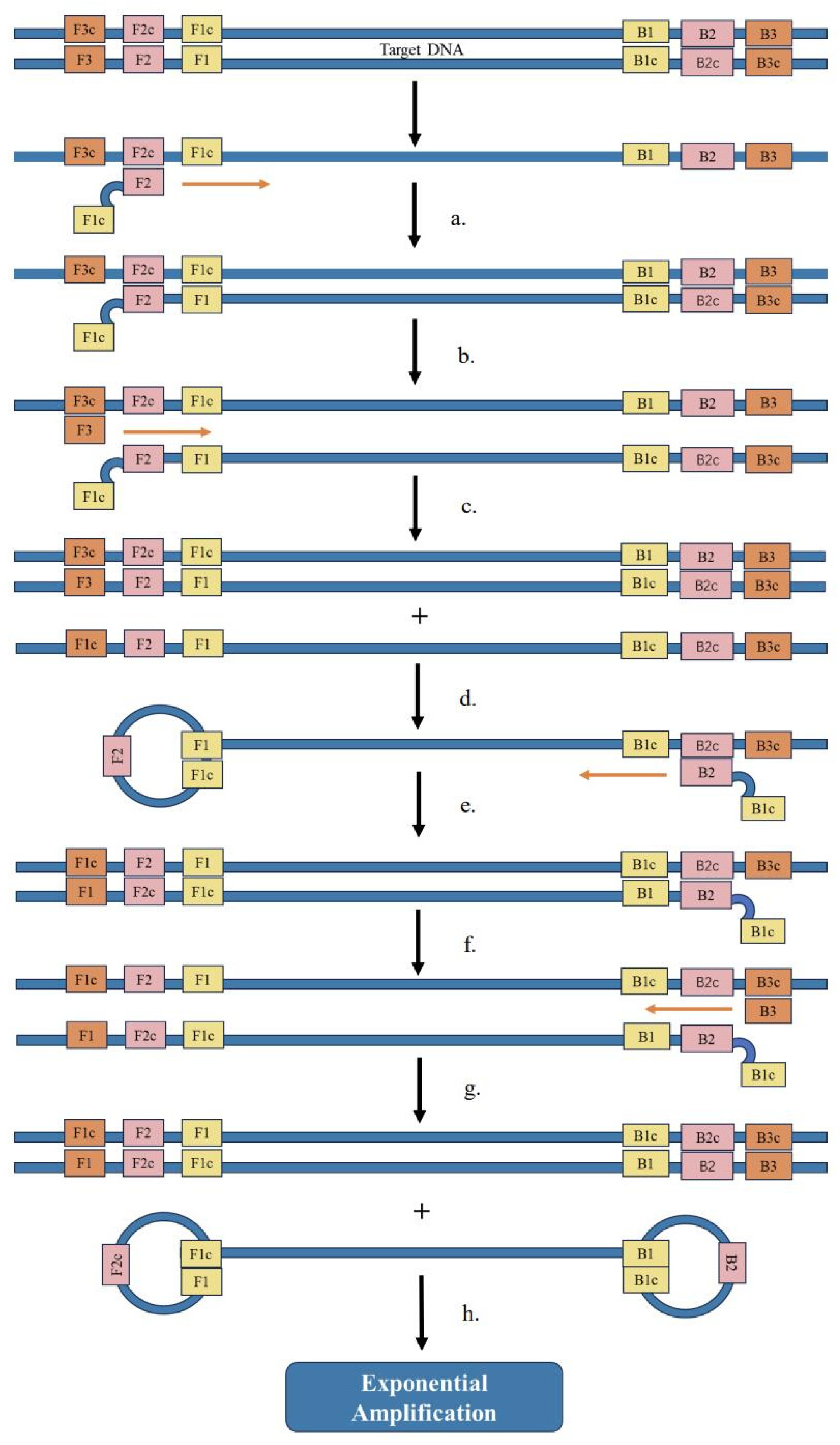
5.2. Recombinase Polymerase Amplification (RPA)
5.3. Loop-Mediated Isothermal Amplification (LAMP)
5.4. Nuclear Acid Sequence-Based Amplification (NASBA)
6. Gene Testing Technology
6.1. Gene Chip
6.2. Next-Generation Sequencing (NGS)
7. Biosensor
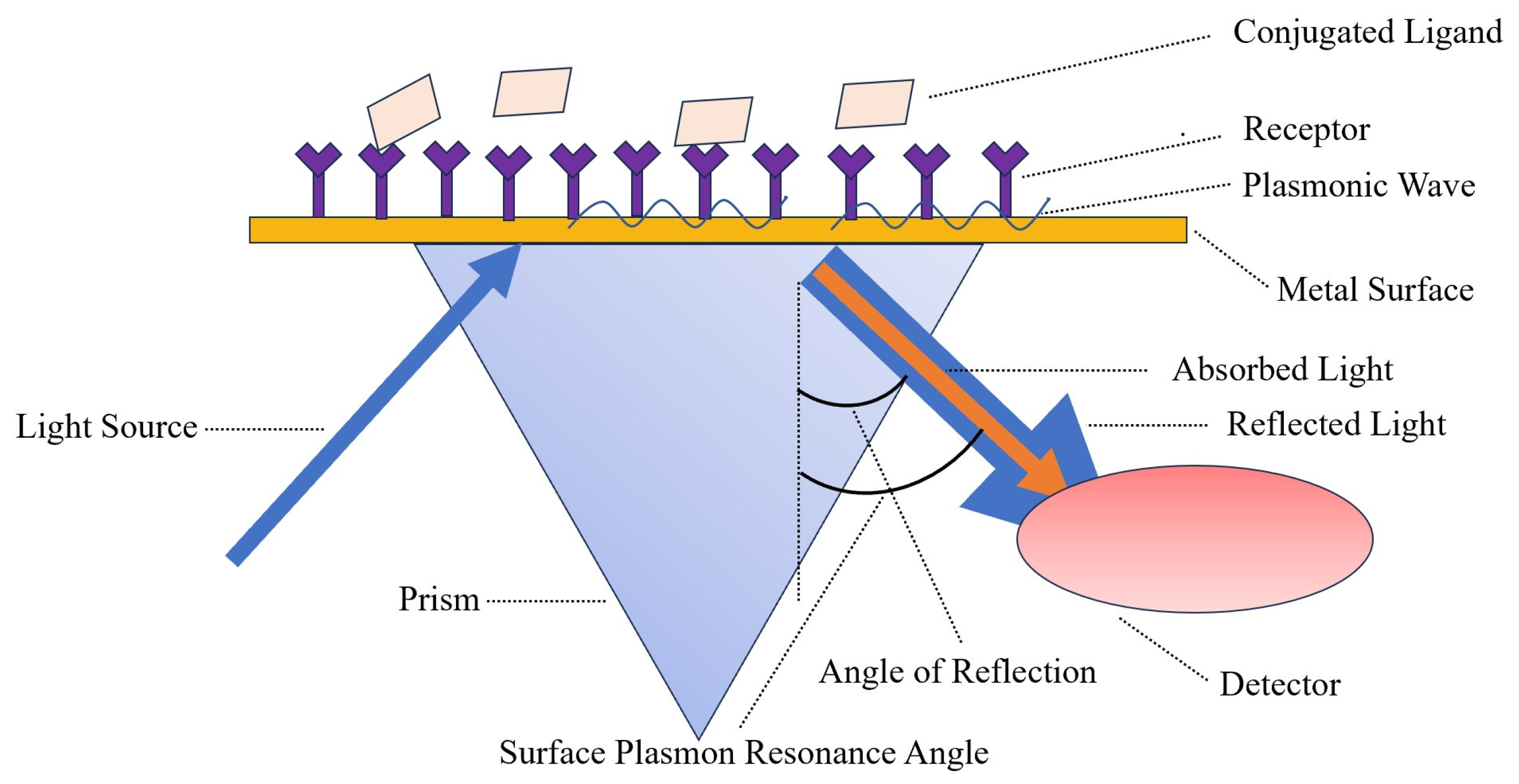
7.1. Surface Plasmon Resonance (SPR)
7.2. Field-Effect Transistor (FET)
7.3. Electrochemical Biosensor
7.4. Gene Biosensor
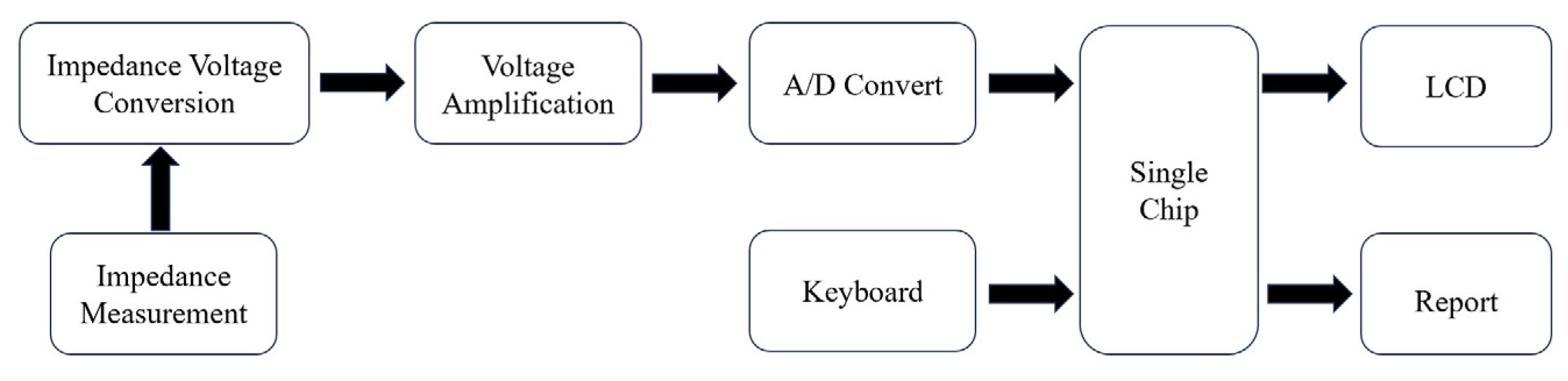
7.5. Impedance Biosensor
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fouchier, R.A.M.; Munster, V.; Wallensten, A.; Bestebroer, T.M.; Herfst, S.; Smith, D.; Rimmelzwaan, G.F.; Olsen, B.; Osterhaus, A.D.M.E. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 2005, 79, 2814–2822. [Google Scholar] [CrossRef] [PubMed]
- Laudert, E.; Sivanandan, V.; Halvorson, D. Effect of an H5N1 avian influenza virus infection on the immune system of mallard ducks. Avian Dis. 1993, 37, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Noda, T.; Kawaoka, Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009, 459, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Tran, K.; Gray, M.; Li, Y.; Ao, Z.J.; Yao, X.J.; Kobasa, D.; Kobinger, G.P. Evaluation of conserved and variable influenza antigens for immunization against different isolates of H5N1 viruses. Vaccine 2009, 27, 3083–3089. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liao, J.; Wang, L.; Luo, P.; Guo, L.; Zheng, W.; Wu, Z.; Ying, Q. Rapid Detection of H5N1 Avian Influenza Virus Based on RAA Fluorescence Assay. China Port Sci. Technol. 2021, 3, 4–9. [Google Scholar]
- Xu, X.; Subbarao, K.; Cox, N.J.; Guo, Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: Similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 1999, 261, 15–19. [Google Scholar] [CrossRef]
- Chen, H.; Smith GJ, D.; Li, K.S.; Wang, J.; Fan, X.H.; Rayner, J.M.; Vijaykrishna, D.; Zhang, J.X.; Zhang, L.J.; Guo, C.T.; et al. Establishment of multiple sublineages of H5N1 influenza virus in Asia: Implications for pandemic control. Proc. Natl. Acad. Sci. USA 2006, 103, 2845–2850. [Google Scholar] [CrossRef]
- Smith, G.J.D.; Fan, X.H.; Wang, J.; Li, K.S.; Qin, K.; Zhang, J.X.; Vijaykrishna, D.; Cheung, C.L.; Huang, K.; Rayner, J.M.; et al. Emergence and predominance of an H5N1 influenza variant in China. Proc. Natl. Acad. Sci. USA 2006, 103, 16936–16941. [Google Scholar] [CrossRef]
- Beyit, A.D.; Meki, I.K.; Barry, Y.; Haki, M.L.; El Ghassem, A.; Hamma, S.M.; Abdelwahab, N.; Doumbia, B.; Benane, H.A.; Daf, D.S.; et al. Avian influenza H5N1 in a great white pelican (Pelecanus onocrotalus), Mauritania 2022. Vet. Res. Commun. 2023, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Furness, R.W.; Gear, S.C.; Camphuysen, K.C.J.; Tyler, G.; de Silva, D.; Warren, C.J.; James, J.; Reid, S.M.; Banyard, A.C. Environmental Samples Test Negative for Avian Influenza Virus H5N1 Four Months after Mass Mortality at A Seabird Colony. Pathogens 2023, 12, 584. [Google Scholar] [CrossRef] [PubMed]
- Caliendo, V.; Lewis, N.S.; Pohlmann, A.; Baillie, S.R.; Banyard, A.C.; Beer, M.; Brown, I.H.; Fouchier, R.A.M.; Hansen, R.D.E.; Lameris, T.K.; et al. Transatlantic spread of highly pathogenic avian influenza H5N1 by wild birds from Europe to North America in 2021. Sci. Rep. 2022, 12, 11729. [Google Scholar] [CrossRef]
- Molini, U.; Yabe, J.; Meki, I.K.; Ben Ali, H.O.A.; Settypalli, T.B.K.; Datta, S.; Coetzee, L.M.; Hamunyela, E.; Khaiseb, S.; Cattoli, G.; et al. Highly pathogenic avian influenza H5N1 virus outbreak among Cape cormorants (Phalacrocorax capensis) in Namibia, 2022. Emerg. Microbes Infect. 2023, 12, 2167610. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.; Alfaro-Núñez, A.; de Mora, D.; Armas, R.; Olmedo, M.; Garcés, J.; Muñoz-López, G.; Garcia-Bereguiain, M.A. First case of human infection with highly pathogenic H5 avian influenza a virus in South America: A new zoonotic pandemic threat for 2023? J. Travel Med. 2023, 30, taad032. [Google Scholar] [CrossRef]
- Hong, S.C.; Murale, D.P.; Jang, S.Y.; Haque, M.M.; Seo, M.; Lee, S.; Woo, D.H.; Kwon, J.; Song, C.S.; Kim, Y.K.; et al. Discrimination of Avian Influenza Virus Subtypes using Host-Cell Infection Fingerprinting by a Sulfinate-based Fluorescence Superoxide Probe. Angew. Chem. 2018, 57, 9716–9721. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef] [PubMed]
- Swayne, D.E.; Kapczynski, D.; Swayne, D.E.; Kapczynski, D. Strategies and challenges for eliciting immunity against avian influenza virus in birds. Immunol. Rev. 2008, 225, 314–331. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.; Ko, L.S.; Collins, R.A.; Wu, Z.; Chen, J.; Chan, K.Y.; Xing, J.; Lau, L.T.; Yu, A.C.H. Comparison of nucleic acid-based detection of avian influenza H5N1 with virus isolation. Biochem. Biophys. Res. Commun. 2003, 302, 377–383. [Google Scholar] [CrossRef]
- Loeffelholz, M.J. Avian Influenza (H5N1) Update: Role of the Clinical Microbiology Laboratory. Lab. Med. 2011, 42, 291–298. [Google Scholar] [CrossRef]
- Li, I.W.S.; Chan, K.H.; To, K.W.K.; Wong, S.S.Y.; Ho, P.L.; Lau, S.K.P.; Woo, P.C.Y.; Tsoi, H.W.; Chan, J.F.W.; Cheng, V.C.C.; et al. Differential susceptibility of different cell lines to swine-origin influenza A H1N1, seasonal human influenza A H1N1, and avian influenza A H5N1 viruses. J. Clin. Virol. 2009, 46, 325–330. [Google Scholar] [CrossRef]
- Schultzcherry, S.; Dybdahlsissoko, N.; Mcgregor, M.; Hinshaw, V.S. Mink Lung Epithelial Cells: Unique Cell Line That Supports Influenza A and B Virus Replication. J. Clin. Microbiol. 1998, 36, 3718–3720. [Google Scholar] [CrossRef]
- Kode, S.S.; Pawar, S.D.; Tare, D.S.; Mullick, J. Application of frozen and stored glutaraldehyde-fixed Turkey red blood cells for hemagglutination and hemagglutination inhibition assays for the detection and identification of influenza viruses. J. Virol. Methods 2020, 289, 114046. [Google Scholar] [CrossRef] [PubMed]
- Pawar, S.D.; Parkhi, S.S.; Koratkar, S.S.; Mishra, A.C. Receptor specificity and erythrocyte binding preferences of avian influenza viruses isolated from India. Virol. J. 2012, 9, 251. [Google Scholar] [CrossRef] [PubMed]
- Wiriyarat, W.; Lerdsamran, H.; Pooruk, P.; Webster, R.G.; Louisirirotchanakul, S.; Ratanakorn, P.; Chaichoune, K.; Nateerom, K.; Puthavathana, P. Erythrocyte binding preference of 16 subtypes of low pathogenic avian influenza and 2009 pandemic influenza A (H1N1) viruses. Vet. Microbiol. 2010, 146, 346–349. [Google Scholar] [CrossRef]
- Comin, A.; Toft, N.; Stegeman, A.; Klinkenberg, D.; Marangon, S. Serological diagnosis of avian influenza in poultry: Is the hemagglutination inhibition test really the ‘gold standard’? Influenza Other Respir. Viruses 2013, 7, 257–264. [Google Scholar] [CrossRef]
- Stelzer-Braid, S.; Wong, B.; Robertson, P.; Lynch, G.W.; Laurie, K.; Shaw, R.; Barr, I.; Selleck, P.W.; Baleriola, C.; Escott, R.; et al. A commercial ELISA detects high levels of human H5 antibody but cross-reacts with influenza A antibodies. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2008, 43, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Rimmelzwaan, G.F.; Verburgh, R.J.; Nieuwkoop, N.J.; Bestebroer, T.M.; Fouchier, R.A.M.; Osterhaus, A.D.M.E. Use of GFP-expressing influenza viruses for the detection of influenza virus A/H5N1 neutralizing antibodies. Vaccine 2011, 29, 3424–3430. [Google Scholar] [CrossRef] [PubMed]
- Rowe, T.; Abernathy, R.A.; Hu-Primmer, J.; Thompson, W.W.; Lu, X.; Lim, W.; Fukuda, K.; Cox, N.J.; Katz, J.M. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 1999, 37, 937–943. [Google Scholar] [CrossRef]
- Bishai, F.R.; Galli, R. Enzyme-linked immunosorbent assay for detection of antibodies to influenza A and B and parainfluenza type 1 in sera of patients. J. Clin. Microbiol. 1978, 8, 648–656. [Google Scholar] [CrossRef]
- Adair, B.M.; Todd, D.; Mckillop, E.R.; McNulty, M.S. Detection of influenza a type-specific antibodies in chicken and turkey sera by enzyme linked immunosorbent assay. Avian Pathol. 1989, 18, 455–463. [Google Scholar] [CrossRef]
- Fatunmbi, O.O.; Newman, J.A.; Sivanandan, V.; Halvorson, D.A. A broad-spectrum avian influenza subtype antigen for indirect enzyme-linked immunosorbent assay. Avian Dis. 1989, 33, 264–269. [Google Scholar] [CrossRef]
- Snyder, D.B.; Marquardt, W.W.; Mallinson, E.T.; Allen, D.A.; Savage, P.K. An enzyme-linked immunosorbent assay method for the simultaneous measurement of antibody titer to multiple viral, bacterial or protein antigens. Vet. Immunol. Immunopathol. 1985, 9, 303–317. [Google Scholar] [CrossRef]
- Shafer, A.L.; Katz, J.B.; Eernisse, K.A. Development and validation of a competitive enzyme-linked immunosorbent assay for detection of type A influenza antibodies in avian sera. Avian Dis. 1998, 42, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Burlington, D.B.; Wright, P.F.; Wyke KL, V.; Phelan, M.A.; Mayner, R.E.; Murphy, B.R. Development of subtype-specific and heterosubtypic antibodies to the influenza A virus hemagglutinin after primary infection in children. J. Clin. Microbiol. 1985, 21, 847–849. [Google Scholar] [CrossRef]
- Remarque, E.J.; de Bruijn, I.A.; Boersma, W.J.; Masurel, N.; Ligthart, G.J. Altered antibody response to influenza H1N1 vaccine in healthy elderly people as determined by HI, ELISA, and neutralization assay. J. Med. Virol. 1998, 55, 82–87. [Google Scholar] [CrossRef]
- Zhang, A.; Jin, M.; Liu, F.; Guo, X.; Hu, Q.; Han, L.; Tan, Y.; Chen, H. Development and evaluation of a DAS-ELISA for rapid detection of avian influenza viruses. Avian Dis. 2006, 50, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.X.; Xu, F.H.; Fan, X.H.; Luo, H.F.; Ge, S.P.; Zheng, Q.B.; Xia, N.S.; Chen, H.L.; Guan, Y.; Zhang, J. Evaluation of a rapid test for detection of H5N1 avian influenza virus. J. Virol. Methods 2008, 154, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.T.; Qian, H.L.; He, F.; Meng, T.; Szyporta, M.; Prabhu, N.; Prabakaran, M.; Chan, K.P.; Kwang, J. Rapid detection of H5N1 subtype influenza viruses by antigen capture enzyme-linked immunosorbent assay using H5- and N1-specific monoclonal antibodies. Clin. Vaccine Immunol. 2009, 16, 726–732. [Google Scholar] [CrossRef]
- Prabakaran, M.; Ho, H.T.; Prabhu, N.; Velumani, S.; Szyporta, M.; He, F.; Chan, K.P.; Chen, L.M.; Matsuoka, Y.; Donis, R.O.; et al. Development of Epitope-Blocking ELISA for Universal Detection of Antibodies to Human H5N1 Influenza Viruses. PLoS ONE 2009, 4, e4566. [Google Scholar] [CrossRef]
- He, F.; Soejoedono, R.D.; Murtini, S.; Goutama, M.; Kwang, J. Complementary monoclonal antibody-based dot ELISA for universal detection of H5 avian influenza virus. BMC Microbiol. 2010, 10, 330. [Google Scholar] [CrossRef]
- Dong, J.H.; Sakurai, A.; Nomura, N.; Park, E.Y.; Shibasaki, F.; Ueda, H. Isolation of Recombinant Phage Antibodies Targeting the Hemagglutinin Cleavage Site of Highly Pathogenic Avian Influenza Virus. PLoS ONE 2013, 8, e61158. [Google Scholar] [CrossRef]
- Ellis, J.S.; Fleming, D.M.; Zambon, M.C. Multiplex reverse transcription-PCR for surveillance of influenza A and B viruses in England and Wales in 1995 and 1996. J. Clin. Microbiol. 1997, 35, 2076–2082. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.; Kawde, A.N.; Daud, M. Designs, formats and applications of lateral flow assay: A literature review. J. Saudi Chem. Soc. 2015, 19, 689–705. [Google Scholar] [CrossRef]
- Li, R.; Li, P.W.; Guo, X.B.; Jin, M.L.; Zhang, W.; Zhang, Q. A chromatographic strip for rapid semi-quantitative detection of H5 subtype avian influenza viruses in poultry. Food Anal. Methods 2013, 6, 1712–1717. [Google Scholar] [CrossRef]
- Yager, P.; Edwards, T.; Fu, E.; Helton, K.; Nelson, K.; Tam, M.R.; Weigl, B.H. Microfluidic diagnostic technologies for global public health. Nature 2006, 442, 412–418. [Google Scholar] [CrossRef]
- Han, M.Y.; Xie, T.A.; Li, J.X.; Chen, H.J.; Yang, X.H.; Guo, X.G. Evaluation of lateral-flow assay for rapid detection of influenza virus. BioMed Res. Int. 2020, 2020, 3969868. [Google Scholar] [CrossRef]
- Durairaj, K.; Than, D.D.; Nguyen AT, V.; Kim, H.S.; Yeo, S.J.; Park, H. Cysteamine-Gold Coated Carboxylated Fluorescent Nanoparticle Mediated Point-of-Care Dual-Modality Detection of the H5N1 Pathogenic Virus. Int. J. Mol. Sci. 2022, 23, 7957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.T.; Wu, D.; Wei, J.H.; Xiao, G.F. A new method for the detection of the H5 influenza virus by magnetic beads capturing quantum dot fluorescent signals. Biotechnol. Lett. 2010, 32, 1933–1937. [Google Scholar] [CrossRef]
- Yeo, S.J.; Kang, H.; Dao, T.D.; Cuc, B.T.; Nguyen, A.T.V.; Tien, T.T.T.; Hang, N.L.K.; Phuong, H.V.M.; Thanh, L.T.; Mai, L.Q.; et al. Development of a smartphone-based rapid dual fluorescent diagnostic system for the simultaneous detection of influenza A and H5 subtype in avian influenza A-infected patients. Theranostics 2018, 8, 6132. [Google Scholar] [CrossRef]
- Sun, J.; Chen, Y.; Wu, F.; Wang, J.J.; Chen, B.; Zheng, X.C.; Tao, H.; Liu, J.L.; Chen, S.K.; Liu, Q.Y.; et al. Development of anovel lateral-flow immunoassay with quantum dots for test of H5 subtype avian influenza virus. Chin. J. Vet. Med. 2019, 55, 5. [Google Scholar]
- Zhao, W.; Zhang, W.P.; Zhang, Z.L.; He, R.L.; Lin, Y.; Xie, M.; Wang, H.Z.; Pang, D.W. Robust and highly sensitive fluorescence approach for point-of-care virus detection based on immunomagnetic separation. Anal. Chem. 2012, 84, 2358–2365. [Google Scholar] [CrossRef]
- Chen, X.Y.; Wen, H.S.; Feng, H.E.; Chen, C.F.; Zhang, J.R.; Jin, G.X.; Shi, B. Partial Sequence Cloning of LHR Gene in Cynoglossus semilaevis and Its Tissue Expression Analysis. Period. Ocean. Univ. China 2010, 40, 71–77. [Google Scholar]
- Doak, S.H.; Zaïr, Z.M. Real-time reverse-transcription polymerase chain reaction: Technical considerations for gene expression analysis. Genet. Toxicol. Princ. Methods 2012, 817, 251–270. [Google Scholar]
- Payungporn, S.; Phakdeewirot, P.; Chutinimitkul, S.; Theamboonlers, A.; Keawcharoen, J.; Oraveerakul, K.; Amonsin, A.; Poovorawan, Y. Single-step multiplex reverse transcription–polymerase chain reaction (RT-PCR) for influenza A virus subtype H5N1 detection. Viral Immunol. 2004, 17, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; He, B.; Li, C.; Zhang, X.; Wu, W.; Yin, X.; Fan, B.; Fan, X.; Wang, J. Real-time RT-PCR for H5N1 avian influenza A virus detection. J. Med. Microbiol. 2007, 56, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.R.; Kim, E.M.; Lee, Y.J.; Yeo1, S.G.; Park, C.K. Multiplex real-time reverse transcription polymerase chain reaction for differential detection of H5, N1, and N8 genes of highly pathogenic avian influenza viruses. Vet. Med. 2017, 62, 211–220. [Google Scholar] [CrossRef]
- Wu, L.; Ding, L.; Pei, Z.; Huo, X.; Wen, G.; Pan, Z. A multiplex reverse transcription-PCR assay for the detection of influenza A virus and differentiation of the H1, H3, H5 and H9 subtypes. J. Virol. Methods 2013, 188, 47–50. [Google Scholar] [CrossRef]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef]
- Kis, Z.; Jones, J.; Creanga, A.; Ferdinand, K.; Donis, R.O. Real-time RT-PCR assay to differentiate clades of H5N1 avian influenza viruses circulating in Vietnam. J. Virol. Methods 2013, 193, 452–458. [Google Scholar] [CrossRef]
- Ellis, J.S.; Smith, J.W.; Braham, S.; Lock, M.; Barlow, K.; Zambon, M.C. Design and validation of an H5 TaqMan real-time one-step reverse transcription-PCR and confirmatory assays for diagnosis and verification of influenza A virus H5 infections in humans. J. Clin. Microbiol. 2007, 45, 1535–1543. [Google Scholar] [CrossRef]
- Lu, Y.Y.; Yan, J.Y.; Feng, Y.; Xu, C.P.; Shi, W.; Mao, H.Y. Rapid detection of H5 avian influenza virus by TaqMan-MGB real-time RT-PCR. Lett. Appl. Microbiol. 2008, 46, 20–25. [Google Scholar] [CrossRef]
- Yin, J.L.; Shackel, N.A.; Zekry, A.; McGuinness, P.H.; Richards, C.; Putten, K.V.; McCaughan, G.W.; Eris, J.M.; Bishop, G.A. Real-time reverse transcriptase–polymerase chain reaction (RT–PCR) for measurement of cytokine growth factor mRNA expression with fluorogenic probes or SYBR Green I. Immunol. Cell Biol. 2001, 79, 213–221. [Google Scholar] [CrossRef]
- Naguib, M.M.; Arafa AS, A.; El-Kady, M.F.; Selim, A.A.; Gunalan, V.; Maurer-Stroh, S.; Goller, K.V.; Hassan, M.K.; Beer, M.; Abdelwhab, E.M.; et al. Evolutionary trajectories and diagnostic challenges of potentially zoonotic avian influenza viruses H5N1 and H9N2 co-circulating in Egypt. Infect. Genet. Evol. 2015, 34, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.B.; Gao, Y.; Wen, L.Y.; Shao, M.; Zou, S.M.; Li, C.G.; Yang, L.; Li, X.Y.; Wang, W.; Shu, Y.L. Development and implementation of the quality control panel of RT-PCR and real-time RT-PCR for avian influenza A (H5N1) surveillance network in mainland China. BMC Infect. Dis. 2011, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.F.; Xu, H.P.; Liu, C.; Peng, L.J.; Khan, H.; Cui, L.B.; Huang, R.; Wu, C.; Shen, S.S.; Wang, S.; et al. CRISPR-Cas13a nanomachine based simple technology for avian influenza A (H7N9) virus on-site detection. J. Biomed. Nanotechnol. 2019, 15, 790–798. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zheng, T.; Gao, L.; Li, F.; Hou, X.D.; Wu, P. Recombinase polymerase amplification: From principle to performance. Chem. J. Chin. Univ.-Chin. 2020, 41, 2587–2597. [Google Scholar] [CrossRef]
- Yehia, N.; Arafa, A.S.; Abd El Wahed, A.; El-Sanousi, A.A.; Weidmann, M.; Shalaby, M.A. Development of reverse transcription recombinase polymerase amplification assay for avian influenza H5N1 HA gene detection. J. Virol. Methods 2015, 223, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Li, Y.; Zhang, F.Y.; Jiang, N.; Zhuang, Q.Y.; Hou, G.Y.; Jiang, L.J.; Yu, J.M.; Yu, X.H.; Liu, H.L.; et al. Reverse transcription recombinase-aided amplification assay for H5 subtype avian influenza virus. Virol. J. 2022, 19, 129. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, P.P.; Zhang, Y.H.; Tian, K.Y.; Bian, C.Z.; Zhao, J. Development of a reverse transcription recombinase polymerase amplification combined with lateral-flow dipstick assay for avian influenza H9N2 HA gene detection. Transbound. Emerg. Dis. 2019, 66, 546–551. [Google Scholar] [CrossRef]
- Yehia, N.; Eldemery, F.; Arafa, A.S.; Wahed, A.A.; El Sanousi, A.; Weidmann, M.; Shalaby, M. Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid Detection of Avian Influenza Virus H9N2 HA Gene. Vet. Sci. 2021, 8, 134. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed]
- Dukes, J.P.; King, D.P.; Alexandersen, S. Novel reverse transcription loop-mediated isothermal amplification for rapid detection of foot-and-mouth disease virus. Arch. Virol. 2006, 151, 1093–1106. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Ninomiya, A.; Minekawa, H.; Notomi, T.; Ishizaki, T.; Tashiro, M.; Odagiri, T. Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine 2006, 24, 6679–6682. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, S.; Cheung, C.Y.; Barr, I.; Chan, K.H.; Chen, H.; Guan, Y.; Peiris, J.S.M.; Poon, L.L.M. Loop-mediated isothermal amplification for influenza A (H5N1) virus. Emerg. Infect. Dis. 2007, 13, 899. [Google Scholar] [CrossRef] [PubMed]
- Dinh, D.T.; Le MT, Q.; Vuong, C.D.; Hasebe, F.; Morita, K. An updated loop-mediated isothermal amplification method for rapid diagnosis of H5N1 avian influenza viruses. Trop. Med. Health 2011, 39, 3–7. [Google Scholar] [CrossRef]
- Jung, J.H.; Oh, S.J.; Kim, Y.T.; Kim, S.Y.; Kim, W.J.; Jung, J.; Seo, T.S. Combination of multiplex reverse-transcription loop-mediated isothermal amplification with an immunochromatographic strip for subtyping influenza A virus. Anal. Chim. Acta 2015, 853, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Yu, X.; Chen, H.; Diao, Y.X. An immunoassay-based reverse-transcription loop-mediated isothermal amplification assay for the rapid detection of avian influenza H5N1 virus viremia. Biosens. Bioelectron. 2016, 86, 255–261. [Google Scholar] [CrossRef]
- Zhang, S.; Shin, J.; Shin, S.; Chung, Y.J. Development of reverse transcription loop-mediated isothermal amplification assays for point-of-care testing of avian influenza virus subtype H5 and H9. Genom. Inform. 2020, 18, e40. [Google Scholar] [CrossRef]
- Ahn, S.J.; Baek, Y.H.; Lloren, K.K.S.; Choi, W.S.; Jeong, J.H.; Antigua, K.J.C.; Kwon, H.I.; Park, S.J.; Kim, E.H.; Kim, Y.I.; et al. Rapid and simple colorimetric detection of multiple influenza viruses infecting humans using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. BMC Infect. Dis. 2019, 19, 676. [Google Scholar] [CrossRef]
- Kievits, T.; van Gemen, B.; van Strijp, D.; Schukkink, R.; Lens, P. NASBATM isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J. Virol. Methods 1991, 35, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Deiman, B.; van Aarle, P.; Sillekens, P. Characteristics and applications of nucleic acid sequence-based amplification (NASBA). Mol. Biotechnol. 2002, 20, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Malek, L.; Sooknanan, R.; Compton, J. Nucleic acid sequence-based amplification (NASBA™). In Protocols for Nucleic Acid Analysis by Nonradioactive Probes; Springer: Berlin/Heidelberg, Germany, 1994; pp. 253–260. [Google Scholar]
- Collins, R.A.; Ko, L.S.; So, K.L.; Ellis, T.; Lau, L.T.; Yu, A.C.H. Detection of highly pathogenic and low pathogenic avian influenza subtype H5 (Eurasian lineage) using NASBA. J. Virol. Methods 2002, 103, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.; Telles, J.N.; Corden, S.; Gao, R.B.; Vernet, G.; Van Aarle, P.; Shu, Y.L. Development and validation of a commercial real-time NASBA assay for the rapid confirmation of influenza A H5N1 virus in clinical samples. J. Virol. Methods 2010, 170, 173–176. [Google Scholar] [CrossRef]
- Shan, S.-H.; Liu, L.-T.; Chen, J.-H.; Wu, Z.-L. Detection of Avian Influenza Virus Subtype H5 Using NASBA. Virol. Sin. 2005, 20, 288–292. [Google Scholar]
- Chantratita, W.; Sukasem, C.; Kaewpongsri, S.; Srichunrusami, C.; Pairoj, W.; Thitithanyanont, A.; Chaichoune, K.; Ratanakron, P.; Songserm, T.; Damrongwatanapokin, S.; et al. Qualitative detection of avian influenza A (H5N1) viruses: A comparative evaluation of four real-time nucleic acid amplification methods. Mol. Cell. Probes 2008, 22, 287–293. [Google Scholar] [CrossRef]
- Teng, X.; Liu, H.; Li, J.; Chen, X. Establishment of nucleic acid sequence-based amplification method for the detection of Zika virus. Chin. J. Front. Health Quar. 2021, 44, 153–155. [Google Scholar]
- Xu, X.; Li, H.; Cui, K.; Yao, X.L.; Ren, H.W.; Zheng, Y.K.; Zhang, Q. Application of three on-site rapid detection technologies in aquatic animal diseases. China Fish. 2013, 2, 70–72. [Google Scholar]
- Kessler, N.; Ferraris, O.; Palmer, K.; Marsh, W.; Steel, A. Use of the DNA flow-thru chip, a three-dimensional biochip, for typing and subtyping of influenza viruses. J. Clin. Microbiol. 2004, 42, 2173–2185. [Google Scholar] [CrossRef]
- Dawson, E.D.; Moore, C.L.; Dankbar, D.M.; Mehlmann, M.; Townsend, M.B.; Smagala, J.A.; Smith, C.B.; Cox, N.J.; Kuchta, R.; Rowlen, K.L. Identification of A/H5N1 influenza viruses using a single gene diagnostic microarray. Anal. Chem. 2007, 79, 378–384. [Google Scholar] [CrossRef]
- Lee, D.H.; Kim, J.H.; Lee, Y.N.; Park, J.K.; Yuk, S.S.; Jung, J.W.; Hwang, S.Y.; Lee, Y.J.; Kang, H.M.; Choi, J.G.; et al. Simultaneous subtyping and pathotyping of the 2010–2011 South Korean HPAI outbreak strain by using a diagnostic microarray. BioChip J. 2011, 5, 369–374. [Google Scholar] [CrossRef]
- Shi, L.; Sun, J.S.; Yang, Z.P.; Bao, H.M.; Jiang, Y.P.; Xiong, Y.Z.; Cao, D.; Yu, X.W.; Chen, H.L.; Zheng, S.M.; et al. Development of a DNA microarray-based multiplex assay of avian influenza virus subtypes H5, H7, H9, N1, and N2. Acta Virol. 2014, 58, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Kwon, N.Y.; Ahn, J.J.; Kim, J.H.; Kim, S.Y.; Lee, J.H.; Kwon, J.H.; Song, C.S.; Hwang, S.Y. Rapid subtyping and pathotyping of avian influenza virus using chip-based RT-PCR. BioChip J. 2019, 13, 333–340. [Google Scholar] [CrossRef]
- Huang, X.; Shi, Y.; Fu, Y.; Jiang, H.; Huang, Z.; Yin, G. Research progress in the application of gene chip technology in animal disease detection. Guizhou J. Anim. Husb. V 2020, 44, 49–51. [Google Scholar]
- Zhu, S.; Chen, R.; Wang, Z. Application progress of high-throughput sequencing technology in infectious diseases. Zhejiang Pract. Med. 2020, 25, 385–389. [Google Scholar]
- Sanger, F.; Air, G.M.; Barrell, B.G.; Brown, N.L.; Coulson, A.R.; Fiddes, J.C.; Hutchison, C.A.; Slocombe, P.M.; Smith, M. Nucleotide sequence of bacteriophage φX174 DNA. Nature 1977, 265, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.; Stenzel, U.; Hofreiter, M. Parallel tagged sequencing on the 454 platform. Nat. Protoc. 2008, 3, 267–278. [Google Scholar] [CrossRef]
- Deyde, V.M.; Gubareva, L.V. Influenza genome analysis using pyrosequencing method: Current applications for a moving target. Expert Rev. Mol. Diagn. 2009, 9, 493–509. [Google Scholar] [CrossRef]
- Spackman, E.; Suarez, D.L. Detection and identification of the H5 hemagglutinin subtype by real-time RT-PCR. Avian Influenza Virus 2008, 436, 27–33. [Google Scholar]
- Nylander, C.; Liedberg, B.; Lind, T. Gas detection by means of surface plasmon resonance. Sens. Actuators 1982, 3, 79–88. [Google Scholar] [CrossRef]
- Cai, Q.; Li, X.; Chen, Y. History, Current Status, and Prospects of Biosensors Based on Surface Plasmon Resonance. Foreign Med.—Biomed. Eng. Div. 1999, 2, 1–7. [Google Scholar]
- Bai, H.; Wang, R.; Hargis, B.; Lu, H.G.; Li, Y.B. A SPR aptasensor for detection of avian influenza virus H5N1. Sensors 2012, 12, 12506–12518. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.L.; Chua, M.; Mittman, H.; Choo, L.X.; Lim, H.Q.; Olivo, M. A phase-intensity surface plasmon resonance biosensor for avian influenza A (H5N1) detection. Sensors 2017, 17, 2363. [Google Scholar] [CrossRef]
- Luo, X.; Xu, J.; Chen, H. Field effect transistor biosensor. Anal. Chem. 2004, 32, 1395–1400. [Google Scholar]
- Yang, J.; Zhang, Y.; Liu, C.; Li, J.; Xiao, Z.; Li, D.; Zhang, L.; Cao, Z. Studying and application of field effect transistor sensors. Chem. Sens. 2018, 38, 1–11. [Google Scholar]
- Gao, A.; Lu, N.; Dai, P.; Li, T.; Pei, H.; Gao, X.L.; Gong, Y.B.; Wang, Y.L.; Fan, C.H. Silicon-nanowire-based CMOS-compatible field-effect transistor nanosensors for ultrasensitive electrical detection of nucleic acids. Nano Lett. 2011, 11, 3974–3978. [Google Scholar] [CrossRef]
- Guo, D.; Zhuo, M.; Zhang, X.; Xu, C.; Jiang, J.; Gao, F.; Wan, Q.; Li, Q.H.; Wang, T.H. Indium-tin-oxide thin film transistor biosensors for label-free detection of avian influenza virus H5N1. Anal. Chim. Acta 2013, 773, 83–88. [Google Scholar] [CrossRef]
- Kwon, J.; Lee, Y.; Lee, T.; Ahn, J.H. Aptamer-based field-effect transistor for detection of avian influenza virus in chicken serum. Anal. Chem. 2020, 92, 5524–5531. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Y.; Song, L.; Su, Z.; Zhang, H. Research progress in the application of field-effect transistor biosensors in biomedical detection. China Biotechnol. 2021, 41, 73–88. [Google Scholar]
- Mo, J.; Zhou, X. Progress in the Development and Application of Bioelectrochemical Sensors. J. Shantou Univ. (Nat. Sci. Ed.) 1993, 1, 80–91. [Google Scholar]
- Zhang, Y.; Jiao, K.; Liu, C. Electrochemical Biosensor. J. Qingdao Inst. Chem. Technol. 1992, 2, 99–105. [Google Scholar]
- Kukol, A.; Li, P.; Estrela, P.; Ko-Ferrigno, P.; Migliorato, P. Label-free electrical detection of DNA hybridization for the example of influenza virus gene sequences. Anal. Biochem. 2008, 374, 143–153. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, Z.; Fan, H.; Ai, S.Y.; Han, R.X. Electrochemical detection of avian influenza virus H5N1 gene sequence using a DNA aptamer immobilized onto a hybrid nanomaterial-modified electrode. Electrochim. Acta 2011, 56, 6266–6270. [Google Scholar] [CrossRef]
- Shi, L.; Chu, Z.; Dong, X.; Jin, W.Q.; Dempsey, E. A highly oriented hybrid microarray modified electrode fabricated by a template-free method for ultrasensitive electrochemical DNA recognition. Nanoscale 2013, 5, 10219–10225. [Google Scholar] [CrossRef] [PubMed]
- Jarocka, U.; Sawicka, R.; Góra-Sochacka, A.; Sirko, A.; Zagórski-Ostoja, W.; Radecki, J.; Radecka, H. Electrochemical immunosensor for detection of antibodies against influenza A virus H5N1 in hen serum. Biosens. Bioelectron. 2014, 55, 301–306. [Google Scholar] [CrossRef]
- Diba, F.S.; Kim, S.; Lee, H.J. Amperometric bioaffinity sensing platform for avian influenza virus proteins with aptamer modified gold nanoparticles on carbon chips. Biosens. Bioelectron. 2015, 72, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.X.; Cao, J.T.; Huang, K.J. A sensitive electrochemical biosensor for specific DNA sequence detection based on flower-like VS2, graphene and Au nanoparticles signal amplification. J. Electroanal. Chem. 2015, 746, 1–8. [Google Scholar] [CrossRef]
- Fu, Y.; Romay, V.; Liu, Y.; Ibarlucea, B.; Baraban, L.; Khavrus, V.; Oswald, S.; Bachmatiuk, A.; Ibrahim, I.; Rümmeli, M.; et al. Chemiresistive biosensors based on carbon nanotubes for label-free detection of DNA sequences derived from avian influenza virus H5N1. Sens. Actuators B Chem. 2017, 249, 691–699. [Google Scholar] [CrossRef]
- Zhao, Z.; Cui, D. Gene Sensor. Electron. Eng. Prod. World 2000, 63–65. [Google Scholar]
- Lee, K.H.; Lee, J.O.; Choi, S.; Yoon, J.B.; Cho, G.H. A CMOS label-free DNA sensor using electrostatic induction of molecular charges. Biosens. Bioelectron. 2012, 31, 343–348. [Google Scholar] [CrossRef]
- Grabowska, I.; Malecka, K.; Stachyra, A.; Góra-Sochacka, A.; Sirko, A.; Zagórski-Ostoja, W.; Radecka, H.; Radecki, J. Single electrode genosensor for simultaneous determination of sequences encoding hemagglutinin and neuraminidase of avian influenza virus type H5N1. Anal. Chem. 2013, 85, 10167–10173. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, I.; Stachyra, A.; Góra-Sochacka, A.; Sirko, A.; Olejniczak, A.B.; Lesnikowski, Z.J.; Radecki, J.; Radecka, H. DNA probe modified with 3-iron bis (dicarbollide) for electrochemical determination of DNA sequence of Avian Influenza Virus H5N1. Biosens. Bioelectron. 2014, 51, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Malecka, K.; Stachyra, A.; Góra-Sochacka, A.; Sirko, A.; Zagorski-Ostoja, W.; Dehaen, W.; Radecka, H.; Radecki, J. New redox-active layer create via epoxy–amine reaction–The base of genosensor for the detection of specific DNA and RNA sequences of avian influenza virus H5N1. Biosens. Bioelectron. 2015, 65, 427–434. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Malecka, K.; Stachyra, A.; Góra-Sochacka, A.; Sirko, A.; Zagórski-Ostoja, W.; Radecka, H.; Radecki, J. Electrochemical genosensor based on disc and screen printed gold electrodes for detection of specific DNA and RNA sequences derived from Avian Influenza Virus H5N1. Sens. Actuators B Chem. 2016, 224, 290–297. [Google Scholar] [CrossRef]
- Ye, Z. A Portable Impedance Biosensor Instrument for Rapid Detection of Chlorpyrifos and Escherichia coli. Ph.D. Thesis, Zhejiang University, Hangzhou, China, 2015. [Google Scholar]
- Yan, X.F.; Wang, M.H.; Wen, X.H.; An, D.; Yarlagadda, P.; Kim, Y.H. Rapid detection of avian influenza virus using immunomagnetic separation and impedance measurement. Appl. Mech. Mater. 2013, 239, 367–371. [Google Scholar] [CrossRef]
- Lin, J.; Lum, J.; Wang, R.; Tung, S.; Hargis, B.; Li, Y.B.; Lu, H.G.; Berghman, L. A portable impedance biosensor instrument for rapid detection of avian influenza virus. In Proceedings of the SENSORS, 2010 IEEE, Waikoloa, HI, USA, 1–4 November 2010; pp. 1558–1563. [Google Scholar]
- Lum, J.; Wang, R.; Lassiter, K.; Srinivasan, B.; Abi-Ghanem, D.; Berghman, L.; Hargis, B.; Tung, S.; Lu, H.G.; Li, Y.B. Rapid detection of avian influenza H5N1 virus using impedance measurement of immuno-reaction coupled with RBC amplification. Biosens. Bioelectron. 2012, 38, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wang, R.; Jiao, P.; Li, Y.T.; Li, Y.B.; Liao, M.; Yu, Y.D.; Wang, M.H. An impedance immunosensor based on low-cost microelectrodes and specific monoclonal antibodies for rapid detection of avian influenza virus H5N1 in chicken swabs. Biosens. Bioelectron. 2015, 67, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Lum, J.; Wang, R.; Hargis, B.; Tung, S.; Bottje, W.; Lu, H.G.; Li, Y.B. An impedance aptasensor with microfluidic chips for specific detection of H5N1 avian influenza virus. Sensors 2015, 15, 18565–18578. [Google Scholar] [CrossRef]
- Cui, S.; Tong, G. A Chromatographic Strip Test for Rapid Detection of One Lineage of the H5 Subtype of Highly Pathogenic Avian Influenza. J. Vet. Diagn. Investig. 2008, 20, 567–571. [Google Scholar] [CrossRef]
- Karash, S.; Wang, R.; Kelso, L.; Lu, H.G.; Huang, T.J.; Li, Y.B. Rapid detection of avian influenza virus H5N1 in chicken tracheal samples using an impedance aptasensor with gold nanoparticles for signal amplification. J. Virol. Methods 2016, 236, 147–156. [Google Scholar] [CrossRef]


| Category | Technology | Sensitivity (%) | Specificity (%) | LOD | Reference |
|---|---|---|---|---|---|
| Serology | ELISA | 95 | 100 | 2.5 × 102 EID50 | [36] |
| Immunology | (1) Collagen Gold Immunochromatography Technology | 100 | 100 | 1 mL of allantoic fluid (HA titer 1:27) continuously diluted 1:100–1:1000 | [132] |
| (2) Fluorescence Immunodetection Technique | 78.57 | 97.37 | 80 HAU/mL | [49] | |
| Molecular biology | (1) RT-PCR | 80 | 100 | 100 copies/reaction | [56] |
| (2) RPA | 97.26 | 100 | 102cRNA copies/μL | [69] | |
| (3) LAMP | 100 | 87.5 | 1 PFU/tube | [75] | |
| (4) NASBA | - | - | 10 copies/reaction | [89] | |
| Genetics | Gene chip | 97 | 100 | - | [93] |
| Biosensor | (1) SPR | - | - | 0.128–12.8 HAU | [105] |
| (2) FET | - | - | 1fM | [109] | |
| (3) Electrochemical biosensor | - | - | 5.2 × 10−14 M | [120] | |
| (4) Gene biosensor | - | - | 100pM | [123] | |
| (5) Impedance biosensor | - | - | 0.25 HAU | [133] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, X.; Wang, Q.; Ma, B.; Zhang, B.; Sun, K.; Yu, X.; Ye, Z.; Zhang, M. Advances in Detection Techniques for the H5N1 Avian Influenza Virus. Int. J. Mol. Sci. 2023, 24, 17157. https://doi.org/10.3390/ijms242417157
Fu X, Wang Q, Ma B, Zhang B, Sun K, Yu X, Ye Z, Zhang M. Advances in Detection Techniques for the H5N1 Avian Influenza Virus. International Journal of Molecular Sciences. 2023; 24(24):17157. https://doi.org/10.3390/ijms242417157
Chicago/Turabian StyleFu, Xianshu, Qian Wang, Biao Ma, Biao Zhang, Kai Sun, Xiaoping Yu, Zihong Ye, and Mingzhou Zhang. 2023. "Advances in Detection Techniques for the H5N1 Avian Influenza Virus" International Journal of Molecular Sciences 24, no. 24: 17157. https://doi.org/10.3390/ijms242417157
APA StyleFu, X., Wang, Q., Ma, B., Zhang, B., Sun, K., Yu, X., Ye, Z., & Zhang, M. (2023). Advances in Detection Techniques for the H5N1 Avian Influenza Virus. International Journal of Molecular Sciences, 24(24), 17157. https://doi.org/10.3390/ijms242417157








