Regulation of Peptidase Activity beyond the Active Site in Human Health and Disease
Abstract
1. Introduction
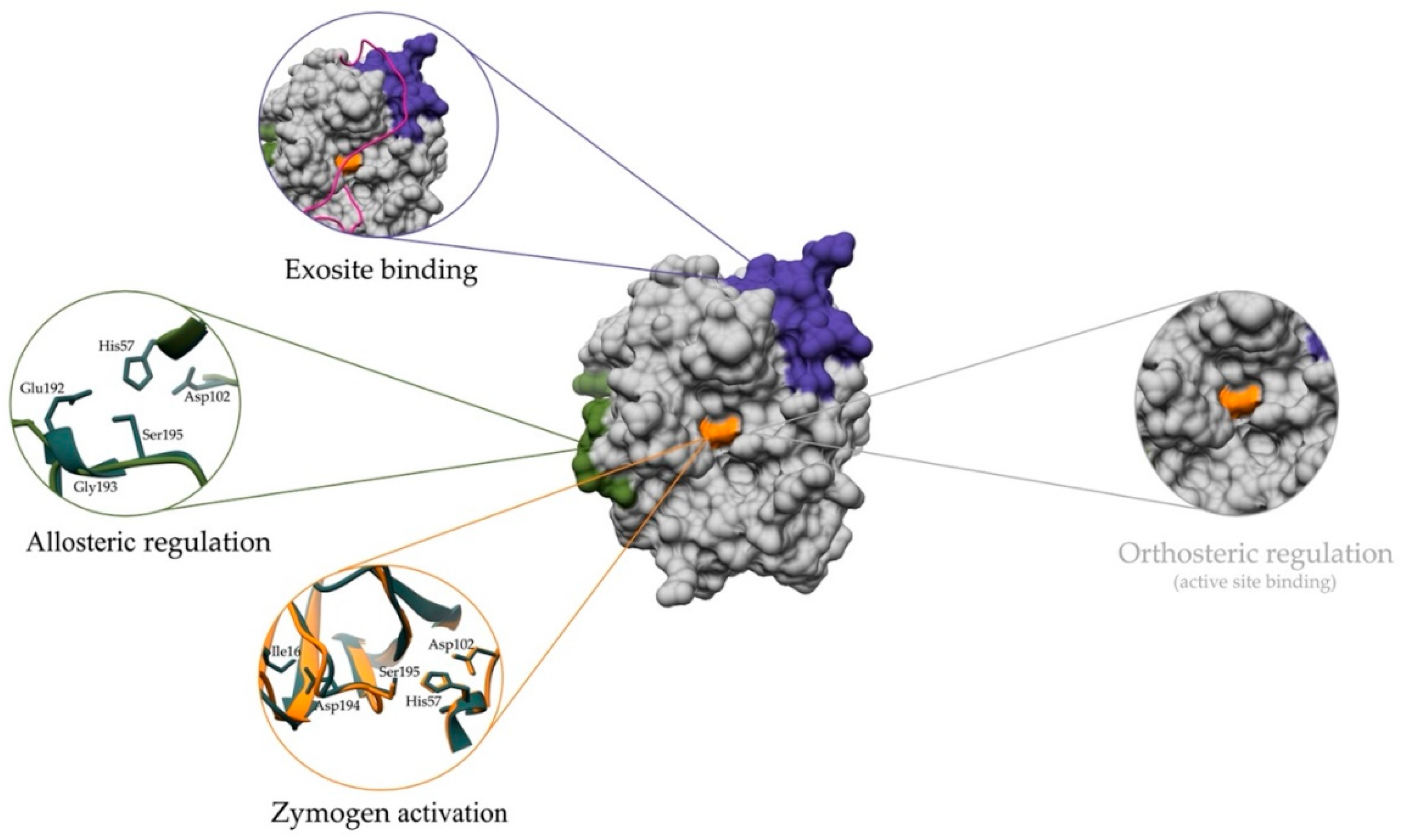
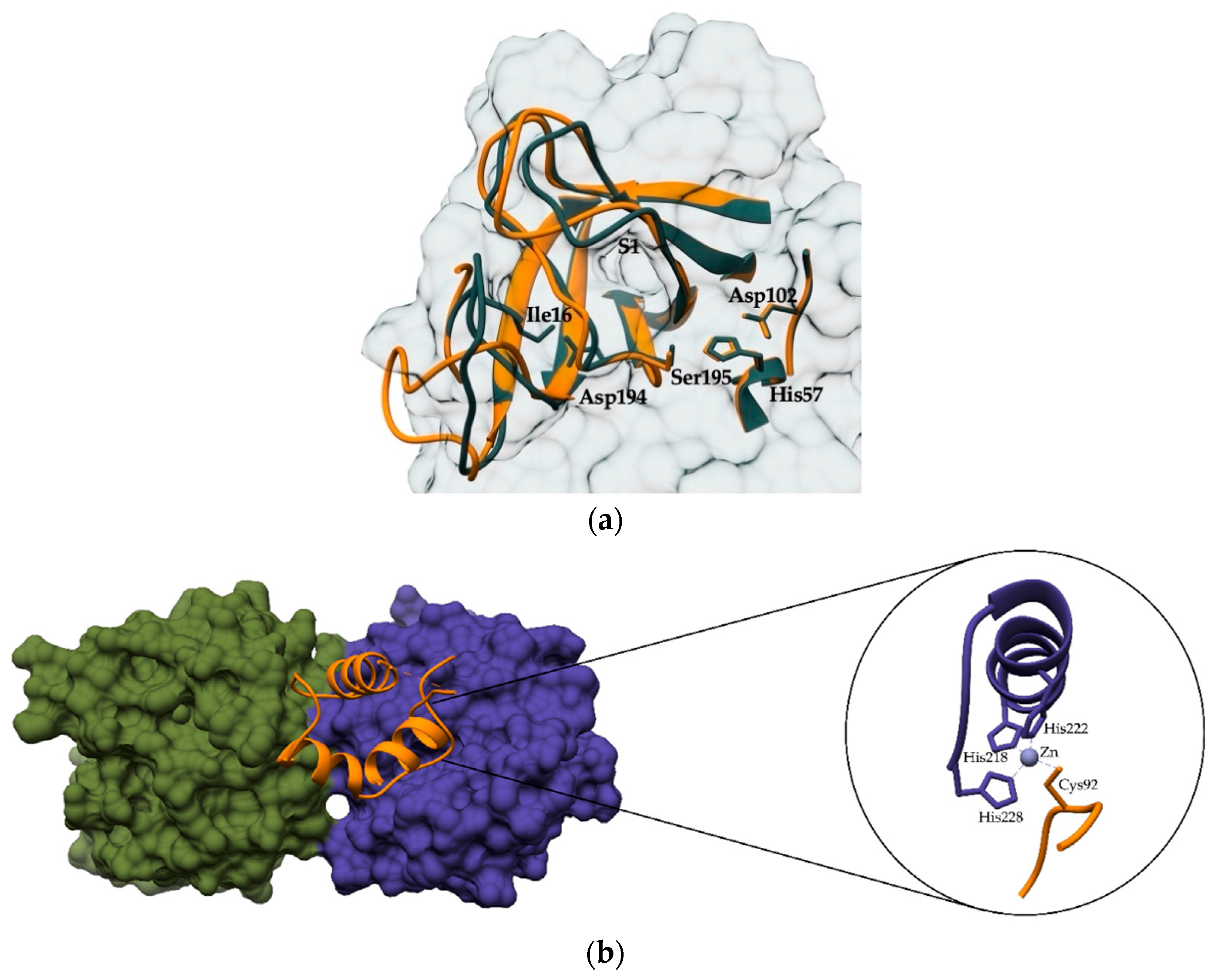
2. Regulating the Activation and Maturation of Zymogens
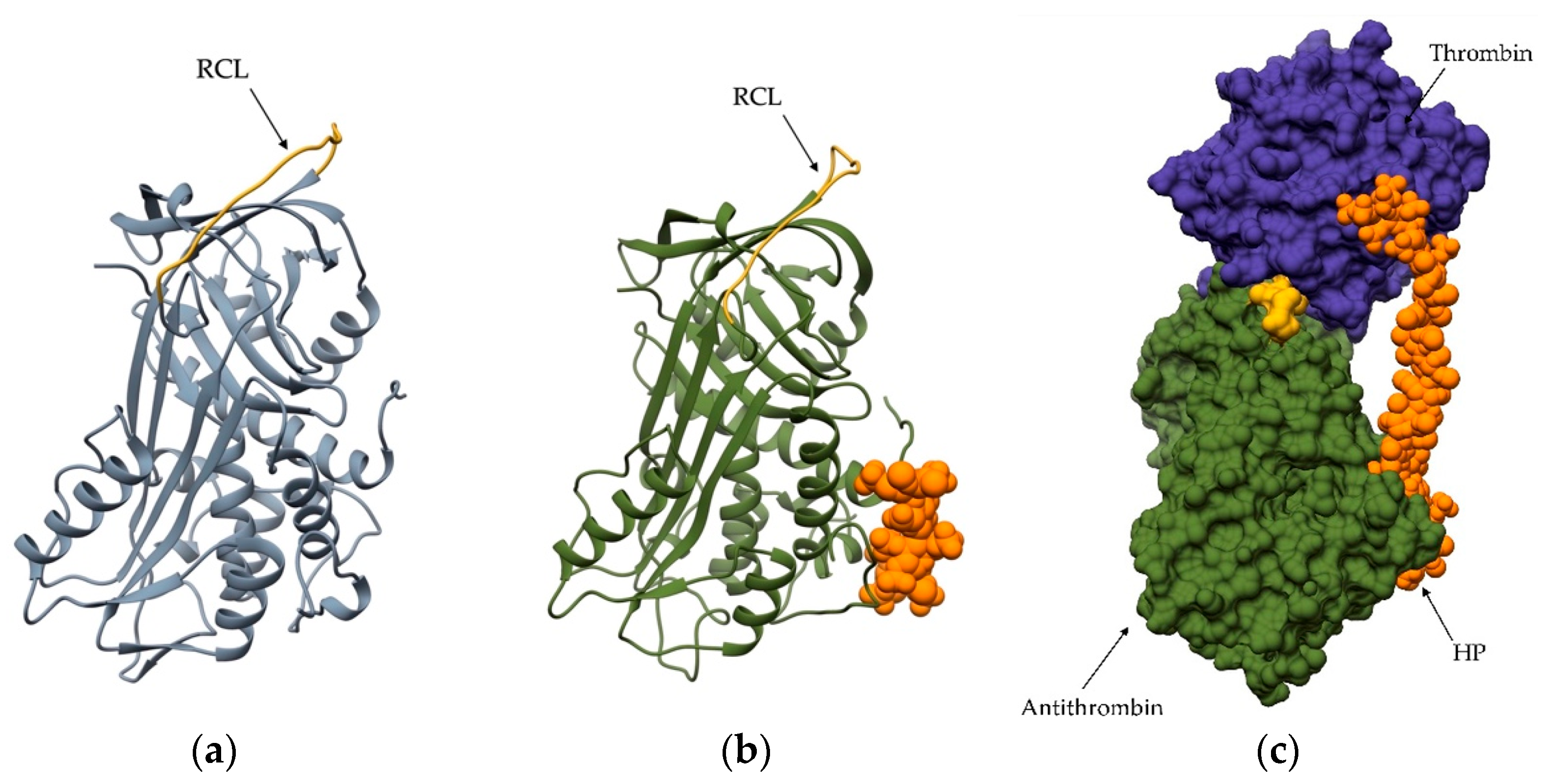
3. Regulation of Peptidase Activity by Glycosaminoglycans
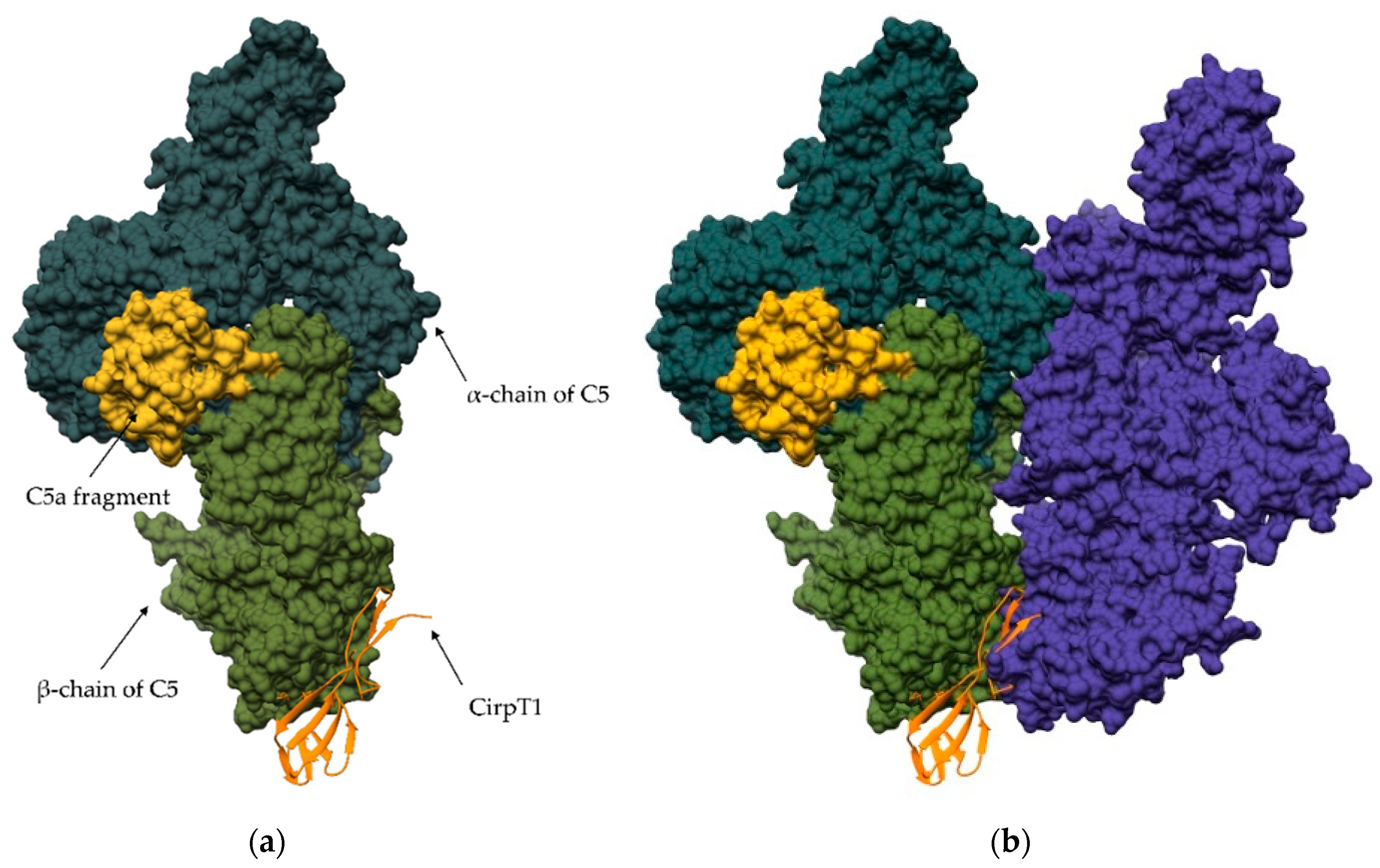
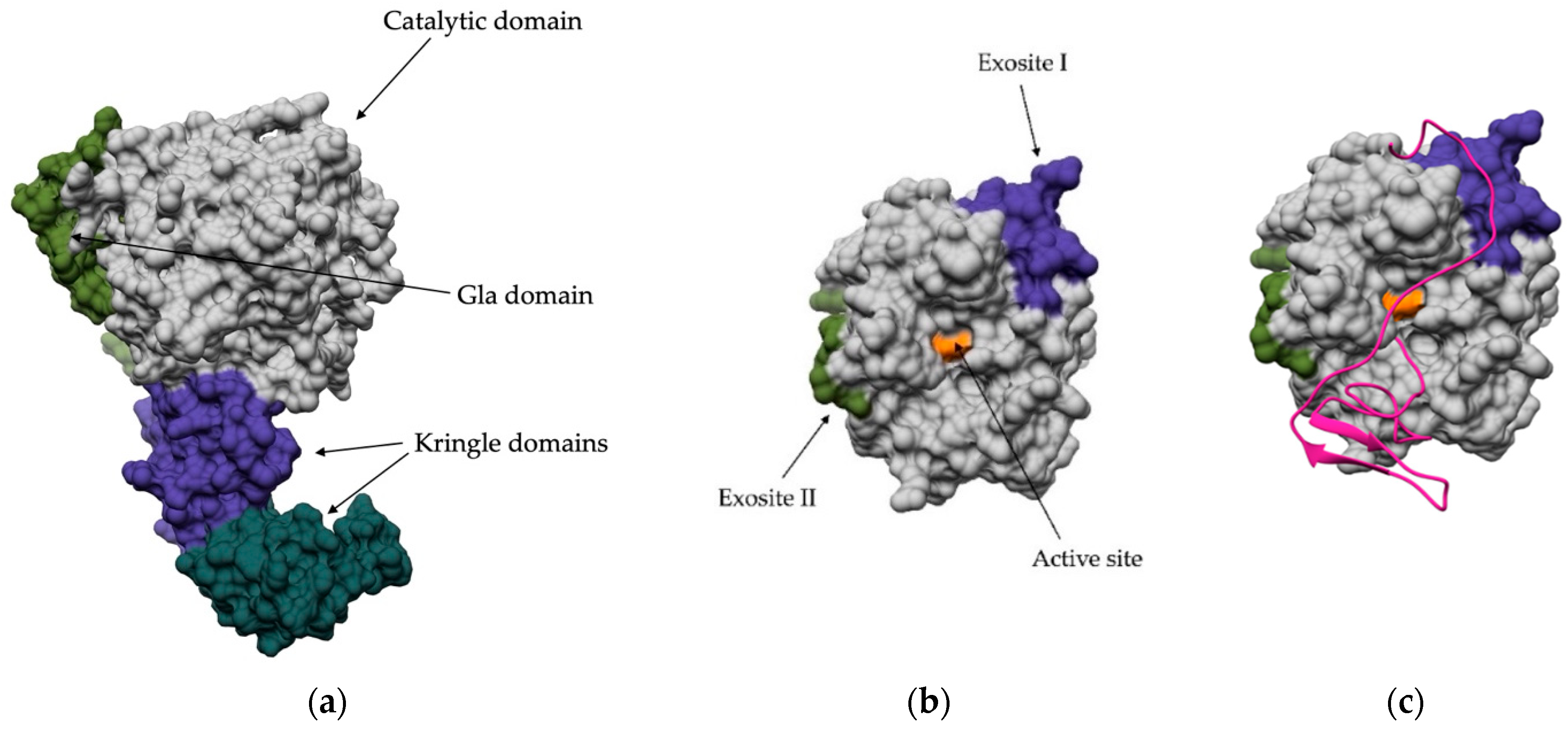
4. Regulation via Exosites
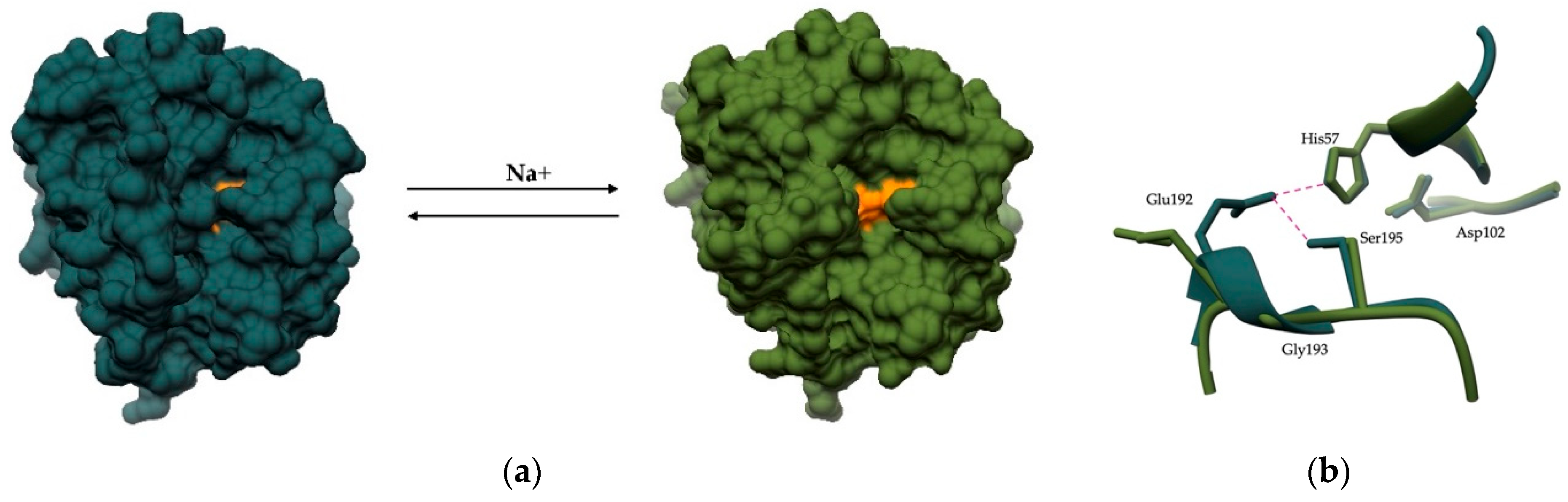
5. Allosteric Regulation by Small Molecule Effectors
6. Other Examples of Peptidase Regulation Outside of the Active Site
7. Non-Inhibitory Binding Partners of Peptidases
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS Database of Proteolytic Enzymes, Their Substrates and Inhibitors in 2017 and a Comparison with Peptidases in the PANTHER Database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.; Lian, X.; Li, F.; Wang, C.; Zhu, F.; Qiu, Y.; Chen, Y. Therapeutic Target Database Update 2022: Facilitating Drug Discovery with Enriched Comparative Data of Targeted Agents. Nucleic Acids Res. 2022, 50, D1398–D1407. [Google Scholar] [CrossRef]
- Tanaka, K.A.; Levy, J.H. Regulation of Thrombin Activity—Pharmacologic and Structural Aspects. Hematol. Oncol. Clin. N. Am. 2007, 21, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Scheen, A.J. The Safety of Gliptins : Updated Data in 2018. Expert Opin. Drug Saf. 2018, 17, 387–405. [Google Scholar] [CrossRef]
- McClung, M.R.; O’Donoghue, M.L.; Papapoulos, S.E.; Bone, H.; Langdahl, B.; Saag, K.G.; Reid, I.R.; Kiel, D.P.; Cavallari, I.; Bonaca, M.P.; et al. Odanacatib for the Treatment of Postmenopausal Osteoporosis: Results of the LOFT Multicentre, Randomised, Double-Blind, Placebo-Controlled Trial and LOFT Extension Study. Lancet Diabetes Endocrinol. 2019, 7, 899–911. [Google Scholar] [CrossRef] [PubMed]
- Freer, S.T.; Kraut, J.; Robertus, J.D.; Wright, H.T.; Xuong, N.H. Chymotrypsinogen: 2.5-Angstrom Crystal Structure, Comparison with Alpha-Chymotrypsin, and Implications for Zymogen Activation. Biochemistry 1970, 9, 1997–2009. [Google Scholar] [CrossRef]
- Bode, W.; Schwager, P.; Huber, R. The Transition of Bovine Trypsinogen to a Trypsin-like State upon Strong Ligand Binding. The Refined Crystal Structures of the Bovine Trypsinogen-Pancreatic Trypsin Inhibitor Complex and of Its Ternary Complex with Ile-Val at 1.9 A Resolution. J. Mol. Biol. 1978, 118, 99–112. [Google Scholar] [CrossRef]
- Birktoft, J.J.; Kraut, J.; Freer, S.T. A Detailed Structural Comparison between the Charge Relay System in Chymotrypsinogen and in Alpha-Chymotrypsin. Biochemistry 1976, 15, 4481–4485. [Google Scholar] [CrossRef]
- Daviet, E.W.; Neurath, H. Identification of a peptide released during autocatalytic activation of trypsinogen. J. Biol. Chem. 1955, 212, 515–529. [Google Scholar] [CrossRef]
- Russo, S.F.; Wahl, E.T. Autocatalytic Activation of Trypsinogen by Trypsin. J. Chem. Educ. 1987, 64, 83–84. [Google Scholar] [CrossRef]
- Yamashina, I. The Action of Enterokinase on Trypsinogen. Biochim. Biophys. Acta 1956, 20, 433–434. [Google Scholar] [CrossRef]
- Drbybr, W.J.; Neurath, H. The Activation of Chymotrypsinogen. J. Am. Chem. Soc. 1955, 77, 814–815. [Google Scholar] [CrossRef]
- Pham, C.T.N.; Ley, T.J. Dipeptidyl Peptidase I Is Required for the Processing and Activation of Granzymes A and B in Vivo. Proc. Natl. Acad. Sci. USA 1999, 96, 8627–8632. [Google Scholar] [CrossRef] [PubMed]
- Adkison, A.M.; Raptis, S.Z.; Kelley, D.G.; Pham, C.T.N. Dipeptidyl Peptidase I Activates Neutrophil-Derived Serine Proteases and Regulates the Development of Acute Experimental Arthritis. J. Clin. Investig. 2002, 109, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Wolters, P.J.; Pham, C.T.N.; Muilenburg, D.J.; Ley, T.J.; Caughey, G.H. Dipeptidyl Peptidase I Is Essential for Activation of Mast Cell Chymases, but Not Tryptases, in Mice. J. Biol. Chem. 2001, 276, 18551–18556. [Google Scholar] [CrossRef]
- Stojkovska Docevska, M.; Novinec, M. Cathepsin C: Structure, Function, and Pharmacological Targeting. Rare Dis. Orphan Drugs J. 2023, 2, 14. [Google Scholar] [CrossRef]
- Cipolla, D.; Zhang, J.; Korkmaz, B.; Chalmers, J.D.; Basso, J.; Lasala, D.; Fernandez, C.; Teper, A.; Mange, K.C.; Perkins, W.R.; et al. Dipeptidyl Peptidase-1 Inhibition with Brensocatib Reduces the Activity of All Major Neutrophil Serine Proteases in Patients with Bronchiectasis: Results from the WILLOW Trial. Respir. Res. 2023, 24, 133. [Google Scholar] [CrossRef] [PubMed]
- Yanamandra, N.; Gumidyala, K.V.; Waldron, K.G.; Gujrati, M.; Olivero, W.C.; Dinh, D.H.; Rao, J.S.; Mohanam, S. Blockade of Cathepsin B Expression in Human Glioblastoma Cells Is Associated with Suppression of Angiogenesis. Oncogene 2004, 23, 2224–2230. [Google Scholar] [CrossRef]
- Moon, H.Y.; Becke, A.; Berron, D.; Becker, B.; Sah, N.; Benoni, G.; Janke, E.; Lubejko, S.T.; Greig, N.H.; Mattison, J.A.; et al. Running-Induced Systemic Cathepsin B Secretion Is Associated with Memory Function. Cell Metab. 2016, 24, 332–340. [Google Scholar] [CrossRef]
- Lakka, S.S.; Gondi, C.S.; Yanamandra, N.; Olivero, W.C.; Dinh, D.H.; Gujrati, M.; Rao, J.S. Inhibition of Cathepsin B and MMP-9 Gene Expression in Glioblastoma Cell Line via RNA Interference Reduces Tumor Cell Invasion, Tumor Growth and Angiogenesis. Oncogene 2004, 23, 4681–4689. [Google Scholar] [CrossRef]
- Veeravalli, K.K.; Chetty, C.; Ponnala, S.; Gondi, C.S.; Lakka, S.S.; Fassett, D.; Klopfenstein, J.D.; Dinh, D.H.; Gujrati, M.; Rao, J.S. MMP-9, UPAR and Cathepsin B Silencing Downregulate Integrins in Human Glioma Xenograft Cells in Vitro and in Vivo in Nude Mice. PLoS ONE 2010, 5, e11583. [Google Scholar] [CrossRef]
- Quraishi, O.; Nägler, D.K.; Fox, T.; Sivaraman, J.; Cygler, M.; Mort, J.S.; Storer, A.C. The Occluding Loop in Cathepsin B Defines the PH Dependence of Inhibition by Its Propeptide. Biochemistry 1999, 38, 5017–5023. [Google Scholar] [CrossRef]
- Pungercar, J.R.; Caglic, D.; Sajid, M.; Dolinar, M.; Vasiljeva, O.; Pozgan, U.; Turk, D.; Bogyo, M.; Turk, V.; Turk, B. Autocatalytic Processing of Procathepsin B Is Triggered by Proenzyme Activity. FEBS J. 2009, 276, 660–668. [Google Scholar] [CrossRef]
- Rozman, J.; Stojan, J.; Kuhelj, R.; Turk, V.; Turk, B. Autocatalytic Processing of Recombinant Human Procathepsin B Is a Bimolecular Process. FEBS Lett. 1999, 459, 358–362. [Google Scholar] [CrossRef]
- McQueney, M.S.; Amegadzie, B.Y.; D’Alessio, K.; Hanning, C.R.; McLaughlin, M.M.; McNulty, D.; Carr, S.A.; Ijames, C.; Kurdyla, J.; Jones, C.S. Autocatalytic Activation of Human Cathepsin K. J. Biol. Chem. 1997, 272, 13955–13960. [Google Scholar] [CrossRef] [PubMed]
- Ishidoh, K.; Kominami, E. Multi-Step Processing of Procathepsin L in Vitro. FEBS Lett. 1994, 352, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Fujisawa, Y. Processing Properties of Recombinant Human Procathepsin L. Biochem. Biophys. Res. Commun. 1997, 230, 143–146. [Google Scholar] [CrossRef]
- Ménard, R.; Carmona, E.; Takebe, S.; Dufour, É.; Plouffe, C.; Mason, P.; Mort, J.S. Autocatalytic Processing of Recombinant Human Procathepsin L. Contribution of Both Intermolecular and Unimolecular Events in the Processing of Procathepsin L in Vitro. J. Biol. Chem. 1998, 273, 4478–4484. [Google Scholar] [CrossRef]
- Kopitar, G.; Dolinar, M.; Štrukelj, B.; Pungerčar, J.; Turk, V. Folding and Activation of Human Procathepsin S from Inclusion Bodies Produced in Escherichia Coli. Eur. J. Biochem. 1996, 236, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Shi, G.P.; Yao, P.M.; Li, Z.; Chapman, H.A.; Brömme, D. Human Cathepsin F. Molecular Cloning, Functional Expression, Tissue Localization, and Enzymatic Characterization. J. Biol. Chem. 1998, 273, 32000–32008. [Google Scholar] [CrossRef]
- Sivaraman, J.; Nägler, D.K.; Zhang, R.; Ménard, R.; Cygler, M. Crystal Structure of Human Procathepsin X: A Cysteine Protease with the Proregion Covalently Linked to the Active Site Cysteine. J. Mol. Biol. 2000, 295, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Nägler, D.K.; Zhang, R.; Tam, W.; Sulea, T.; Purisima, E.O.; Ménard, R. Human Cathepsin X: A Cysteine Protease with Unique Carboxypeptidase Activity. Biochemistry 1999, 38, 12648–12654. [Google Scholar] [CrossRef] [PubMed]
- Turk, D.; Janjic, V.; Stern, I.; Podobnik, M.; Lamba, D.; Dahl, S.W.; Lauritzen, C.; Pedersen, J.; Turk, V.; Turk, B. Structure of Human Dipeptidyl Peptidase I (Cathepsin C): Exclusion Domain Added to an Endopeptidase Framework Creates the Machine for Activation of Granular Serine Proteases. EMBO J. 2001, 20, 6570–6582. [Google Scholar] [CrossRef]
- Rebernik, M.; Lenarčič, B.; Novinec, M. The Catalytic Domain of Cathepsin C (Dipeptidyl-Peptidase I) Alone Is a Fully Functional Endoprotease. Protein Expr. Purif. 2019, 157, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Dahl, S.W.; Halkier, T.; Lauritzen, C.; Dolenc, I.; Pedersen, J.; Turk, V.; Turk, B. Human Recombinant Pro-Dipeptidyl Peptidase I (Cathepsin C) Can Be Activated by Cathepsins L and S but Not by Autocatalytic Processing. Biochemistry 2001, 40, 1671–1678. [Google Scholar] [CrossRef]
- Gunčar, G.; Podobnik, M.; Pungerčar, J.; Štrukelj, B.; Turk, V.; Turk, D. Crystal Structure of Porcine Cathepsin H Determined at 2.1 å Resolution: Location of the Mini-Chain C-Terminal Carboxyl Group Defines Cathepsin H Aminopeptidase Function. Structure 1998, 6, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Purtha, W.; Cortesio, C.; Rui, H.; Gu, Y.; Chen, H.; Sickmier, E.A.; Manzanillo, P.; Huang, X. Crystal Structures of Human Procathepsin H. PLoS ONE 2018, 13, e0200374. [Google Scholar] [CrossRef]
- Van Wart, H.E.; Birkedal-Hansen, H. The Cysteine Switch: A Principle of Regulation of Metalloproteinase Activity with Potential Applicability to the Entire Matrix Metalloproteinase Gene Family. Proc. Natl. Acad. Sci. USA 1990, 87, 5578–5582. [Google Scholar] [CrossRef]
- Chen, L.C.; Noelken, M.E.; Nagase, H. Disruption of the Cysteine-75 and Zinc Ion Coordination Is Not Sufficient to Activate the Precursor of Human Matrix Metalloproteinase 3 (Stromelysin 1). Biochemistry 1993, 32, 10289–10295. [Google Scholar] [CrossRef]
- Nagase, H.; Enghild, J.J.; Suzuki, K.; Salvesen, G. Stepwise Activation Mechanisms of the Precursor of Matrix Metalloproteinase 3 (Stromelysin) by Proteinases and (4-Aminophenyl)Mercuric Acetate. Biochemistry 1990, 29, 5783–5789. [Google Scholar] [CrossRef]
- Peppin, G.J.; Weiss, S.J. Activation of the Endogenous Metalloproteinase, Gelatinase, by Triggered Human Neutrophils. Proc. Natl. Acad. Sci. USA 1986, 83, 4322–4326. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.J.; Peppin, G.; Ortiz, X.; Ragsdale, C.; Test, S.T. Oxidative Autoactivation of Latent Collagenase by Human Neutrophils. Science 1985, 227, 747–749. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Kassimm, S.Y.; Parks, W.C.; Heinecke, J.W. Hypochlorous Acid Oxygenates the Cysteine Switch Domain of Pro-Matrilysin (MMP-7). A Mechanism for Matrix Metalloproteinase Activation and Atherosclerotic Plaque Rupture by Myeloperoxidase. J. Biol. Chem. 2001, 276, 41279–41287. [Google Scholar] [CrossRef] [PubMed]
- Perlmann, G.E. The Optical Rotatory Properties of Pepsinogen. J. Mol. Biol. 1963, 6, 452–464. [Google Scholar] [CrossRef] [PubMed]
- James, M.N.G.; Sielecki, A.R. Molecular Structure of an Aspartic Proteinase Zymogen, Porcine Pepsinogen, at 1.8 A Resolution. Nature 1986, 319, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Al-Janabi, J.; Hartsuck, J.A.; Tang, J. Kinetics and Mechanism of Pepsinogen Activation. J. Biol. Chem. 1972, 247, 4628–4632. [Google Scholar] [CrossRef]
- Derkx, F.H.; Schalekamp, M.P.; Schalekamp, M.A. Two-Step Prorenin-Renin Conversion. Isolation of an Intermediary Form of Activated Prorenin. J. Biol. Chem. 1987, 262, 2472–2477. [Google Scholar] [CrossRef]
- Gieselmann, V.; Hasilik, A.; Von Figura, K. Processing of Human Cathepsin D in Lysosomes in Vitro. J. Biol. Chem. 1985, 260, 3215–3220. [Google Scholar] [CrossRef]
- Hasilik, A.; von Figura, K.; Conzelmann, E.; Nehrkorn, H.; Sandhoff, K. Lysosomal Enzyme Precursors in Human Fibroblasts. Activation of Cathepsin D Precursor in Vitro and Activity of Beta-Hexosaminidase A Precursor towards Ganglioside GM2. Eur. J. Biochem. 1982, 125, 317–321. [Google Scholar] [CrossRef]
- Richo, G.R.; Conner, G.E. Structural Requirements of Procathepsin D Activation and Maturation. J. Biol. Chem. 1994, 269, 14806–14812. [Google Scholar] [CrossRef]
- Wittlin, S.; Ro, È.; Sel, J.; Hofmann, F.; Stover, D.R. Mechanisms and Kinetics of Procathepsin D Activation. Eur. J. Biochem. 1999, 265, 384–393. [Google Scholar] [CrossRef]
- Beckman, M.; Freeman, C.; Parish, C.R.; Small, D.H. Activation of Cathepsin D by Glycosaminoglycans. FEBS J. 2009, 276, 7343–7352. [Google Scholar] [CrossRef]
- Fox, T.; Storer, A.C.; de Miguel, E.; Mort, J.S. Potent Slow-Binding Inhibition of Cathepsin B by Its Propeptide. Biochemistry 1992, 31, 12571–12576. [Google Scholar] [CrossRef]
- Carmona, E.; Dufour, É.; Plouffe, C.; Takebe, S.; Mason, P.; Mort, J.S.; Ménard, R. Potency and Selectivity of the Cathepsin L Propeptide as an Inhibitor of Cysteine Proteases. Biochemistry 1996, 35, 8149–8157. [Google Scholar] [CrossRef] [PubMed]
- Maubach, G.; Schilling, K.; Rommerskirch, W.; Wenz, I.; Schultz, J.E.; Weber, E.; Wiederanders, B. The Inhibition of Cathepsin S by Its Propeptide--Specificity and Mechanism of Action. Eur. J. Biochem. 1997, 250, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Guay, J.; Falgueyret, J.P.; Ducret, A.; Percival, M.D.; Mancini, J.A. Potency and Selectivity of Inhibition of Cathepsin K, L and S by Their Respective Propeptides. Eur. J. Biochem. 2000, 267, 6311–6318. [Google Scholar] [CrossRef] [PubMed]
- Reichhardt, M.P.; Johnson, S.; Tang, T.; Morgan, T.; Tebeka, N.; Popitsch, N.; Deme, J.C.; Jore, M.M.; Lea, S.M. An Inhibitor of Complement C5 Provides Structural Insights into Activation. Proc. Natl. Acad. Sci. USA 2020, 117, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Chen, Y.W.; Battu, A.; Wilder, P.; Weber, D.; Yu, W.; MacKerell, A.D.; Chen, L.M.; Chai, K.X.; Johnson, M.D.; et al. Targeting Zymogen Activation to Control the Matriptase-Prostasin Proteolytic Cascade. J. Med. Chem. 2011, 54, 7567–7578. [Google Scholar] [CrossRef]
- Lee, S.L.; Dickson, R.B.; Lin, C.Y. Activation of Hepatocyte Growth Factor and Urokinase/Plasminogen Activator by Matriptase, an Epithelial Membrane Serine Protease. J. Biol. Chem. 2000, 275, 36720–36725. [Google Scholar] [CrossRef]
- List, K.; Szabo, R.; Molinolo, A.; Sriuranpong, V.; Redeye, V.; Murdock, T.; Burke, B.; Nielsen, B.S.; Gutkind, J.S.; Bugge, T.H. Deregulated Matriptase Causes Ras-Independent Multistage Carcinogenesis and Promotes Ras-Mediated Malignant Transformation. Genes Dev. 2005, 19, 1934–1950. [Google Scholar] [CrossRef]
- Andreasen, P.A.; Egelund, R.; Petersen, H.H. The Plasminogen Activation System in Tumor Growth, Invasion, and Metastasis. Cell. Mol. Life Sci. 2000, 57, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Blouse, G.E.; Bøotkjaer, K.A.; Deryugina, E.; Byszuk, A.A.; Jensen, J.M.; Mortensen, K.K.; Quigley, J.P.; Andreasen, P.A. A Novel Mode of Intervention with Serine Protease Activity: Targeting Zymogen Activation. J. Biol. Chem. 2009, 284, 4647–4657. [Google Scholar] [CrossRef]
- Bergers, G.; Brekken, R.; McMahon, G.; Vu, T.H.; Itoh, T.; Tamaki, K.; Tanzawa, K.; Thorpe, P.; Itohara, S.; Werb, Z.; et al. Matrix Metalloproteinase-9 Triggers the Angiogenic Switch during Carcinogenesis. Nat. Cell Biol. 2000, 2, 737–744. [Google Scholar] [CrossRef]
- Song, Z.; Wang, J.; Su, Q.; Luan, M.; Chen, X.; Xu, X. The Role of MMP-2 and MMP-9 in the Metastasis and Development of Hypopharyngeal Carcinoma. Braz. J. Otorhinolaryngol. 2021, 87, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Mehner, C.; Hockla, A.; Miller, E.; Ran, S.; Radisky, D.C.; Radisky, E.S. Tumor Cell-Produced Matrix Metalloproteinase 9 (MMP-9) Drives Malignant Progression and Metastasis of Basal-like Triple Negative Breast Cancer. Oncotarget 2014, 5, 2736–2749. [Google Scholar] [CrossRef] [PubMed]
- Scannevin, R.H.; Alexander, R.; Haarlander, T.M.; Burke, S.L.; Singer, M.; Huo, C.; Zhang, Y.M.; Maguire, D.; Spurlino, J.; Deckman, I.; et al. Discovery of a Highly Selective Chemical Inhibitor of Matrix Metalloproteinase-9 (MMP-9) That Allosterically Inhibits Zymogen Activation. J. Biol. Chem. 2017, 292, 17963–17974. [Google Scholar] [CrossRef]
- Muzio, M.; Stockwell, B.R.; Stennicke, H.R.; Salvesen, G.S.; Dixit, V.M. An Induced Proximity Model for Caspase-8 Activation. J. Biol. Chem. 1998, 273, 2926–2930. [Google Scholar] [CrossRef]
- Li, P.; Nijhawan, D.; Budihardjo, I.; Srinivasula, S.M.; Ahmad, M.; Alnemri, E.S.; Wang, X. Cytochrome c and DATP-Dependent Formation of Apaf-1/Caspase-9 Complex Initiates an Apoptotic Protease Cascade. Cell 1997, 91, 479–489. [Google Scholar] [CrossRef]
- Pop, C.; Chen, Y.R.; Smith, B.; Bose, K.; Bobay, B.; Tripathy, A.; Franzen, S.; Clark, A.C. Removal of the Pro-Domain Does Not Affect the Conformation of the Procaspase-3 Dimer. Biochemistry 2001, 40, 14224–14235. [Google Scholar] [CrossRef]
- Stennicke, H.R.; Jürgensmeier, J.M.; Shin, H.; Deveraux, Q.; Wolf, B.B.; Yang, X.; Zhou, Q.; Ellerby, H.M.; Ellerby, L.M.; Bredesen, D.; et al. Pro-Caspase-3 Is a Major Physiologic Target of Caspase-8. J. Biol. Chem. 1998, 273, 27084–27090. [Google Scholar] [CrossRef]
- Chai, J.; Wu, Q.; Shiozaki, E.; Srinivasula, S.M.; Alnemri, E.S.; Shi, Y. Crystal Structure of a Procaspase-7 Zymogen: Mechanisms of Activation and Substrate Binding. Cell 2001, 107, 399–407. [Google Scholar] [CrossRef]
- Peterson, Q.P.; Goode, D.R.; West, D.C.; Ramsey, K.N.; Lee, J.J.Y.; Hergenrother, P.J. PAC-1 Activates Procaspase-3 in Vitro through Relief of Zinc-Mediated Inhibition. J. Mol. Biol. 2009, 388, 144. [Google Scholar] [CrossRef] [PubMed]
- Putt, K.S.; Chen, G.W.; Pearson, J.M.; Sandhorst, J.S.; Hoagland, M.S.; Kwon, J.T.; Hwang, S.K.; Jin, H.; Churchwell, M.I.; Cho, M.H.; et al. Small-Molecule Activation of Procaspase-3 to Caspase-3 as a Personalized Anticancer Strategy. Nat. Chem. Biol. 2006, 2, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Wolan, D.W.; Zorn, J.A.; Gray, D.C.; Wells, J.A. Small-Molecule Activators of a Proenzyme. Science 2009, 326, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Zorn, J.A.; Wille, H.; Wolan, D.W.; Wells, J.A. Self-Assembling Small Molecules Form Nanofibrils That Bind Procaspase-3 to Promote Activation. J. Am. Chem. Soc. 2011, 133, 19630–19633. [Google Scholar] [CrossRef] [PubMed]
- Zorn, J.A.; Wolan, D.W.; Agard, N.J.; Wells, J.A. Fibrils Colocalize Caspase-3 with Procaspase-3 to Foster Maturation. J. Biol. Chem. 2012, 287, 33781–33795. [Google Scholar] [CrossRef] [PubMed]
- Botham, R.C.; Fan, T.M.; Im, I.; Borst, L.B.; Dirikolu, L.; Hergenrother, P.J. Dual Small-Molecule Targeting of Procaspase-3 Dramatically Enhances Zymogen Activation and Anticancer Activity. J. Am. Chem. Soc. 2014, 136, 1312–1319. [Google Scholar] [CrossRef]
- O’Farrell, P.A.; Gonzalez, F.; Zheng, W.; Johnston, S.A.; Joshua-Tor, L. Crystal Structure of Human Bleomycin Hydrolase, a Self-Compartmentalizing Cysteine Protease. Structure 1999, 7, 619–627. [Google Scholar] [CrossRef]
- Schmidt, U.; Darke, P.L. Dimerization and Activation of the Herpes Simplex Virus Type 1 Protease. J. Biol. Chem. 1997, 272, 7732–7735. [Google Scholar] [CrossRef]
- Wlodawer, A.; Miller, M.; Jaskólski, M.; Sathyanarayana, B.K.; Baldwin, E.; Weber, I.T.; Selk, L.M.; Clawson, L.; Schneider, J.; Kent, S.B.H. Conserved Folding in Retroviral Proteases: Crystal Structure of a Synthetic HIV-1 Protease. Science 1989, 245, 616–621. [Google Scholar] [CrossRef]
- Darke, P.L.; Cole, J.L.; Waxman, L.; Hall, D.L.; Sardana, M.K.; Kuo, L.C. Active Human Cytomegalovirus Protease Is a Dimer. J. Biol. Chem. 1996, 271, 7445–7449. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal Structure of SARS-CoV-2 Main Protease Provides a Basis for Design of Improved α-Ketoamide Inhibitors. Science 2020, 368, 409. [Google Scholar] [CrossRef] [PubMed]
- Majerová, Á.; Novotný, P. Precursors of Viral Proteases as Distinct Drug Targets. Viruses 2021, 13, 1981. [Google Scholar] [CrossRef] [PubMed]
- Zephyr, J.; Kurt Yilmaz, N.; Schiffer, C.A. Viral Proteases: Structure, Mechanism and Inhibition. Enzymes 2021, 50, 301. [Google Scholar] [CrossRef]
- Bannwarth, L.; Rose, T.; Dufau, L.; Vanderesse, R.; Dumond, J.; Jamart-Grégoire, B.; Pannecouque, C.; De Clercq, E.; Le Reboud-Ravaux, M. Dimer Disruption and Monomer Sequestration by Alkyl Tripeptides Are Successful Strategies for Inhibiting Wild-Type and Multidrug-Resistant Mutated HIV-1 Proteases. Biochemistry 2008, 48, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Zutshi, R.; Franciskovich, J.; Shultz, M.; Schweitzer, B.; Bishop, P.; Wilson, M.; Chmielewski, J. Targeting the Dimerization Interface of HIV-1 Protease: Inhibition with Cross-Linked Interfacial Peptides. J. Am. Chem. Soc. 1997, 119, 4841–4845. [Google Scholar] [CrossRef]
- Goyal, B.; Goyal, D. Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy. ACS Comb. Sci. 2020, 22, 297–305. [Google Scholar] [CrossRef]
- Vallet, S.D.; Clerc, O.; Ricard-Blum, S. Glycosaminoglycan-Protein Interactions: The First Draft of the Glycosaminoglycan Interactome. J. Histochem. Cytochem. 2021, 69, 93–104. [Google Scholar] [CrossRef]
- Abildgaard, U. Binding of Thrombin to Antithrombin III. Scand. J. Clin. Lab. Investig. 1969, 24, 23–27. [Google Scholar] [CrossRef]
- Rosenberg, R.D.; Damus, P.S. The Purification and Mechanism of Action of Human Antithrombin-Heparin Cofactor. J. Biol. Chem. 1973, 248, 6490–6505. [Google Scholar] [CrossRef]
- Jin, L.; Abrahams, J.P.; Skinner, R.; Petitou, M.; Pike, R.N.; Carrell, R.W. The Anticoagulant Activation of Antithrombin by Heparin. Proc. Natl. Acad. Sci. USA 1997, 94, 14683–14688. [Google Scholar] [CrossRef] [PubMed]
- Craig, P.A.; Olson, S.T.; Shore, J.D. Transient Kinetics of Heparin-Catalyzed Protease Inactivation by Antithrombin III: Characterization of Assembly, Product Formation, and Heparin Dissociation Steps in the Factor Xa Reaction. J. Biol. Chem. 1989, 264, 5452–5461. [Google Scholar] [CrossRef] [PubMed]
- Gozzo, A.J.; Nunes, V.A.; Nader, H.B.; Dietrich, C.P.; Carmona, A.K.; Sampaio, M.U.; Sampaio, C.A.M.; Araújo, M.S. Glycosaminoglycans Affect the Interaction of Human Plasma Kallikrein with Plasminogen, Factor XII and Inhibitors. Braz. J. Med. Biol. Res. 2003, 36, 1055–1059. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Burrowes, C.E.; Habal, F.M.; Movat, H.Z. The Inhibition of Human Plasma Kallikrein by Antithrombin III. Thromb. Res. 1975, 7, 175–183. [Google Scholar] [CrossRef]
- Highsmith, R.F.; Rosenberg, R.D. The Inhibition of Human Plasmin by Human Antithrombin-Heparin Cofactor. J. Biol. Chem. 1974, 249, 4335–4338. [Google Scholar] [CrossRef]
- Lawson, J.H.; Butenas, S.; Ribarik, N.; Mann, K.G. Complex-Dependent Inhibition of Factor VIIa by Antithrombin III and Heparin. J. Biol. Chem. 1993, 268, 767–770. [Google Scholar] [CrossRef]
- Hallgren, J.; Lindahl, S.; Pejler, G. Structural Requirements and Mechanism for Heparin-Dependent Activation and Tetramerization of Human BetaI- and BetaII-Tryptase. J. Mol. Biol. 2005, 345, 129–139. [Google Scholar] [CrossRef]
- José Barbosa Pereira, P.; Bergner, A.; Macedo-Ribeiro, S.; Huber, R.; Matschiner, G.; Fritz, H.; Sommerhoff, C.P.; Bode, W. Human Beta-Tryptase Is a Ring-like Tetramer with Active Sites Facing a Central Pore. Nature 1998, 392, 306–311. [Google Scholar] [CrossRef]
- Fajardo, I.; Pejler, G. Formation of Active Monomers from Tetrameric Human Beta-Tryptase. Biochem. J. 2003, 369, 603. [Google Scholar] [CrossRef]
- Lindstedt, L.; Lee, M.; Kovanen, P.T. Chymase Bound to Heparin Is Resistant to Its Natural Inhibitors and Capable of Proteolyzing High Density Lipoproteins in Aortic Intimal Fluid. Atherosclerosis 2001, 155, 87–97. [Google Scholar] [CrossRef]
- Walter, M.; Plotnick, M.; Schechter, N.M. Inhibition of Human Mast Cell Chymase by Secretory Leukocyte Proteinase Inhibitor: Enhancement of the Interaction by Heparin. Arch. Biochem. Biophys. 1996, 327, 81–88. [Google Scholar] [CrossRef]
- Ermolieff, J.; Boudier, C.; Laine, A.; Meyer, B.; Bieth, J.G. Heparin Protects Cathepsin G against Inhibition by Protein Proteinase Inhibitors. J. Biol. Chem. 1994, 269, 29502–29508. [Google Scholar] [CrossRef] [PubMed]
- Ermolieff, J.; Duranton, J.; Petitou, M.; Bieth, J.G. Heparin Accelerates the Inhibition of Cathepsin G by Mucus Proteinase Inhibitor: Potent Effect of O-Butyrylated Heparin. Biochem. J. 1998, 330 Pt 3, 1369–1374. [Google Scholar] [CrossRef]
- Cadene, M.; Boudier, C.; De Marcillac, G.D.; Bieth, J.G. Influence of Low Molecular Mass Heparin on the Kinetics of Neutrophil Elastase Inhibition by Mucus Proteinase Inhibitor. J. Biol. Chem. 1995, 270, 13204–13209. [Google Scholar] [CrossRef] [PubMed]
- Faller, B.; Bieth, J.G.; Mely, Y.; Gerard, D. Heparin-Induced Conformational Change and Activation of Mucus Proteinase Inhibitor. Biochemistry 1992, 31, 8285–8290. [Google Scholar] [CrossRef]
- Frommherz, K.J.; Faller, B.; Bieth, J.G. Heparin Strongly Decreases the Rate of Inhibition of Neutrophil Elastase by Alpha 1-Proteinase Inhibitor. J. Biol. Chem. 1991, 266, 15356–15362. [Google Scholar] [CrossRef] [PubMed]
- Ledoux, D.; Merciris, D.; Barritault, D.; Caruelle, J.P. Heparin-like Dextran Derivatives as Well as Glycosaminoglycans Inhibit the Enzymatic Activity of Human Cathepsin G. FEBS Lett. 2003, 537, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, V.; Piccardoni, P.; Maugeri, N.; De Gaetano, G.; Cerletti, C. Inhibition by Heparin of Platelet Activation Induced by Neutrophil-Derived Cathepsin G. Eur. J. Pharmacol. 1992, 216, 401–405. [Google Scholar] [CrossRef]
- Spencer, J.L.; Stone, P.J.; Nugent, M.A. New Insights into the Inhibition of Human Neutrophil Elastase by Heparin. Biochemistry 2006, 45, 9104–9120. [Google Scholar] [CrossRef]
- Baici, A.; Bradamante, P. Interaction between Human Leukocyte Elastase and Chondroitin Sulfate. Chem. Biol. Interact. 1984, 51, 1–11. [Google Scholar] [CrossRef]
- Kostoulas, G.; Hörler, D.; Naggi, A.; Casu, B.; Baici, A. Electrostatic Interactions between Human Leukocyte Elastase and Sulfated Glycosaminoglycans: Physiological Implications. Biol. Chem. 1997, 378, 1481–1489. [Google Scholar] [CrossRef]
- Schenker, P.; Baici, A. Paradoxical Interactions between Modifiers and Elastase-2. FEBS J. 2010, 277, 2486–2495. [Google Scholar] [CrossRef]
- Dewald, B.; Rindler-Ludwig, R.; Bretz, U.; Baggiolini, M. Subcellular Localization and Heterogeneity of Neutral Proteases in Neutrophilic Polymorphonuclear Leukocytes. J. Exp. Med. 1975, 141, 709–723. [Google Scholar] [CrossRef]
- Li, Z.; Yasuda, Y.; Li, W.; Bogyo, M.; Katz, N.; Gordon, R.E.; Fields, G.B.; Brömme, D. Regulation of Collagenase Activities of Human Cathepsins by Glycosaminoglycans. J. Biol. Chem. 2004, 279, 5470–5479. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hou, W.S.; Escalante-Torres, C.R.; Gelb, B.D.; Bromme, D. Collagenase Activity of Cathepsin K Depends on Complex Formation with Chondroitin Sulfate. J. Biol. Chem. 2002, 277, 28669–28676. [Google Scholar] [CrossRef] [PubMed]
- Cherney, M.M.; Lecaille, F.; Kienitz, M.; Nallaseth, F.S.; Li, Z.; James, M.N.G.; Brömme, D. Structure-Activity Analysis of Cathepsin K/Chondroitin 4-Sulfate Interactions. J. Biol. Chem. 2011, 286, 8988–8998. [Google Scholar] [CrossRef]
- Aguda, A.H.; Panwar, P.; Du, X.; Nguyen, N.T.; Brayer, G.D.; Brömme, D. Structural Basis of Collagen Fiber Degradation by Cathepsin K. Proc. Natl. Acad. Sci. USA 2014, 111, 17474–17479. [Google Scholar] [CrossRef]
- Li, Z.; Kienetz, M.; Cherney, M.M.; James, M.N.G.; Brömme, D. The Crystal and Molecular Structures of a Cathepsin K:Chondroitin Sulfate Complex. J. Mol. Biol. 2008, 383, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hou, W.S.; Brömme, D. Collagenolytic Activity of Cathepsin K Is Specifically Modulated by Cartilage-Resident Chondroitin Sulfates. Biochemistry 2000, 39, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Novinec, M.; Kovačič, L.; Lenarĉič, B.; Baici, A. Conformational Flexibility and Allosteric Regulation of Cathepsin K. Biochem. J. 2010, 429, 379–389. [Google Scholar] [CrossRef]
- Sage, J.; Mallèvre, F.; Barbarin-Costes, F.; Samsonov, S.A.; Gehrcke, J.P.; Pisabarro, M.T.; Perrier, E.; Schnebert, S.; Roget, A.; Livache, T.; et al. Binding of Chondroitin 4-Sulfate to Cathepsin S Regulates Its Enzymatic Activity. Biochemistry 2013, 52, 6487–6498. [Google Scholar] [CrossRef] [PubMed]
- Denamur, S.; Chazeirat, T.; Maszota-Zieleniak, M.; Vivès, R.R.; Saidi, A.; Zhang, F.; Linhardt, R.J.; Labarthe, F.; Samsonov, S.A.; Lalmanach, G.; et al. Binding of Heparan Sulfate to Human Cystatin C Modulates Inhibition of Cathepsin L: Putative Consequences in Mucopolysaccharidosis. Carbohydr. Polym. 2022, 293, 119734. [Google Scholar] [CrossRef] [PubMed]
- Higgins, W.J.; Fox, D.M.; Kowalski, P.S.; Nielsen, J.E.; Worrall, D.M. Heparin Enhances Serpin Inhibition of the Cysteine Protease Cathepsin L. J. Biol. Chem. 2010, 285, 3722–3729. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, P.A.; Huang, L.; Zhuo, Y.; Lu, J.; Bahnck, C.; Stachel, S.J.; Carroll, S.S.; Duong, L.T. Chondroitin Sulfate Promotes Activation of Cathepsin K. J. Biol. Chem. 2014, 289, 21562–21572. [Google Scholar] [CrossRef]
- Caglič, D.; Pungerčar, J.R.; Pejler, G.; Turk, V.; Turk, B. Glycosaminoglycans Facilitate Procathepsin B Activation through Disruption of Propeptide-Mature Enzyme Interactions. J. Biol. Chem. 2007, 282, 33076–33085. [Google Scholar] [CrossRef] [PubMed]
- Vasiljeva, O.; Dolinar, M.; Pungerčar, J.R.; Turk, V.; Turk, B. Recombinant Human Procathepsin S Is Capable of Autocatalytic Processing at Neutral PH in the Presence of Glycosaminoglycans. FEBS Lett. 2005, 579, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Echtermeyer, F.; Bertrand, J.; Dreier, R.; Meinecke, I.; Neugebauer, K.; Fuerst, M.; Lee, Y.J.; Song, Y.W.; Herzog, C.; Theilmeier, G.; et al. Syndecan-4 Regulates ADAMTS-5 Activation and Cartilage Breakdown in Osteoarthritis. Nat. Med. 2009, 15, 1072–1076. [Google Scholar] [CrossRef]
- Crabbe, T.; Ioannou, C.; Docherty, A.J.P. Human Progelatinase A Can Be Activated by Autolysis at a Rate That Is Concentration-Dependent and Enhanced by Heparin Bound to the C-Terminal Domain. Eur. J. Biochem. 1993, 218, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Crabbe, T.; O’Connell, J.P.; Smith, B.J.; Docherty, A.J.P. Reciprocated Matrix Metalloproteinase Activation: A Process Performed by Interstitial Collagenase and Progelatinase A. Biochemistry 1994, 33, 14419–14425. [Google Scholar] [CrossRef]
- Gao, G.; Plaas, A.; Thompson, V.P.; Jin, S.; Zuo, F.; Sandy, J.D. ADAMTS4 (Aggrecanase-1) Activation on the Cell Surface Involves C-Terminal Cleavage by Glycosylphosphatidyl Inositol-Anchored Membrane Type 4-Matrix Metalloproteinase and Binding of the Activated Proteinase to Chondroitin Sulfate and Heparan Sulfate on Syndecan-1. J. Biol. Chem. 2004, 279, 10042–10051. [Google Scholar] [CrossRef]
- Sørensen, H.P.; Vivès, R.R.; Manetopoulos, C.; Albrechtsen, R.; Lydolph, M.C.; Jacobsen, J.; Couchman, J.R.; Wewer, U.M. Heparan Sulfate Regulates ADAM12 through a Molecular Switch Mechanism. J. Biol. Chem. 2008, 283, 31920–31932. [Google Scholar] [CrossRef]
- Pozzi, N.; Chen, Z.; Gohara, D.W.; Niu, W.; Heyduk, T.; Di Cera, E. Crystal Structure of Prothrombin Reveals Conformational Flexibility and Mechanism of Activation. J. Biol. Chem. 2013, 288, 22734–22744. [Google Scholar] [CrossRef] [PubMed]
- Stubbs, M.T.; Oschkinat, H.; Mayr, I.; Huber, R.; Angliker, H.; Stone, S.R.; Bode, W. The Interaction of Thrombin with Fibrinogen. A Structural Basis for Its Specificity. Eur. J. Biochem. 1992, 206, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Pechik, I.; Madrazo, J.; Mosesson, M.W.; Hernandez, I.; Gilliland, G.L.; Medved, L. Crystal Structure of the Complex between Thrombin and the Central “E” Region of Fibrin. Proc. Natl. Acad. Sci. USA 2004, 101, 2718–2723. [Google Scholar] [CrossRef] [PubMed]
- Baglin, T.P.; Carrell, R.W.; Church, F.C.; Esmon, C.T.; Huntington, J.A. Crystal Structures of Native and Thrombin-Complexed Heparin Cofactor II Reveal a Multistep Allosteric Mechanism. Proc. Natl. Acad. Sci. USA 2002, 99, 11079–11084. [Google Scholar] [CrossRef]
- Yun, T.H.; Baglia, F.A.; Myles, T.; Navaneetham, D.; López, J.A.; Walsh, P.N.; Leung, L.L.K. Thrombin Activation of Factor XI on Activated Platelets Requires the Interaction of Factor XI and Platelet Glycoprotein Ib Alpha with Thrombin Anion-Binding Exosites I and II, Respectively. J. Biol. Chem. 2003, 278, 48112–48119. [Google Scholar] [CrossRef]
- Fuentes-Prior, P.; Iwanaga, Y.; Huber, R.; Pagila, R.; Rumennik, G.; Seto, M.; Morser, J.; Light, D.R.; Bode, W. Structural Basis for the Anticoagulant Activity of the Thrombin-Thrombomodulin Complex. Nature 2000, 404, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Carter, W.J.; Cama, E.; Huntington, J.A. Crystal Structure of Thrombin Bound to Heparin. J. Biol. Chem. 2005, 280, 2745–2749. [Google Scholar] [CrossRef]
- Sheehan, J.P.; Sadler, J.E. Molecular Mapping of the Heparin-Binding Exosite of Thrombin. Proc. Natl. Acad. Sci. USA 1994, 91, 5518–5522. [Google Scholar] [CrossRef]
- Li, C.Q.; Vindigni, A.; Sadler, J.E.; Wardell, M.R. Platelet Glycoprotein Ib Alpha Binds to Thrombin Anion-Binding Exosite II Inducing Allosteric Changes in the Activity of Thrombin. J. Biol. Chem. 2001, 276, 6161–6168. [Google Scholar] [CrossRef]
- Naskis, M.C.; Fenton, J.W.; Maraganoren, J.M.; Olson, S.T.; Shafer, J.A. The COOH-Terminal Domain of Hirudin. J. Biol. Chem. 1990, 265, 13484–13489. [Google Scholar]
- Krstenansky, J.L.; Mao, S.J. Antithrombin Properties of C-Terminus of Hirudin Using Synthetic Unsulfated N Alpha-Acetyl-Hirudin45-65. FEBS Lett. 1987, 211, 10–16. [Google Scholar] [CrossRef]
- Arocas, V.; Zingali, R.B.; Guillin, M.C.; Bon, C.; Jandrot-Perrus, M. Bothrojaracin: A Potent Two-Site-Directed Thrombin Inhibitor. Biochemistry 1996, 35, 9083–9089. [Google Scholar] [CrossRef] [PubMed]
- Zingali, R.B.; Jandrot-Perrus, M.; Guillin, M.C.; Bon, C. Bothrojaracin, a New Thrombin Inhibitor Isolated from Bothrops Jararaca Venom: Characterization and Mechanism of Thrombin Inhibition. Biochemistry 1993, 32, 10794–10802. [Google Scholar] [CrossRef] [PubMed]
- AR, R. Identification of Basic Residues in the Heparin-Binding Exosite of Factor Xa Critical for Heparin and Factor Va Binding. J. Biol. Chem. 2000, 275, 3320–3327. [Google Scholar] [CrossRef]
- Yang, L.; Manithody, C.; Rezaie, A.R. Localization of the Heparin Binding Exosite of Factor IXa. J. Biol. Chem. 2002, 277, 50756–50760. [Google Scholar] [CrossRef] [PubMed]
- Banner, D.W.; D’Arcy, A.; Chène, C.; Winkler, F.K.; Guha, A.; Konigsberg, W.H.; Nemerson, Y.; Kirchhofer, D. The Crystal Structure of the Complex of Blood Coagulation Factor VIIa with Soluble Tissue Factor. Nature 1996, 380, 41–46. [Google Scholar] [CrossRef]
- Francischetti, I.M.B.; Valenzuela, J.G.; Andersen, J.F.; Mather, T.N.; Ribeiro, J.M.C. Ixolaris, a Novel Recombinant Tissue Factor Pathway Inhibitor (TFPI) from the Salivary Gland of the Tick, Ixodes Scapularis: Identification of Factor X and Factor Xa as Scaffolds for the Inhibition of Factor VIIa/Tissue Factor Complex. Blood 2002, 99, 3602–3612. [Google Scholar] [CrossRef]
- Du, X.; Chen, N.L.H.; Wong, A.; Craik, C.S.; Brömme, D. Elastin Degradation by Cathepsin V Requires Two Exosites. J. Biol. Chem. 2013, 288, 34871–34881. [Google Scholar] [CrossRef]
- Sharma, V.; Panwar, P.; O’Donoghue, A.J.; Cui, H.; Guido, R.V.C.; Craik, C.S.; Brömme, D. Structural Requirements for the Collagenase and Elastase Activity of Cathepsin K and Its Selective Inhibition by an Exosite Inhibitor. Biochem. J. 2015, 465, 163–173. [Google Scholar] [CrossRef]
- Panwar, P.; Law, S.; Jamroz, A.; Azizi, P.; Zhang, D.; Ciufolini, M.; Brömme, D. Tanshinones That Selectively Block the Collagenase Activity of Cathepsin K Provide a Novel Class of Ectosteric Antiresorptive Agents for Bone. Br. J. Pharmacol. 2018, 175, 902. [Google Scholar] [CrossRef] [PubMed]
- Panwar, P.; Xue, L.; Søe, K.; Srivastava, K.; Law, S.; Delaisse, J.M.; Brömme, D. An Ectosteric Inhibitor of Cathepsin K Inhibits Bone Resorption in Ovariectomized Mice. J. Bone Miner. Res. 2017, 32, 2415–2430. [Google Scholar] [CrossRef] [PubMed]
- Robichaud, T.K.; Steffensen, B.; Fieldss, G.B. Exosite Interactions Impact Matrix Metalloproteinase Collagen Specificities. J. Biol. Chem. 2011, 286, 37535–37542. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Anderson, P.J.; Majerus, E.M.; Tuley, E.A.; Sadler, J.E. Exosite Interactions Contribute to Tension-Induced Cleavage of von Willebrand Factor by the Antithrombotic ADAMTS13 Metalloprotease. Proc. Natl. Acad. Sci. USA 2006, 103, 19099–19104. [Google Scholar] [CrossRef]
- Santamaria, S.; Yamamoto, K.; Teraz-Orosz, A.; Koch, C.; Apte, S.S.; de Groot, R.; Lane, D.A.; Ahnström, J. Exosites in Hypervariable Loops of ADAMTS Spacer Domains Control Substrate Recognition and Proteolysis. Sci. Rep. 2019, 9, 10914. [Google Scholar] [CrossRef]
- Knapinska, A.M.; Dreymuller, D.; Ludwig, A.; Smith, L.; Golubkov, V.; Sohail, A.; Fridman, R.; Giulianotti, M.; LaVoi, T.M.; Houghten, R.A.; et al. SAR Studies of Exosite-Binding Substrate-Selective Inhibitors of A Disintegrin And Metalloprotease 17 (ADAM17) and Application as Selective in Vitro Probes. J. Med. Chem. 2015, 58, 5808. [Google Scholar] [CrossRef]
- Strisovsky, K.; Sharpe, H.J.; Freeman, M. Sequence-Specific Intramembrane Proteolysis: Identification of a Recognition Motif in Rhomboid Substrates. Mol. Cell 2009, 36, 1048. [Google Scholar] [CrossRef] [PubMed]
- Woskowicz, A.M.; Weaver, S.A.; Shitomi, Y.; Ito, N.; Itoh, Y. MT-LOOP-Dependent Localization of Membrane Type I Matrix Metalloproteinase (MT1-MMP) to the Cell Adhesion Complexes Promotes Cancer Cell Invasion. J. Biol. Chem. 2013, 288, 35149–35158. [Google Scholar] [CrossRef]
- Arnold, L.H.; Butt, L.E.; Prior, S.H.; Read, C.M.; Fields, G.B.; Pickford, A.R. The Interface between Catalytic and Hemopexin Domains in Matrix Metalloproteinase-1 Conceals a Collagen Binding Exosite. J. Biol. Chem. 2011, 286, 45073–45082. [Google Scholar] [CrossRef]
- Mehana, E.S.E.; Khafaga, A.F.; El-Blehi, S.S. The Role of Matrix Metalloproteinases in Osteoarthritis Pathogenesis: An Updated Review. Life Sci. 2019, 234, 116786. [Google Scholar] [CrossRef]
- Gresele, P.; Falcinelli, E.; Sebastiano, M.; Momi, S. Matrix Metalloproteinases and Platelet Function. Prog. Mol. Biol. Transl. Sci. 2017, 147, 133–165. [Google Scholar] [CrossRef] [PubMed]
- Mead, T.J.; Apte, S.S. ADAMTS Proteins in Human Disorders. Matrix Biol. 2018, 71–72, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Mao, C.; Jia, Y.; Fu, Y.; Kong, W. Tissue Remodeling: From Regeneration to Fibrosis: Extracellular Matrix Dynamics in Vascular Remodeling. Am. J. Physiol. Cell Physiol. 2020, 319, C481. [Google Scholar] [CrossRef] [PubMed]
- Cui, N.; Hu, M.; Khalil, R.A. Biochemical and Biological Attributes of Matrix Metalloproteinases. Prog. Mol. Biol. Transl. Sci. 2017, 147, 1–73. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, S.; Yamamoto, K.; Botkjaer, K.; Tape, C.; Dyson, M.R.; McCafferty, J.; Murphy, G.; Nagase, H. Antibody-Based Exosite Inhibitors of ADAMTS-5 (Aggrecanase-2). Biochem. J. 2015, 471, 391. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, S.; Cuffaro, D.; Nuti, E.; Ciccone, L.; Tuccinardi, T.; Liva, F.; D’Andrea, F.; de Groot, R.; Rossello, A.; Ahnström, J. Exosite Inhibition of ADAMTS-5 by a Glycoconjugated Arylsulfonamide. Sci. Rep. 2021, 11, 949. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K. Aptamer for ADAMTS5 and Use for Aptamer for ADAMTS5. U.S. Patent No. 11,473,089, 18 October 2022. [Google Scholar]
- Neuhold, L.A.; Killar, L.; Zhao, W.; Sung, M.L.A.; Warner, L.; Kulik, J.; Turner, J.; Wu, W.; Billinghurst, C.; Meijers, T.; et al. Postnatal Expression in Hyaline Cartilage of Constitutively Active Human Collagenase-3 (MMP-13) Induces Osteoarthritis in Mice. J. Clin. Investig. 2001, 107, 35–44. [Google Scholar] [CrossRef]
- Spicer, T.P.; Jiang, J.; Taylor, A.B.; Choi, J.Y.; Hart, P.J.; Roush, W.R.; Fields, G.B.; Hodder, P.S.; Minond, D. Characterization of Selective Exosite-Binding Inhibitors of Matrix Metalloproteinase 13 That Prevent Articular Cartilage Degradation in Vitro. J. Med. Chem. 2014, 57, 9598–9611. [Google Scholar] [CrossRef]
- Engel, C.K.; Pirard, B.; Schimanski, S.; Kirsch, R.; Habermann, J.; Klingler, O.; Schlotte, V.; Weithmann, K.U.; Wendt, K.U. Structural Basis for the Highly Selective Inhibition of MMP-13. Chem. Biol. 2005, 12, 181–189. [Google Scholar] [CrossRef]
- Johnson, A.R.; Pavlovsky, A.G.; Ortwine, D.F.; Prior, F.; Man, C.F.; Bornemeier, D.A.; Banotai, C.A.; Mueller, W.T.; McConnell, P.; Yan, C.; et al. Discovery and Characterization of a Novel Inhibitor of Matrix Metalloprotease-13 That Reduces Cartilage Damage in Vivo without Joint Fibroplasia Side Effects. J. Biol. Chem. 2007, 282, 27781–27791. [Google Scholar] [CrossRef]
- Piecha, D.; Weik, J.; Kheil, H.; Becher, G.; Timmermann, A.; Jaworski, A.; Burger, M.; Hofmann, M.W. Novel Selective MMP-13 Inhibitors Reduce Collagen Degradation in Bovine Articular and Human Osteoarthritis Cartilage Explants. Inflamm. Res. 2010, 59, 379–389. [Google Scholar] [CrossRef]
- Baragi, V.M.; Becher, G.; Bendele, A.M.; Biesinger, R.; Bluhm, H.; Boer, J.; Deng, H.; Dodd, R.; Essers, M.; Feuerstein, T.; et al. A New Class of Potent Matrix Metalloproteinase 13 Inhibitors for Potential Treatment of Osteoarthritis: Evidence of Histologic and Clinical Efficacy without Musculoskeletal Toxicity in Rat Models. Arthritis Rheum. 2009, 60, 2008–2018. [Google Scholar] [CrossRef] [PubMed]
- Udi, Y.; Grossman, M.; Solomonov, I.; Dym, O.; Rozenberg, H.; Moreno, V.; Cuniasse, P.; Dive, V.; Arroyo, A.G.; Sagi, I. Inhibition Mechanism of Membrane Metalloprotease by an Exosite-Swiveling Conformational Antibody. Structure 2015, 23, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Ingvarsen, S.; Madsen, D.H.; Hillig, T.; Lund, L.R.; Holmbeck, K.; Behrendt, N.; Engelholm, L.H. Dimerization of Endogenous MT1-MMP Is a Regulatory Step in the Activation of the 72-KDa Gelatinase MMP-2 on Fibroblasts and Fibrosarcoma Cells. Biol. Chem. 2008, 389, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Basu, B.; Correa De Sampaio, P.; Mohammed, H.; Fogarasi, M.; Corrie, P.; Watkins, N.A.; Smethurst, P.A.; English, W.R.; Ouwehand, W.H.; Murphy, G. Inhibition of MT1-MMP Activity Using Functional Antibody Fragments Selected against Its Hemopexin Domain. Int. J. Biochem. Cell Biol. 2012, 44, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, Z.; Wang, Y.; Bonewald, L.; Steffensen, B. Inhibition of MMP-2 Gelatinolysis by Targeting Exodomain–Substrate Interactions. Biochem. J. 2007, 406, 147. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, S.; de Groot, R. Monoclonal Antibodies against Metzincin Targets. Br. J. Pharmacol. 2019, 176, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Cuffaro, D.; Ciccone, L.; Rossello, A.; Nuti, E.; Santamaria, S. Targeting Aggrecanases for Osteoarthritis Therapy: From Zinc Chelation to Exosite Inhibition. J. Med. Chem. 2022, 65, 13505–13532. [Google Scholar] [CrossRef]
- Li, K.; Tay, F.R.; Yiu, C.K.Y. The Past, Present and Future Perspectives of Matrix Metalloproteinase Inhibitors. Pharmacol. Ther. 2020, 207, 107465. [Google Scholar] [CrossRef]
- Blasi, J.; Chapman, E.R.; Link, E.; Binz, T.; Yamasaki, S.T.C.; Niemann, H.; Jahn, R. Botulinum Neurotoxin A Selectively Cleaves the Synaptic Protein SNAP-25. Nature 1993, 365, 160–163. [Google Scholar] [CrossRef]
- Xue, S.; Javor, S.; Hixon, M.S.; Janda, K.D. Probing BoNT/A Protease Exosites: Implications for Inhibitor Design and Light Chain Longevity. Biochemistry 2014, 53, 6820–6824. [Google Scholar] [CrossRef]
- Hu, X.; Legler, P.M.; Southall, N.; Maloney, D.J.; Simeonov, A.; Jadhav, A. Structural Insight into Exosite Binding and Discovery of Novel Exosite Inhibitors of Botulinum Neurotoxin Serotype A through in Silico Screening. J. Comput. Aided Mol. Des. 2014, 28, 765–778. [Google Scholar] [CrossRef][Green Version]
- Xue, S.; Seki, H.; Remes, M.; Šilhár, P.; Janda, K. Examination of α-Exosite Inhibitors against Botulinum Neurotoxin A Protease through Structure-Activity Relationship Studies of Chicoric Acid. Bioorg Med. Chem. Lett. 2017, 27, 4956. [Google Scholar] [CrossRef] [PubMed]
- Bremer, P.T.; Xue, S.; Janda, K.D. Picolinic Acids as β-Exosite Inhibitors of Botulinum Neurotoxin A Light Chain. Chem. Commun. 2016, 52, 12521–12524. [Google Scholar] [CrossRef]
- Dong, J.; Thompson, A.A.; Fan, Y.; Lou, J.; Conrad, F.; Ho, M.; Pires-Alves, M.; Wilson, B.A.; Stevens, R.C.; Marks, J.D. A Single-Domain Llama Antibody Potently Inhibits the Enzymatic Activity of Botulinum Neurotoxin by Binding to the Non-Catalytic Alpha-Exosite Binding Region. J. Mol. Biol. 2010, 397, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Geren, I.N.; Dong, J.; Lou, J.; Wen, W.; Conrad, F.; Smith, T.J.; Smith, L.A.; Ho, M.; Pires-Alves, M.; et al. Monoclonal Antibodies Targeting the Alpha-Exosite of Botulinum Neurotoxin Serotype/A Inhibit Catalytic Activity. PLoS ONE 2015, 10, e0135306. [Google Scholar] [CrossRef] [PubMed]
- Bannwarth, L.; Goldberg, A.B.; Chen, C.; Turk, B.E. Identification of Exosite-Targeting Inhibitors of Anthrax Lethal Factor by High Throughput Screening. Chem. Biol. 2012, 19, 875. [Google Scholar] [CrossRef] [PubMed]
- Monod, J.; Wyman, J.; Changeux, J.P. On the Nature of Allosteric Transitions: A Plausible Model. J. Mol. Biol. 1965, 12, 88–118. [Google Scholar] [CrossRef] [PubMed]
- Koshland, D.E.; Nemethy, J.G.; Filmer, D. Comparison of Experimental Binding Data and Theoretical Models in Proteins Containing Subunits. Biochemistry 1966, 5, 365–385. [Google Scholar] [CrossRef]
- Nussinov, R.; Tsai, C.J. Allostery in Disease and in Drug Discovery. Cell 2013, 153, 293–305. [Google Scholar] [CrossRef]
- Esmon, N.L.; Owen, W.G.; Esmon, C.T. Isolation of a Membrane-Bound Cofactor for Thrombin-Catalyzed Activation of Protein C. J. Biol. Chem. 1982, 257, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.M.; Di Cera, E. Thrombin Is a Sodium Ion Activated Enzyme. Biochemistry 1992, 31, 11721–11730. [Google Scholar] [CrossRef] [PubMed]
- Dang, O.D.; Vindigni, A.; Di Cera, E. An Allosteric Switch Controls the Procoagulant and Anticoagulant Activities of Thrombin. Proc. Natl. Acad. Sci. USA 1995, 92, 5977–5981. [Google Scholar] [CrossRef] [PubMed]
- Orthner, C.L.; Kosow, D.P. Evidence That Human α-Thrombin Is a Monovalent Cation-Activated Enzyme. Arch. Biochem. Biophys. 1980, 202, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Ayala, Y.; Cera, E. Di Molecular Recognition by Thrombin. Role of the Slows→Fast Transition, Site-Specific Ion Binding Energetics and Thermodynamic Mapping of Structural Components. J. Mol. Biol. 1994, 235, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Huntington, J.A. How Na+ Activates Thrombin—A Review of the Functional and Structural Data. Biol. Chem. 2008, 389, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Bode, W.; Huber, R. Induction of the Bovine Trypsinogen-Trypsin Transition by Peptides Sequentially Similar to the N-Terminus of Trypsin. FEBS Lett. 1976, 68, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Castelliwo, F.J. Activation of Human Plasminogen by Equimolar Levels of Streptokinase. J. Biol. Chem. 1977, 252, 492–498. [Google Scholar] [CrossRef]
- Hemker, H.C.; Bas, B.M.; Muller, A.D. Activation of a Pro-Enzyme by a Stoichiometric Reaction with Another Protein: The Reaction between Prothrombin and Staphylocoagulase. Biochim. Biophys. Acta (BBA)-Protein Struct. 1975, 379, 180–188. [Google Scholar] [CrossRef]
- Friedrich, R.; Panizzi, P.; Fuentes-Prior, P.; Richter, K.; Verhamme, I.; Anderson, P.J.; Kawabata, S.I.; Huber, R.; Bode, W.; Bock, P.E. Staphylocoagulase Is a Prototype for the Mechanism of Cofactor-Induced Zymogen Activation. Nature 2003, 425, 535–539. [Google Scholar] [CrossRef]
- Novinec, M.; Korenč, M.; Caflisch, A.; Ranganathan, R.; Lenarčič, B.; Baici, A. A Novel Allosteric Mechanism in the Cysteine Peptidase Cathepsin K Discovered by Computational Methods. Nat. Commun. 2014, 5, 3287. [Google Scholar] [CrossRef]
- Novinec, M.; Lenarčič, B.; Baici, A. Probing the Activity Modification Space of the Cysteine Peptidase Cathepsin K with Novel Allosteric Modifiers. PLoS ONE 2014, 9, e106642. [Google Scholar] [CrossRef]
- Novinec, M. Computational Investigation of Conformational Variability and Allostery in Cathepsin K and Other Related Peptidases. PLoS ONE 2017, 12, e0182387. [Google Scholar] [CrossRef]
- Shiozaki, E.N.; Chai, J.; Rigotti, D.J.; Riedl, S.J.; Li, P.; Srinivasula, S.M.; Alnemri, E.S.; Fairman, R.; Shi, Y. Mechanism of XIAP-Mediated Inhibition of Caspase-9. Mol. Cell 2003, 11, 519–527. [Google Scholar] [CrossRef]
- Datta, D.; Scheer, J.M.; Romanowski, M.J.; Wells, J.A. An Allosteric Circuit in Caspase-1. J. Mol. Biol. 2008, 381, 1157–1167. [Google Scholar] [CrossRef][Green Version]
- Schweizer, A.; Roschitzki-Voser, H.; Amstutz, P.; Briand, C.; Gulotti-Georgieva, M.; Prenosil, E.; Binz, H.K.; Capitani, G.; Baici, A.; Plückthun, A.; et al. Inhibition of Caspase-2 by a Designed Ankyrin Repeat Protein: Specificity, Structure, and Inhibition Mechanism. Structure 2007, 15, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.A.; Lam, J.; Nguyen, J.T.; O’Brien, T.; Wells, J.A. Discovery of an Allosteric Site in the Caspases. Proc. Natl. Acad. Sci. USA 2004, 101, 12461. [Google Scholar] [CrossRef] [PubMed]
- Feldman, T.; Kabaleeswaran, V.; Jang, S.B.; Antczak, C.; Djaballah, H.; Wu, H.; Jiang, X. A Class of Allosteric, Caspase Inhibitors Identified by High-Throughput Screening. Mol. Cell 2012, 47, 585. [Google Scholar] [CrossRef] [PubMed]
- Flütsch, A.; Schroeder, T.; Barandun, J.; Ackermann, R.; Bühlmann, M.; Grütter, M.G.; Flütsch, A.; Schroeder, T.; Barandun, J.; Ackermann, R.; et al. Specific Targeting of Human Caspases Using Designed Ankyrin Repeat Proteins. Biol. Chem. 2014, 395, 1243–1252. [Google Scholar] [CrossRef]
- Van Es, J.H.; Van Gijn, M.E.; Riccio, O.; Van Den Born, M.; Vooijs, M.; Begthel, H.; Cozijnsen, M.; Robine, S.; Winton, D.J.; Radtke, F.; et al. Notch/Gamma-Secretase Inhibition Turns Proliferative Cells in Intestinal Crypts and Adenomas into Goblet Cells. Nature 2005, 435, 959–963. [Google Scholar] [CrossRef] [PubMed]
- Weggen, S.; Eriksen, J.L.; Das, P.; Sagi, S.A.; Wang, R.; Pietrzik, C.U.; Findlay, K.A.; Smith, T.E.; Murphy, M.P.; Bulter, T.; et al. A Subset of NSAIDs Lower Amyloidogenic Abeta42 Independently of Cyclooxygenase Activity. Nature 2001, 414, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Mayer, S.C.; Kreft, A.F.; Harrison, B.; Abou-Gharbia, M.; Antane, M.; Aschmies, S.; Atchison, K.; Chlenov, M.; Cole, D.C.; Comery, T.; et al. Discovery of Begacestat, a Notch-1-Sparing Gamma-Secretase Inhibitor for the Treatment of Alzheimer’s Disease. J. Med. Chem. 2008, 51, 7348–7351. [Google Scholar] [CrossRef] [PubMed]
- Netzer, W.J.; Dou, F.; Cai, D.; Veach, D.; Jean, S.; Li, Y.; Bornmann, W.G.; Clarkson, B.; Xu, H.; Greengard, P. Gleevec Inhibits Beta-Amyloid Production but Not Notch Cleavage. Proc. Natl. Acad. Sci. USA 2003, 100, 12444–12449. [Google Scholar] [CrossRef] [PubMed]
- Shelton, C.C.; Zhu, L.; Chau, D.; Yang, L.; Wang, R.; Djaballah, H.; Zheng, H.; Li, Y.M. Modulation of γ-Secretase Specificity Using Small Molecule Allosteric Inhibitors. Proc. Natl. Acad. Sci. USA 2009, 106, 20228. [Google Scholar] [CrossRef] [PubMed]
- Campagna, J.; Vadivel, K.; Jagodzinska, B.; Jun, M.; Bilousova, T.; Spilman, P.; John, V. Evaluation of an Allosteric BACE Inhibitor Peptide to Identify Mimetics That Can Interact with the Loop F Region of the Enzyme and Prevent APP Cleavage. J. Mol. Biol. 2018, 430, 1566–1576. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.J.; Angelina, E.; Gyebrovszki, A.; Fülöp, L.; Peruchena, N.; Baldoni, H.A.; Penke, B.; Enriz, R.D. New Small-Size Peptides Modulators of the Exosite of BACE1 Obtained from a Structure-Based Design. J. Biomol. Struct. Dyn. 2017, 35, 413–426. [Google Scholar] [CrossRef]
- Ugbaja, S.C.; Lawal, I.A.; Abubakar, B.H.; Mushebenge, A.G.; Lawal, M.M.; Kumalo, H.M. Allostery Inhibition of BACE1 by Psychotic and Meroterpenoid Drugs in Alzheimer’s Disease Therapy. Molecules 2022, 27, 4372. [Google Scholar] [CrossRef] [PubMed]
- Atwal, J.K.; Chen, Y.; Chiu, C.; Mortensen, D.L.; Meilandt, W.J.; Liu, Y.; Heise, C.E.; Hoyte, K.; Luk, W.; Lu, Y.; et al. A Therapeutic Antibody Targeting BACE1 Inhibits Amyloid-β Production in Vivo. Sci. Transl. Med. 2011, 3, 84ra43. [Google Scholar] [CrossRef]
- Sela-Passwell, N.; Rosenblum, G.; Shoham, T.; Sagi, I. Structural and Functional Bases for Allosteric Control of MMP Activities: Can It Pave the Path for Selective Inhibition? Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2010, 1803, 29–38. [Google Scholar] [CrossRef]
- Kumar, L.; Nash, A.; Harms, C.; Planas-Iglesias, J.; Wright, D.; Klein-Seetharaman, J.; Sarkar, S.K. Allosteric Communications between Domains Modulate the Activity of Matrix Metalloprotease-1. Biophys. J. 2020, 119, 360–374. [Google Scholar] [CrossRef]
- Kamboj, S.; Harms, C.; Wright, D.; Nash, A.; Kumar, L.; Klein-Seetharaman, J.; Sarkar, S.K. Identification of Allosteric Fingerprints of Alpha-Synuclein Aggregates in Matrix Metalloprotease-1 and Substrate-Specific Virtual Screening with Single Molecule Insights. Sci. Rep. 2022, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lubbe, L.; Sewell, B.T.; Woodward, J.D.; Sturrock, E.D. Cryo-EM Reveals Mechanisms of Angiotensin I-converting Enzyme Allostery and Dimerization. EMBO J. 2022, 41, 110550. [Google Scholar] [CrossRef] [PubMed]
- Mabanglo, M.F.; Houry, W.A. Recent Structural Insights into the Mechanism of ClpP Protease Regulation by AAA+ Chaperones and Small Molecules. J. Biol. Chem. 2022, 298, 101781. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Neher, S.B.; Kim, Y.I.; Sauer, R.T.; Baker, T.A. Proteomic Discovery of Cellular Substrates of the ClpXP Protease Reveals Five Classes of ClpX-Recognition Signals. Mol. Cell 2003, 11, 671–683. [Google Scholar] [CrossRef] [PubMed]
- El-Baba, T.J.; Lutomski, C.A.; Kantsadi, A.L.; Malla, T.R.; John, T.; Mikhailov, V.; Bolla, J.R.; Schofield, C.J.; Zitzmann, N.; Vakonakis, I.; et al. Allosteric Inhibition of the SARS-CoV-2 Main Protease: Insights from Mass Spectrometry Based Assays. Angew. Chem. Int. Ed. 2020, 59, 23544–23548. [Google Scholar] [CrossRef]
- Günther, S.; Reinke, P.Y.A.; Fernández-Garciá, Y.; Lieske, J.; Lane, T.J.; Ginn, H.M.; Koua, F.H.M.; Ehrt, C.; Ewert, W.; Oberthuer, D.; et al. X-Ray Screening Identifies Active Site and Allosteric Inhibitors of SARS-CoV-2 Main Protease. Science 2021, 372, 642–646. [Google Scholar] [CrossRef]
- Du, R.; Cooper, L.; Chen, Z.; Lee, H.; Rong, L.; Cui, Q. Discovery of Chebulagic Acid and Punicalagin as Novel Allosteric Inhibitors of SARS-CoV-2 3CLpro. Antivir. Res. 2021, 190, 105075. [Google Scholar] [CrossRef]
- Mirković, B.; Renko, M.; Turk, S.; Sosič, I.; Jevnikar, Z.; Obermajer, N.; Turk, D.; Gobec, S.; Kos, J. Novel Mechanism of CathepsinB Inhibition by Antibiotic Nitroxoline and Related Compounds. ChemMedChem 2011, 6, 1351–1356. [Google Scholar] [CrossRef]
- Sosič, I.; Mitrović, A.; Ćurić, H.; Knez, D.; Brodnik Žugelj, H.; Štefane, B.; Kos, J.; Gobec, S. Cathepsin B Inhibitors: Further Exploration of the Nitroxoline Core. Bioorg. Med. Chem. Lett. 2018, 28, 1239–1247. [Google Scholar] [CrossRef]
- Ulčakar, L.; Novinec, M. Inhibition of Human Cathepsins B and L by Caffeic Acid and Its Derivatives. Biomolecules 2021, 11, 31. [Google Scholar] [CrossRef]
- Goričan, T.; Ciber, L.; Petek, N.; Svete, J.; Novinec, M. Synthesis and Kinetic Characterization of Hyperbolic Inhibitors of Human Cathepsins K and S Based on a Succinimide Scaffold. Bioorg. Chem. 2021, 115, 105213. [Google Scholar] [CrossRef] [PubMed]
- Verespy, S.; Mehta, A.Y.; Afosah, D.; Al-Horani, R.A.; Desai, U.R. Allosteric Partial Inhibition of Monomeric Proteases. Sulfated Coumarins Induce Regulation, Not Just Inhibition, of Thrombin. Sci. Rep. 2016, 6, 24043. [Google Scholar] [CrossRef]
- Novinec, M.; Bembič, P.; Janković, M.; Kisilak, M.; Kljun, J.; Turel, I. Kinetic Characterization of Cerium and Gallium Ions as Inhibitors of Cysteine Cathepsins L, K, and S. Int. J. Mol. Sci. 2022, 23, 8993. [Google Scholar] [CrossRef] [PubMed]
- Kramer, L.; Renko, M.; Završnik, J.; Turk, D.; Seeger, M.A.; Vasiljeva, O.; Grütter, M.G.; Turk, V.; Turk, B. Non-Invasive in Vivo Imaging of Tumour-Associated Cathepsin B by a Highly Selective Inhibitory DARPin. Theranostics 2017, 7, 2806. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Murphy, G.; Troeberg, L. Extracellular Regulation of Metalloproteinases. Matrix Biol. 2015, 44–46, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Dumin, J.A.; Dickeson, S.K.; Stricker, T.P.; Bhattacharyya-Pakrasi, M.; Roby, J.D.; Santoro, S.A.; Parks, W.C. Pro-Collagenase-1 (Matrix Metalloproteinase-1) Binds the A2β1 Integrin upon Release from Keratinocytes Migrating on Type I Collagen. J. Biol. Chem. 2001, 276, 29368–29374. [Google Scholar] [CrossRef]
- Piccard, H.; Van den Steen, P.E.; Opdenakker, G. Hemopexin Domains as Multifunctional Liganding Modules in Matrix Metalloproteinases and Other Proteins. J. Leukoc. Biol. 2007, 81, 870–892. [Google Scholar] [CrossRef]
- Lafleur, M.A.; Xu, D.; Hemler, M.E. Tetraspanin Proteins Regulate Membrane Type-1 Matrix Metalloproteinase-Dependent Pericellular Proteolysis. Mol. Biol. Cell 2009, 20, 2030–2040. [Google Scholar] [CrossRef]
- Takino, T.; Miyamori, H.; Kawaguchi, N.; Uekita, T.; Seiki, M.; Sato, H. Tetraspanin CD63 Promotes Targeting and Lysosomal Proteolysis of Membrane-Type 1 Matrix Metalloproteinase. Biochem. Biophys. Res. Commun. 2003, 304, 160–166. [Google Scholar] [CrossRef]
- Bharadwaj, A.G.; Holloway, R.W.; Miller, V.A.; Waisman, D.M. Plasmin and Plasminogen System in the Tumor Microenvironment: Implications for Cancer Diagnosis, Prognosis, and Therapy. Cancers 2021, 13, 1838. [Google Scholar] [CrossRef]
- Zhai, B.T.; Tian, H.; Sun, J.; Zou, J.B.; Zhang, X.F.; Cheng, J.X.; Shi, Y.J.; Fan, Y.; Guo, D.Y. Urokinase-Type Plasminogen Activator Receptor (UPAR) as a Therapeutic Target in Cancer. J. Transl. Med. 2022, 20, 135. [Google Scholar] [CrossRef]
- Leth, J.M.; Ploug, M. Targeting the Urokinase-Type Plasminogen Activator Receptor (UPAR) in Human Diseases With a View to Non-Invasive Imaging and Therapeutic Intervention. Front. Cell Dev. Biol. 2021, 9, 732015. [Google Scholar] [CrossRef]
- Turk, B.; Dolenc, I.; Žerovnik, E.; Turk, D.; Gubenšek, F.; Turk, V. Human Cathepsin B Is a Metastable Enzyme Stabilized by Specific Ionic Interactions Associated with the Active Site. Biochemistry 1994, 33, 14800–14806. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, N.; Sloane, B.F. Cathepsin B: Multiple Roles in Cancer. Proteom. Clin. Appl. 2014, 8, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Cavallo-Medved, D.; Mai, J.; Dosescu, J.; Sameni, M.; Sloane, B.F. Caveolin-1 Mediates the Expression and Localization of Cathepsin B, pro-Urokinase Plasminogen Activator and Their Cell-Surface Receptors in Human Colorectal Carcinoma Cells. J. Cell Sci. 2005, 118, 1493–1503. [Google Scholar] [CrossRef] [PubMed]
- Cavallo-Medved, D.; Rudy, D.; Blum, G.; Bogyo, M.; Caglic, D.; Sloane, B.F. Live-Cell Imaging Demonstrates Extracellular Matrix Degradation in Association with Active Cathepsin B in Caveolae of Endothelial Cells during Tube Formation. Exp. Cell Res. 2009, 315, 1234–1246. [Google Scholar] [CrossRef] [PubMed]
- Mai, J.; Finley, R.L.; Waisman, D.M.; Sloane, B.F. Human Procathepsin B Interacts with the Annexin II Tetramer on the Surface of Tumor Cells. J. Biol. Chem. 2000, 275, 12806–12812. [Google Scholar] [CrossRef]
- Cui, Z.; Zeng, C.; Huang, F.; Yuan, F.; Yan, J.; Zhao, Y.; Zhou, Y.; Hankey, W.; Jin, V.X.; Huang, J.; et al. Cas13d Knockdown of Lung Protease Ctsl Prevents and Treats SARS-CoV-2 Infection. Nat. Chem. Biol. 2022, 18, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Kameoka, J.; Tanaka, T.; Nojima, Y.; Schlossman, S.F.; Morimoto, C. Direct Association of Adenosine Deaminase with a T Cell Activation Antigen, CD26. Science 1993, 261, 466–469. [Google Scholar] [CrossRef]
- Ishii, T.; Ohnuma, K.; Murakami, A.; Takasawa, N.; Kobayashi, S.; Dang, N.H.; Schlossman, S.F.; Morimoto, C. CD26-Mediated Signaling for T Cell Activation Occurs in Lipid Rafts through Its Association with CD45RO. Proc. Natl. Acad. Sci. USA 2001, 98, 12138–12143. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.W.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.A.; Zaki, A.; Fouchier, R.A.M.; et al. Dipeptidyl Peptidase 4 Is a Functional Receptor for the Emerging Human Coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasllieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greeneugh, T.C.; et al. Angiotensin-Converting Enzyme 2 Is a Functional Receptor for the SARS Coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.L.; Jaiswal, R.K. Neutralizing Antibody: A Savior in the Covid-19 Disease. Mol. Biol. Rep. 2022, 49, 2465–2474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lv, P.; Jiang, J.; Liu, Y.; Yan, R.; Shu, S.; Hu, B.; Xiao, H.; Cai, K.; Yuan, S.; et al. Advances in Developing ACE2 Derivatives against SARS-CoV-2. Lancet Microbe 2023, 4, e369–e378. [Google Scholar] [CrossRef] [PubMed]
- Jarocki, V.M.; Tacchi, J.L.; Djordjevic, S.P. Non-Proteolytic Functions of Microbial Proteases Increase Pathological Complexity. Proteomics 2015, 15, 1075–1088. [Google Scholar] [CrossRef] [PubMed]
- Hytönen, J.; Haataja, S.; Gerlach, D.; Podbielski, A.; Finne, J. The SpeB Virulence Factor of Streptococcus Pyogenes, a Multifunctional Secreted and Cell Surface Molecule with Strepadhesin, Laminin-Binding and Cysteine Protease Activity. Mol. Microbiol. 2001, 39, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, C.; Waggoner, J.D.; Harris, T.O.; Tamura, G.S.; Rubens, C.E. Identification of Novel Adhesins from Group B Streptococci by Use of Phage Display Reveals That C5a Peptidase Mediates Fibronectin Binding. Infect. Immun. 2002, 70, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Rudzińska, M.; Daglioglu, C.; Savvateeva, L.V.; Kaci, F.N.; Antoine, R.; Zamyatnin, A.A. Current Status and Perspectives of Protease Inhibitors and Their Combination with Nanosized Drug Delivery Systems for Targeted Cancer Therapy. Drug Des. Dev. Ther. 2021, 15, 9–20. [Google Scholar] [CrossRef]
- Turk, B. Targeting Proteases: Successes, Failures and Future Prospects. Nat. Rev. Drug Dis. 2006, 5, 785–799. [Google Scholar] [CrossRef]
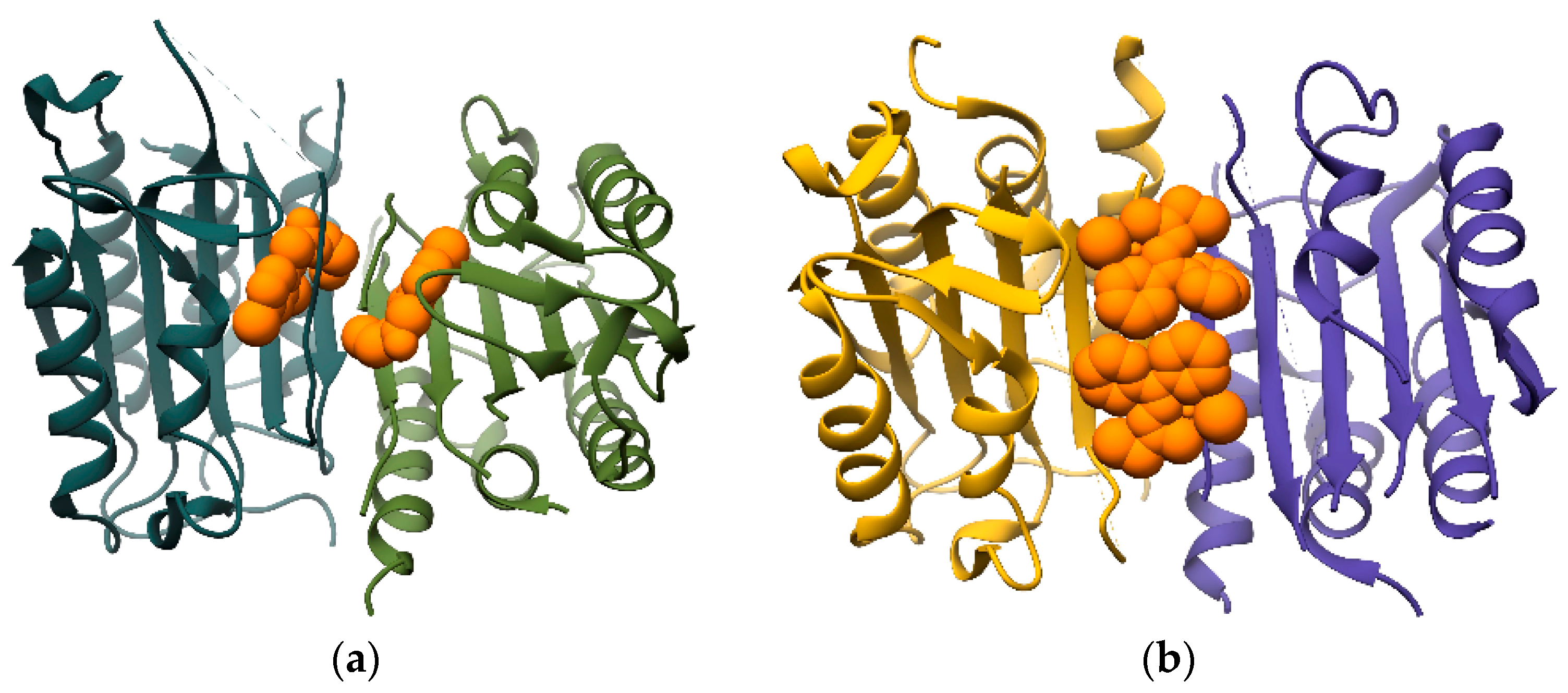
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Obaha, A.; Novinec, M. Regulation of Peptidase Activity beyond the Active Site in Human Health and Disease. Int. J. Mol. Sci. 2023, 24, 17120. https://doi.org/10.3390/ijms242317120
Obaha A, Novinec M. Regulation of Peptidase Activity beyond the Active Site in Human Health and Disease. International Journal of Molecular Sciences. 2023; 24(23):17120. https://doi.org/10.3390/ijms242317120
Chicago/Turabian StyleObaha, Ana, and Marko Novinec. 2023. "Regulation of Peptidase Activity beyond the Active Site in Human Health and Disease" International Journal of Molecular Sciences 24, no. 23: 17120. https://doi.org/10.3390/ijms242317120
APA StyleObaha, A., & Novinec, M. (2023). Regulation of Peptidase Activity beyond the Active Site in Human Health and Disease. International Journal of Molecular Sciences, 24(23), 17120. https://doi.org/10.3390/ijms242317120





