New Insights into the TIFY Gene Family of Brassica napus and Its Involvement in the Regulation of Shoot Branching
Abstract
:1. Introduction
2. Results
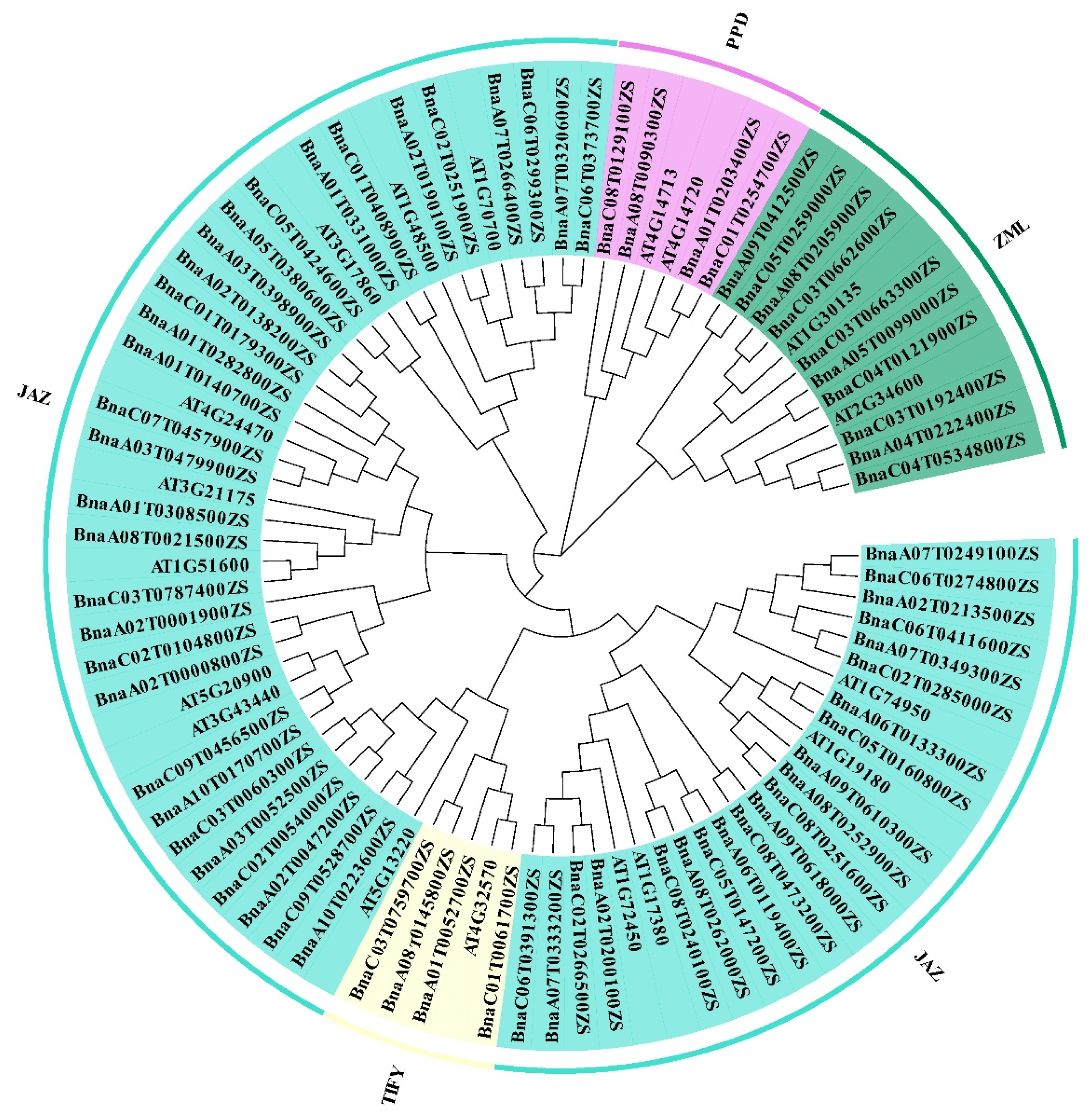
2.1. Identification of the TIFY Members and Phylogenetic Analyses of the TIFY Family Genes in Brassica napus
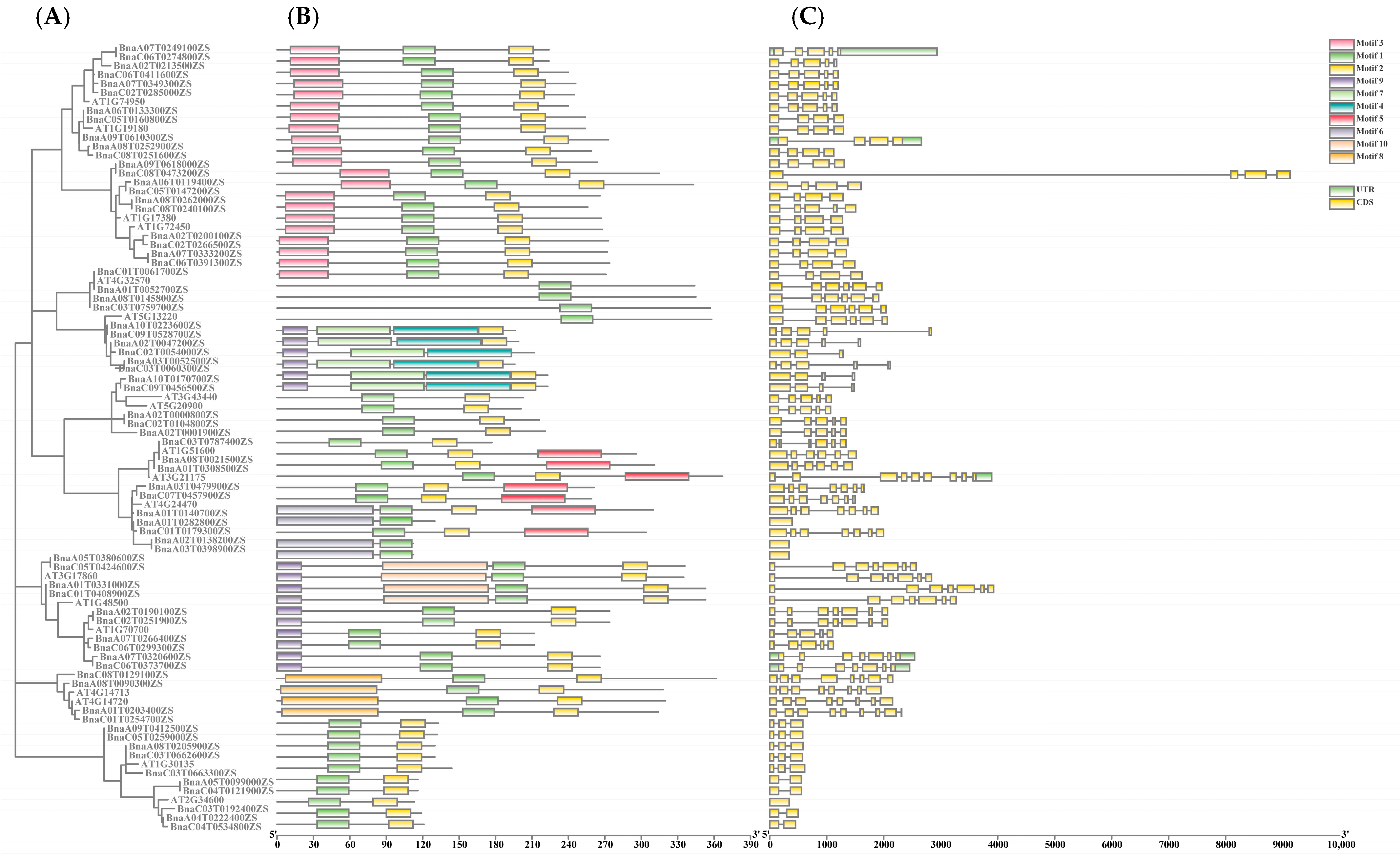
2.2. Gene Structural Analysis and Motif Composition of the Rapeseed TIFY Family Genes
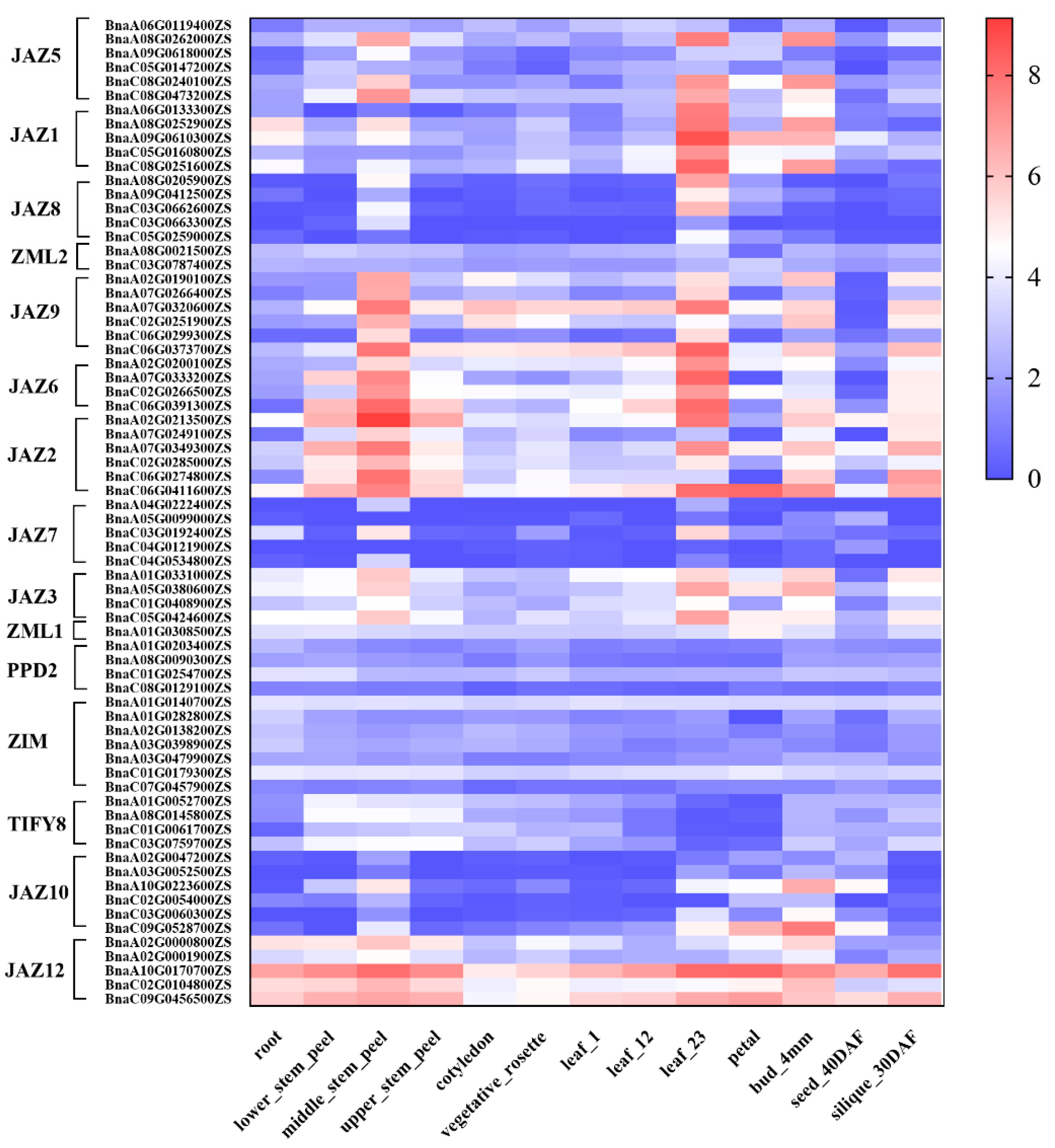
2.3. The Tissue Expression Profile of TIFY Family Genes in Rapeseed
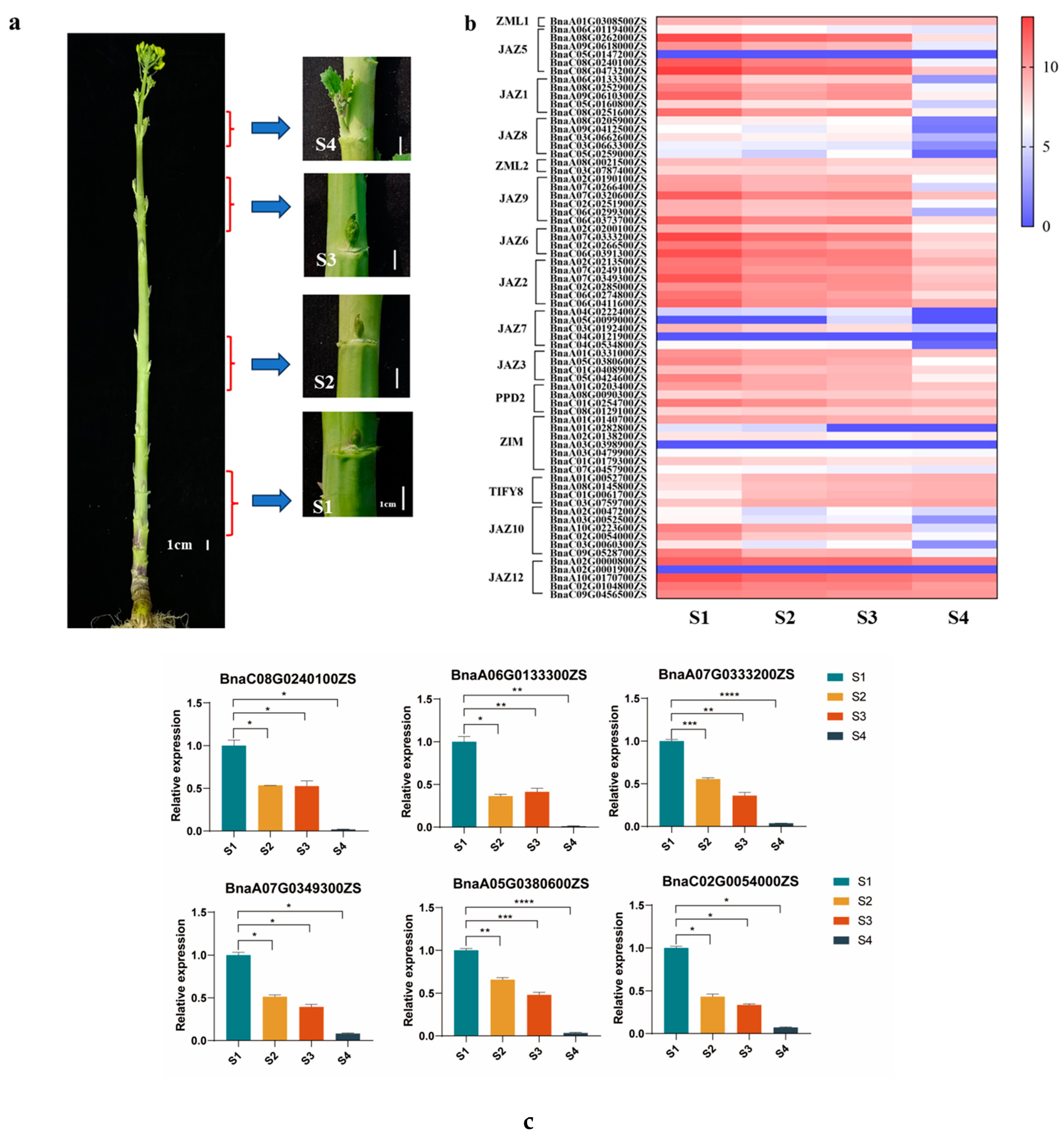
2.4. The Transcripts of TIFY Family Genes Highly Accumulate in Dormant Axillary Buds and Significantly Decrease in Outgrowing Axillary Buds
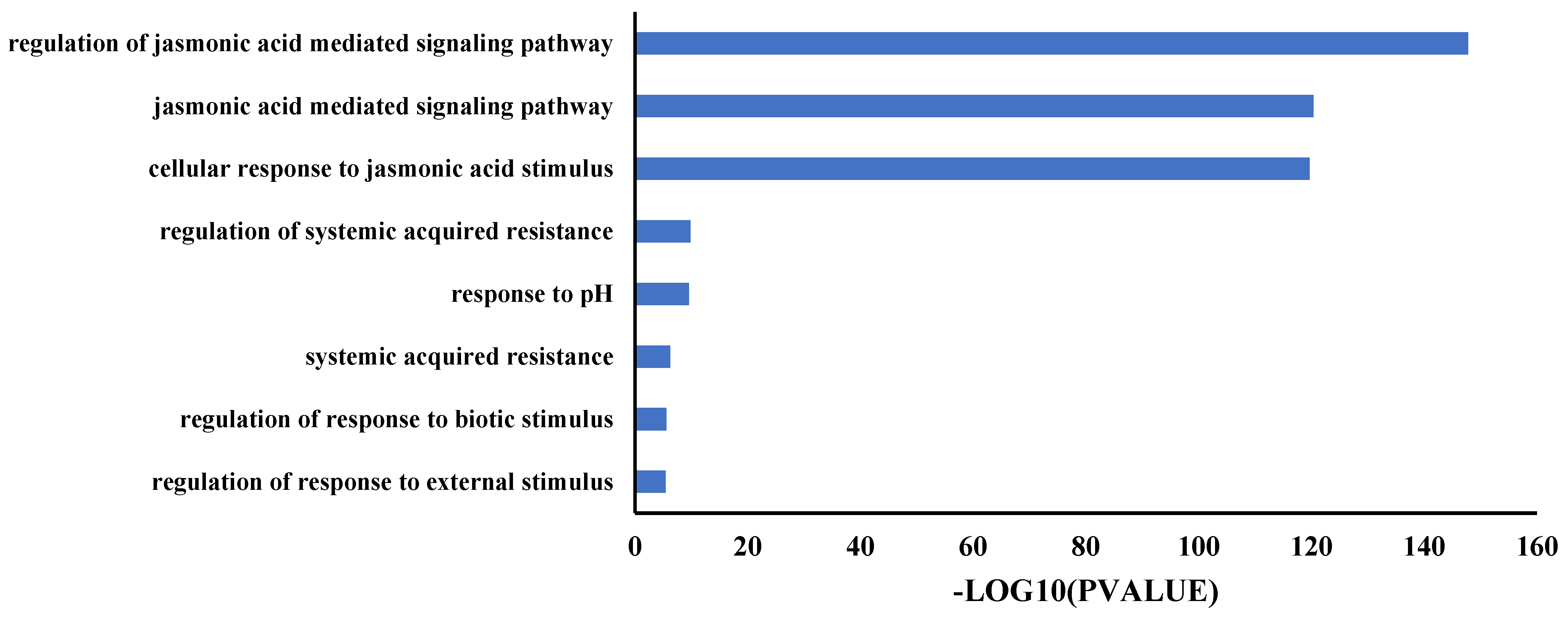
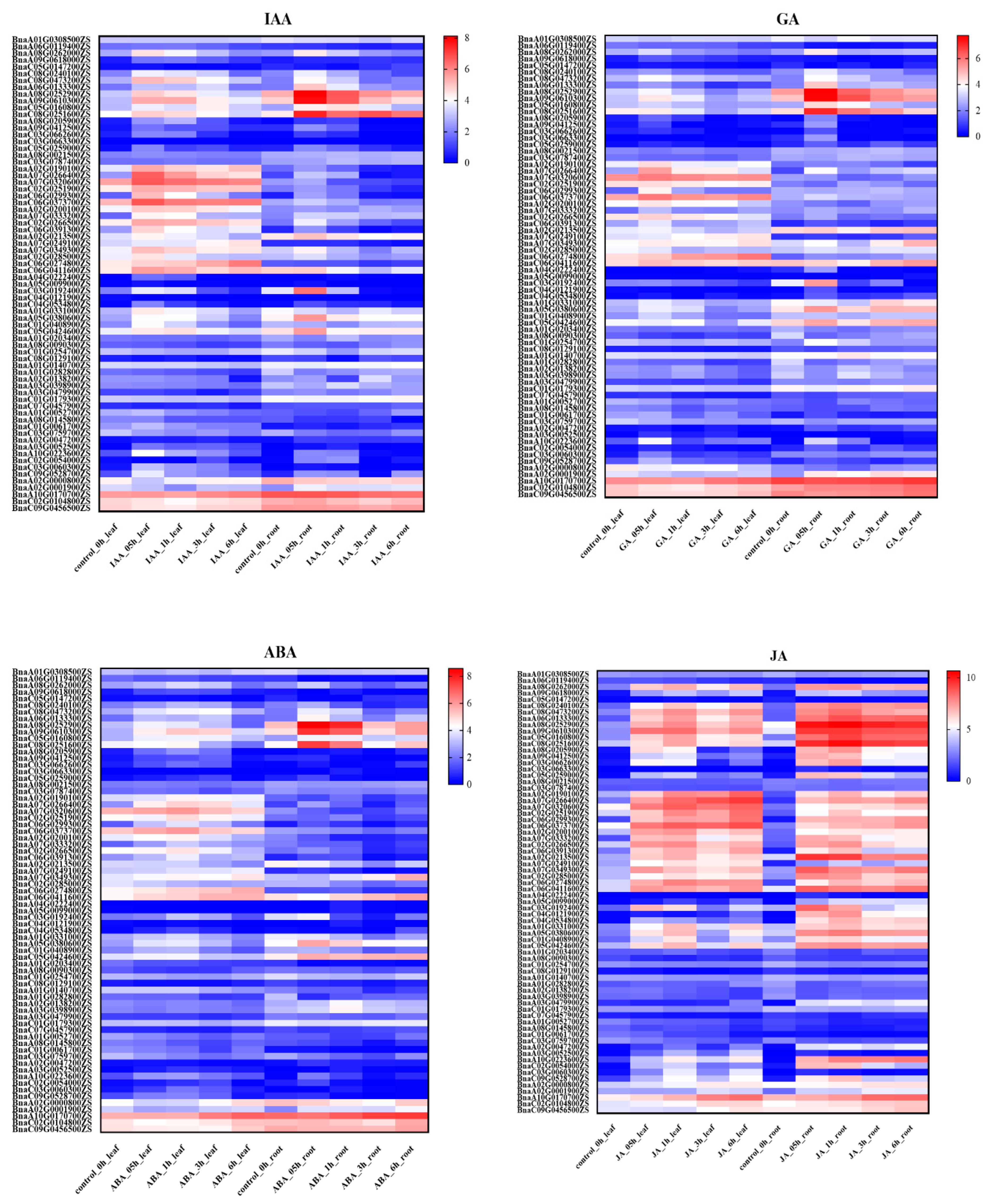
2.5. TIFY Family Genes Were Involved in the Response of Jasmonic Acid Signaling
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Sampling, RNA Extraction, and RNA-Seq
4.2. Identification of TIFY Gene Family in Brassica napus
4.3. Sequence Alignments and Phylogenetic Analyses
4.4. Chromosomal Location
4.5. Exon/Intron Structure Analyses and Conserved Motif Identification
4.6. Expression Analyses and Heatmap Analyses
4.7. Gene Ontology (GO) Enrichment Analysis
4.8. qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shikata, M.; Matsuda, Y.; Ando, K.; Nishii, A.; Takemura, M.; Yokota, A.; Kohchi, T. Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 2004, 55, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14 (Suppl. 1), S153–S164. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Meng, Y.; Huang, D.; Qi, Y.; Chen, M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 2011, 98, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; You, J.; Chan, Z. Identification and characterization of TIFY family genes in Brachypodium distachyon. J. Plant Res. 2015, 128, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Luthe, D. Identification and evolution analysis of the JAZ gene family in maize. BMC Genom. 2021, 22, 256. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Howe, G.A. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 2009, 21, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Shikata, M.; Takemura, M.; Yokota, A.; Kohchi, T. Arabidopsis ZIM, a plant-specific GATA factor, can function as a transcriptional activator. Biosci. Biotechnol. Biochem. 2003, 67, 2495–2497. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, B.; Grunewald, W.; Bateman, A.; Kohchi, T.; Gheysen, G. The tify family previously known as ZIM. Trends Plant Sci. 2007, 12, 239–244. [Google Scholar] [CrossRef]
- Chung, H.S.; Niu, Y.; Browse, J.; Howe, G.A. Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry 2009, 70, 1547–1559. [Google Scholar] [CrossRef]
- Staswick, P.E. JAZing up jasmonate signaling. Trends Plant Sci. 2008, 13, 66–71. [Google Scholar] [CrossRef]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. Cell Mol. Biol. 2008, 55, 979–988. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhu, L.; Wassan, G.M.; Wang, Y.; Miao, Y.; Shaban, M.; Hu, H.; Sun, H.; Zhang, X. GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171. Mol. Plant Pathol. 2018, 19, 896–908. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signaling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; He, X.; Tu, L.; Zhu, L.; Zhu, S.; Ge, Z.; Zhang, X. GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant J. Cell Mol. Biol. 2016, 88, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Ding, L.; Jiang, J.; Jia, D.; Li, S.; Jin, L.; Zhao, W.; Zhang, X.; Song, A.; Chen, S.; et al. The TIFY family protein CmJAZ1-like negatively regulates petal size via interaction with the bHLH transcription factor CmBPE2 in Chrysanth. Morifolium. Plant J. Cell Mol. Biol. 2022, 112, 1489–1506. [Google Scholar] [CrossRef] [PubMed]
- Heidari, P.; Faraji, S.; Ahmadizadeh, M.; Ahmar, S.; Mora-Poblete, F. New insights into Structure and function of TIFY genes in Zea mays and Solanum lycopersicum: A Genome-wide comprehensive analysis. Front. Genet. 2021, 12, 657970. [Google Scholar] [CrossRef]
- Zhao, Z.; Meng, G.; Zamin, I.; Wei, T.; Ma, D.; An, L.; Yue, X. Genome-wide identification and functional snalysis of the TIFY family genes in response to abiotic stresses and hormone treatments in Tartary buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 2023, 24, 10916. [Google Scholar] [CrossRef]
- Zhu, D.; Bai, X.; Chen, C.; Chen, Q.; Cai, H.; Li, Y.; Ji, W.; Zhai, H.; Lv, D.; Luo, X.; et al. GsTIFY10, a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Mol. Biol. 2011, 77, 285–297. [Google Scholar] [CrossRef]
- Liu, B.; Seong, K.; Pang, S.; Song, J.; Gao, H.; Wang, C.; Zhai, J.; Zhang, Y.; Gao, S.; Li, X.; et al. Functional specificity, diversity, and redundancy of Arabidopsis JAZ family repressors in jasmonate and COI1-regulated growth, development, and defense. New Phytol. 2021, 231, 1525–1545. [Google Scholar] [CrossRef]
- Singh, A.P.; Pandey, B.K.; Deveshwar, P.; Narnoliya, L.; Parida, S.K.; Giri, J. JAZ repressors: Potential involvement in nutrients deficiency response in rice and chickpea. Front. Plant Sci. 2015, 6, 975. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, M.; Singer, S.D.; Fei, Z.; Wang, H.; Wang, X. Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE 2012, 7, e44465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, X.; Yu, R.; Han, M.; Wu, Z. Isolation, structural analysis, and expression characteristics of the maize TIFY gene family. Mol. Genet. Genom. MGG 2015, 290, 1849–1858. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; He, X.; Luo, X.; Xu, L.; Liu, L.; Min, L.; Jin, L.; Zhu, L.; Zhang, X. Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 2014, 166, 2179–2194. [Google Scholar] [CrossRef]
- Zhu, D.; Cai, H.; Luo, X.; Bai, X.; Deyholos, M.K.; Chen, Q.; Chen, C.; Ji, W.; Zhu, Y. Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 2012, 426, 273–279. [Google Scholar] [CrossRef]
- Oh, Y.; Baldwin, I.T.; Gális, I. NaJAZh regulates a subset of defense responses against herbivores and spontaneous leaf necrosis in Nicotiana attenuata plants. Plant Physiol. 2012, 159, 769–788. [Google Scholar] [CrossRef]
- Oh, Y.; Baldwin, I.T.; Galis, I. A jasmonate ZIM-domain protein NaJAZd regulates floral jasmonic acid levels and counteracts flower abscission in Nicotiana attenuata plants. PLoS ONE 2013, 8, e57868. [Google Scholar] [CrossRef]
- Manfield, I.W.; Devlin, P.F.; Jen, C.H.; Westhead, D.R.; Gilmartin, P.M. Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol. 2007, 143, 941–958. [Google Scholar] [CrossRef]
- Lacatus, G.; Sunter, G. The Arabidopsis PEAPOD2 transcription factor interacts with geminivirus AL2 protein and the coat protein promoter. Virology 2009, 392, 196–202. [Google Scholar] [CrossRef]
- Shaikhali, J.; de Dios Barajas-Lopéz, J.; Ötvös, K.; Kremnev, D.; Garcia, A.S.; Srivastava, V.; Wingsle, G.; Bako, L.; Strand, Å. The CRYPTOCHROME1-dependent response to excess light is mediated through the transcriptional activators ZINC FINGER PROTEIN EXPRESSED IN INFLORESCENCE MERISTEM LIKE1 and ZML2 in Arabidopsis. Plant Cell 2012, 24, 3009–3025. [Google Scholar] [CrossRef]
- Vélez-Bermúdez, I.C.; Salazar-Henao, J.E.; Fornalé, S.; López-Vidriero, I.; Franco-Zorrilla, J.M.; Grotewold, E.; Gray, J.; Solano, R.; Schmidt, W.; Pagés, M.; et al. A MYB/ZML complex regulates wound-Induced lignin genes in Maize. Plant Cell 2015, 27, 3245–3259. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, S.; Wei, L.; Huang, Y.; Liu, D.; Jia, Y.; Luo, C.; Lin, Y.; Liang, C.; Hu, Y.; et al. BnIR: A multi-omics database with various tools for Brassica napus research and breeding. Mol. Plant 2023, 16, 775–789. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Song, J.M.; Guan, Z.; Hu, J.; Guo, C.; Yang, Z.; Wang, S.; Liu, D.; Wang, B.; Lu, S.; Zhou, R.; et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 2020, 6, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Fan, G.; Hu, Q.; Zhou, Y.; Guan, M.; Tong, C.; Li, J.; Du, D.; Qi, C.; Jiang, L.; et al. The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. Cell Mol. Biol. 2017, 92, 452–468. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tong, C.; Zhang, X.; Song, A.; Hu, M.; Dong, W.; Chen, F.; Wang, Y.; Tu, J.; Liu, S.; et al. A high-quality Brassica napus genome reveals expansion of transposable elements, subgenome evolution and disease resistance. Plant Biotechnol. J. 2021, 19, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yu, L.; Wei, L.; Yu, P.; Wang, J.; Zhao, H.; Zhang, Y.; Zhang, S.; Yang, Z.; Chen, G.; et al. BnTIR: An online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus. Plant Biotechnol. J. 2021, 19, 1895–1897. [Google Scholar] [CrossRef]
- Singh, P.; Mukhopadhyay, K. Comprehensive molecular dissection of TIFY transcription factors reveal their dynamic responses to biotic and abiotic stress in wheat (Triticum aestivum L.). Sci. Rep. 2021, 11, 9739. [Google Scholar] [CrossRef]
- Yi, R.; Shan, X. Post-translational modifications: Emerging regulators manipulating jasmonate biosynthesis and signaling. Plant Cell Rep. 2023, 42, 215–222. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef]
- Köllmer, I.; Novák, O.; Strnad, M.; Schmülling, T.; Werner, T. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. Cell Mol. Biol. 2014, 78, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. Perspect. Biol. 2010, 2, a001594. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Hansen, C.H.; Wittstock, U.; Halkier, B.A. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 2000, 275, 33712–33717. [Google Scholar] [CrossRef] [PubMed]
- Hull, A.K.; Celenza, J.L. Bacterial expression and purification of the Arabidopsis NADPH-cytochrome P450 reductase ATR2. Protein Expr. Purif. 2000, 18, 310–315. [Google Scholar] [CrossRef]
- Hull, A.K.; Vij, R.; Celenza, J.L. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 2379–2384. [Google Scholar] [CrossRef]
- Kindl, H. Oxydasen and Oxygenasen in höheren Pflanzen, I. Uber das Vorkommen von Indolyl-(3)-acetaldehydoxim und seine Bildung aus L-Tryptophan [Oxidases and oxygenases in higher plants, I. On the occurrence of indolyl-(3)-acetaldehyde oxime and its formation from L-tryptophan]. Hoppe-Seyler’s Z. Physiol. Chem. 1968, 349, 519–520. [Google Scholar]
- Hussain, S.; Kim, S.H.; Bahk, S.; Ali, A.; Nguyen, X.C.; Yun, D.J.; Chung, W.S. The auxin signaling repressor IAA8 promotes seed germination through down-regulation of ABI3 transcription in Arabidopsis. Front. Plant Sci. 2020, 11, 111. [Google Scholar] [CrossRef]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, F.; Chen, D.; Chu, W.; Liu, H.; Xiang, Y. Genome-wide identification and analysis of the Populus trichocarpa TIFY gene family. Plant Physiol. Biochem. PPB 2017, 115, 360–371. [Google Scholar] [CrossRef]
- Sun, Q.; Wang, G.; Zhang, X.; Zhang, X.; Qiao, P.; Long, L.; Yuan, Y.; Cai, Y. Genome-wide identification of the TIFY gene family in three cultivated Gossypium species and the expression of JAZ genes. Sci. Rep. 2017, 7, 42418. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Pan, X.; Yu, Y.; Zhu, Y.; Kong, F.; Sun, X.; Wang, F. Overexpression of a TIFY family gene, GsJAZ2, exhibits enhanced tolerance to alkaline stress in soybean. Mol. Breed. 2020, 40, 33. [Google Scholar] [CrossRef]
- Sun, P.; Shi, Y.; Valerio, A.G.O.; Borrego, E.J.; Luo, Q.; Qin, J.; Liu, K.; Yan, Y. An updated census of the maize TIFY family. PLoS ONE 2021, 16, e0247271. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, F.; Yang, G.; Liu, X.; Peng, S. Identification of TIFY gene family in walnut and analysis of its expression under abiotic stresses. BMC Genom. 2022, 23, 190. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benning, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–E10777. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Kim, Y.S.; Park, S.H.; Koo, Y.J.; Choi, Y.D.; Chung, Y.Y.; Lee, I.J.; Kim, J.K. Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice. Plant Physiol. 2009, 149, 1751–1760. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Wu, S.; Zhu, Z.; Liu, F.; Fu, Y.; Cai, H.; Sun, X.; Gu, P.; Xie, D.; Tan, L.; et al. NOG1 increases grain production in rice. Nat. Commun. 2017, 8, 1497. [Google Scholar] [CrossRef]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014, 5, 3476. [Google Scholar] [CrossRef]
- You, X.; Zhu, S.; Zhang, W.; Zhang, J.; Wang, C.; Jing, R.; Chen, W.; Wu, H.; Cai, Y.; Feng, Z.; et al. OsPEX5 regulates rice spikelet development through modulating jasmonic acid biosynthesis. New Phytol. 2019, 224, 712–724. [Google Scholar] [CrossRef]
- Hu, S.; Yang, H.; Gao, H.; Yan, J.; Xie, D. Control of seed size by jasmonate. Sci. China. Life Sci. 2021, 64, 1215–1226. [Google Scholar] [CrossRef]
- He, X.; Kang, Y.; Li, W.; Liu, W.; Xie, P.; Liao, L.; Huang, L.; Yao, M.; Qian, L.; Liu, Z.; et al. Genome-wide identification and functional analysis of the TIFY gene family in the response to multiple stresses in Brassica napus L. BMC Genom. 2020, 21, 736. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liang, C.; Qiu, Z.; Zhou, S.; Liu, J.; Yang, Y.; Wang, R.; Yin, J.; Ma, C.; Cui, Z.; et al. Jasmonic acid negatively regulates branch growth in pear. Front. Plant Sci. 2023, 14, 1105521. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Chen, Z.; Zhang, Q.; Wan, Y.; Hu, R.; Shen, S.; Chen, S.; Yin, N.; Tang, Y.; Liang, Y.; et al. Genome-wide identification of the TIFY gene family in Brassiceae and its potential association with heavy metal stress in rapeseed. Plants (Basel Switz.) 2022, 11, 667. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxf. Engl.) 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Birney, E.; Andrews, T.D.; Bevan, P.; Caccamo, M.; Chen, Y.; Clarke, L.; Coates, G.; Cuff, J.; Curwen, V.; Cutts, T.; et al. An overview of Ensemble. Genome Res. 2004, 14, 925–928. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics (Oxf. Engl.) 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]



| Gene Name | Chromosome Location | Direction | AtGI | At Name | Protein Length (aa) | Molecular Weight (kDa) | Isoelectric Point (pI) | Subcellular Prediction |
|---|---|---|---|---|---|---|---|---|
| BnaA01G0052700ZS | A01:2859984…2861895 | + | AT4G32570 | TIFY8 | 310 | 33.56 | 6.51 | nucl |
| BnaA01G0140700ZS | A01:8392342…8394246 | + | AT4G24470 | GATA25 | 320 | 35.09 | 9.49 | nucl |
| BnaA01G0203400ZS | A01:12583796…12585951 | + | AT4G14720 | TIFY4B | 130 | 13.73 | 4.29 | nucl |
| BnaA01G0282800ZS | A01:26329701…26330093 | + | AT4G24470 | GATA25 | 367 | 40.68 | 7.35 | chlo |
| BnaA01G0308500ZS | A01:28824803…28828698 | − | AT3G21175 | GATA24 | 353 | 37.58 | 9.5 | nucl |
| BnaA01G0331000ZS | A01:30506077…30510006 | − | AT3G17860 | TIFY6B | 345 | 37.44 | 10.12 | nucl |
| BnaA02G0000800ZS | A02:293262…294602 | − | AT5G20900 | TIFY3B | 216 | 22.47 | 4.7 | cyto |
| BnaA02G0001900ZS | A02:350665…352002 | + | AT5G20900 | TIFY3B | 112 | 11.96 | 4.35 | cysk |
| BnaA02G0047200ZS | A02:2647926…2649211 | + | AT5G13220 | TIFY9 | 274 | 29.34 | 10.43 | nucl |
| BnaA02G0138200ZS | A02:7624707…7625045 | − | AT4G24470 | GATA25 | 273 | 30.16 | 9.64 | nucl |
| BnaA02G0190100ZS | A02:11734586…11736653 | + | AT1G70700 | TIFY7 | 240 | 26.20 | 9.6 | nucl |
| BnaA02G0200100ZS | A02:12545103…12546473 | − | AT1G72450 | TIFY11B | 177 | 19.36 | 10.52 | cyto |
| BnaA02G0213500ZS | A02:13411661…13412862 | − | AT1G74950 | TIFY10B | 212 | 23.99 | 10.64 | golg |
| BnaA03G0052500ZS | A03:2524449…2525938 | + | AT5G13220 | TIFY9 | 112 | 11.96 | 4.35 | cysk |
| BnaA03G0398900ZS | A03:21529159…21529497 | − | AT4G24470 | GATA25 | 261 | 28.28 | 6.41 | nucl |
| BnaA03G0479900ZS | A03:26610896…26612549 | + | AT4G24470 | GATA25 | 223 | 25.25 | 10.44 | chlo |
| BnaA04G0222400ZS | A04:21368091…21368591 | + | AT2G34600 | TIFY 5B | 119 | 13.70 | 10.24 | nucl |
| BnaA05G0099000ZS | A05:5800314…5800869 | − | AT2G34600 | TIFY 5B | 116 | 13.06 | 8.89 | nucl |
| BnaA05G0380600ZS | A05:37865369…37867939 | − | AT3G17860 | TIFY6B | 336 | 35.87 | 9.9 | nucl |
| BnaA06G0119400ZS | A06:6986490…6987776 | − | AT1G17380 | TIFY11A | 266 | 29.80 | 9.7 | nucl |
| BnaA06G0133300ZS | A06:7845736…7847029 | + | AT1G19180 | TIFY10A | 254 | 27.30 | 10.51 | chlo |
| BnaA07G0249100ZS | A07:24141740…24144675 | + | AT1G74950 | TIFY10B | 224 | 24.59 | 9.56 | nucl |
| BnaA07G0266400ZS | A07:25167684…25168787 | − | AT1G70700 | TIFY7 | 212 | 23.41 | 9.12 | mito |
| BnaA07G0320600ZS | A07:28617818…28620360 | + | AT1G70700 | TIFY7 | 266 | 28.64 | 10.45 | nucl |
| BnaA07G0333200ZS | A07:29319157…29320650 | − | AT1G72450 | TIFY11B | 274 | 30.60 | 9.62 | nucl |
| BnaA07G0349300ZS | A07:30135515…30136690 | − | AT1G74950 | TIFY10B | 245 | 26.76 | 9.62 | nucl |
| BnaA08G0021500ZS | A08:1797523…1798971 | − | AT1G51600 | GATA28 | 318 | 35.01 | 9.58 | nucl |
| BnaA08G0090300ZS | A08:15229435…15231385 | − | AT4G14720 | TIFY4B | 357 | 38.75 | 9.11 | nucl |
| BnaA08G0145800ZS | A08:19339471…19341512 | + | AT4G32570 | TIFY8 | 130 | 14.97 | 10.39 | nucl |
| BnaA08G0205900ZS | A08:22909636…22910221 | − | AT1G30135 | TIFY5A | 311 | 33.96 | 6.45 | nucl |
| BnaA08G0252900ZS | A08:25292226…25293349 | − | AT1G19180 | TIFY10A | 259 | 28.31 | 9.99 | nucl |
| BnaA08G0262000ZS | A08:25690409…25691687 | + | AT1G17380 | TIFY11A | 267 | 29.71 | 9.57 | nucl |
| BnaA09G0412500ZS | A09:47302666…47303247 | − | AT1G30135 | TIFY5A | 133 | 15.23 | 9.83 | nucl |
| BnaA09G0610300ZS | A09:60116951…60119612 | − | AT1G19180 | TIFY10A | 273 | 29.82 | 10 | nucl |
| BnaA09G0618000ZS | A09:60520671…60529802 | + | AT1G17380 | TIFY11A | 315 | 35.08 | 10.34 | chlo |
| BnaA10G0170700ZS | A10:20196301…20197382 | − | AT5G20900 | TIFY3B | 203 | 21.43 | 7.58 | nucl |
| BnaA10G0223600ZS | A10:22916557…22919394 | − | AT5G13220 | TIFY9 | 196 | 21.82 | 10.65 | nucl |
| BnaC01G0061700ZS | C01:3490282…3492250 | + | AT4G32570 | TIFY8 | 304 | 33.06 | 6.64 | nucl |
| BnaC01G0179300ZS | C01:13276298…13278294 | + | AT4G24470 | GATA25 | 314 | 34.53 | 8.44 | nucl |
| BnaC01G0254700ZS | C01:20032455…20034770 | − | AT4G14720 | TIFY4B | 353 | 37.52 | 9.38 | nucl |
| BnaC01G0408900ZS | C01:47295575…47298846 | − | AT3G17860 | TIFY6B | 344 | 37.27 | 10.12 | nucl |
| BnaC02G0054000ZS | C02:3401968…3404083 | + | AT5G13220 | TIFY9 | 221 | 23.08 | 4.72 | nucl |
| BnaC02G0104800ZS | C02:6810525…6811864 | + | AT5G20900 | TIFY3B | 274 | 29.41 | 10.51 | cyto |
| BnaC02G0251900ZS | C02:23433804…23435871 | + | AT1G70700 | TIFY7 | 272 | 29.95 | 7.71 | nucl |
| BnaC02G0266500ZS | C02:25372570…25373915 | − | AT1G72450 | TIFY11B | 240 | 26.20 | 9.75 | nucl |
| BnaC02G0285000ZS | C02:27147851…27149030 | − | AT1G74950 | TIFY10B | 196 | 21.80 | 10.68 | nucl |
| BnaC03G0060300ZS | C03:3153036…3154515 | + | AT5G13220 | TIFY9 | 113 | 13.03 | 8.78 | chlo |
| BnaC03G0192400ZS | C03:11016512…11016853 | + | AT2G34600 | TIFY 5B | 223 | 25.20 | 10.44 | nucl |
| BnaC03G0662600ZS | C03:64076368…64076942 | + | AT1G30135 | TIFY5A | 130 | 14.97 | 10.39 | nucl |
| BnaC03G0663300ZS | C03:64131128…64131740 | + | AT1G30135 | TIFY5A | 144 | 16.74 | 8.64 | nucl |
| BnaC03G0759700ZS | C03:73567925…73569989 | + | AT4G32570 | TIFY8 | 358 | 38.78 | 9.17 | nucl |
| BnaC03G0787400ZS | C03:76176548…76178066 | + | AT1G51600 | GATA28 | 296 | 32.35 | 6.18 | nucl |
| BnaC04G0121900ZS | C04:11013009…11013565 | − | AT2G34600 | TIFY 5B | 116 | 13.13 | 9.33 | nucl |
| BnaC04G0534800ZS | C04:65634871…65635324 | + | AT2G34600 | TIFY 5B | 121 | 13.65 | 9.26 | nucl |
| BnaC05G0147200ZS | C05:9303032…9304540 | − | AT1G17380 | TIFY11A | 256 | 28.40 | 10.36 | nucl |
| BnaC05G0160800ZS | C05:10437306…10438599 | + | AT1G19180 | TIFY10A | 254 | 27.26 | 10.3 | nucl |
| BnaC05G0259000ZS | C05:21020991…21021569 | + | AT1G30135 | TIFY5A | 132 | 15.24 | 9.83 | nucl |
| BnaC05G0424600ZS | C05:47654460…47657298 | − | AT3G17860 | TIFY6B | 335 | 35.76 | 9.85 | nucl |
| BnaC06G0274800ZS | C06:38103129…38104304 | + | AT1G74950 | TIFY10B | 224 | 24.46 | 9.77 | nucl |
| BnaC06G0299300ZS | C06:40408436…40409553 | − | AT1G70700 | TIFY7 | 212 | 23.32 | 9.12 | nucl |
| BnaC06G0373700ZS | C06:47272615…47275070 | + | AT1G70700 | TIFY7 | 266 | 28.53 | 10.12 | nucl |
| BnaC06G0391300ZS | C06:48490374…48491996 | − | AT1G72450 | TIFY11B | 271 | 30.32 | 10.11 | nucl |
| BnaC06G0411600ZS | C06:49662272…49663471 | − | AT1G74950 | TIFY10B | 246 | 27.24 | 9.17 | nucl |
| BnaC07G0457900ZS | C07:55670963…55672463 | + | AT4G24470 | GATA25 | 259 | 28.08 | 6.23 | nucl |
| BnaC08G0129100ZS | C08:23055859…23058016 | − | AT4G14720 | TIFY4B | 362 | 39.79 | 8.43 | ER |
| BnaC08G0240100ZS | C08:33426315…33427601 | − | AT1G17380 | TIFY11A | 268 | 29.79 | 9.1 | nucl |
| BnaC08G0251600ZS | C08:34242963…34244270 | + | AT1G19180 | TIFY10A | 264 | 28.74 | 10.22 | nucl |
| BnaC08G0473200ZS | C08:50094384…50095986 | + | AT1G17380 | TIFY11A | 343 | 38.00 | 10.11 | nucl |
| BnaC09G0456500ZS | C09:56962395…56963463 | − | AT5G20900 | TIFY3B | 201 | 21.25 | 6.95 | cyto |
| BnaC09G0528700ZS | C09:62610296…62611889 | − | AT5G13220 | TIFY9 | 199 | 22.46 | 10.73 | nucl |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhang, Q.; Wang, L.; Wang, X.; Qiao, J.; Wang, H. New Insights into the TIFY Gene Family of Brassica napus and Its Involvement in the Regulation of Shoot Branching. Int. J. Mol. Sci. 2023, 24, 17114. https://doi.org/10.3390/ijms242317114
Li Y, Zhang Q, Wang L, Wang X, Qiao J, Wang H. New Insights into the TIFY Gene Family of Brassica napus and Its Involvement in the Regulation of Shoot Branching. International Journal of Molecular Sciences. 2023; 24(23):17114. https://doi.org/10.3390/ijms242317114
Chicago/Turabian StyleLi, Yarong, Qian Zhang, Luman Wang, Xinfa Wang, Jiangwei Qiao, and Hanzhong Wang. 2023. "New Insights into the TIFY Gene Family of Brassica napus and Its Involvement in the Regulation of Shoot Branching" International Journal of Molecular Sciences 24, no. 23: 17114. https://doi.org/10.3390/ijms242317114
APA StyleLi, Y., Zhang, Q., Wang, L., Wang, X., Qiao, J., & Wang, H. (2023). New Insights into the TIFY Gene Family of Brassica napus and Its Involvement in the Regulation of Shoot Branching. International Journal of Molecular Sciences, 24(23), 17114. https://doi.org/10.3390/ijms242317114







