Dual Effects of 3-epi-betulin from Daphniphyllum glaucescens in Suppressing SARS-CoV-2-Induced Inflammation and Inhibiting Virus Entry
Abstract
:1. Introduction
2. Results
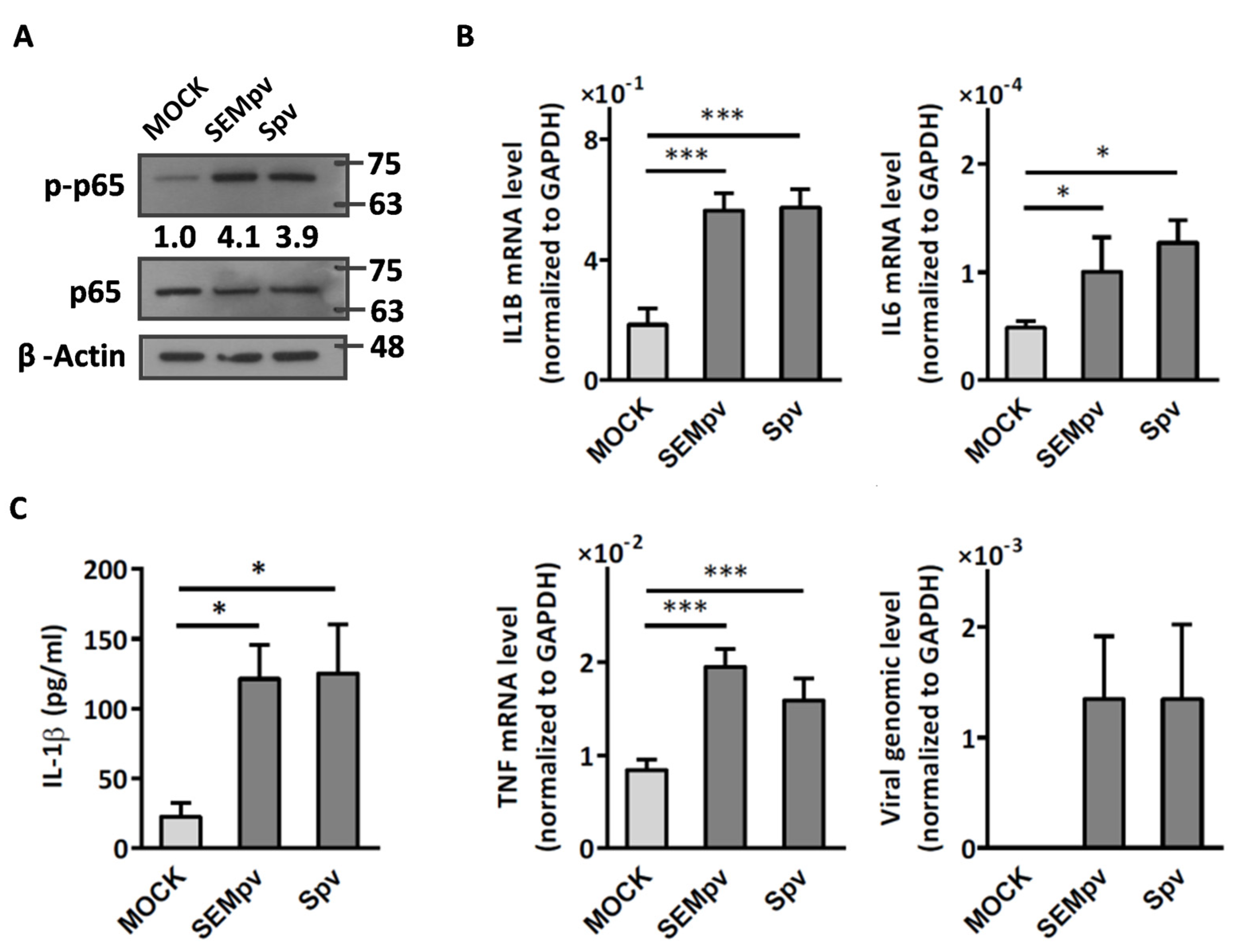
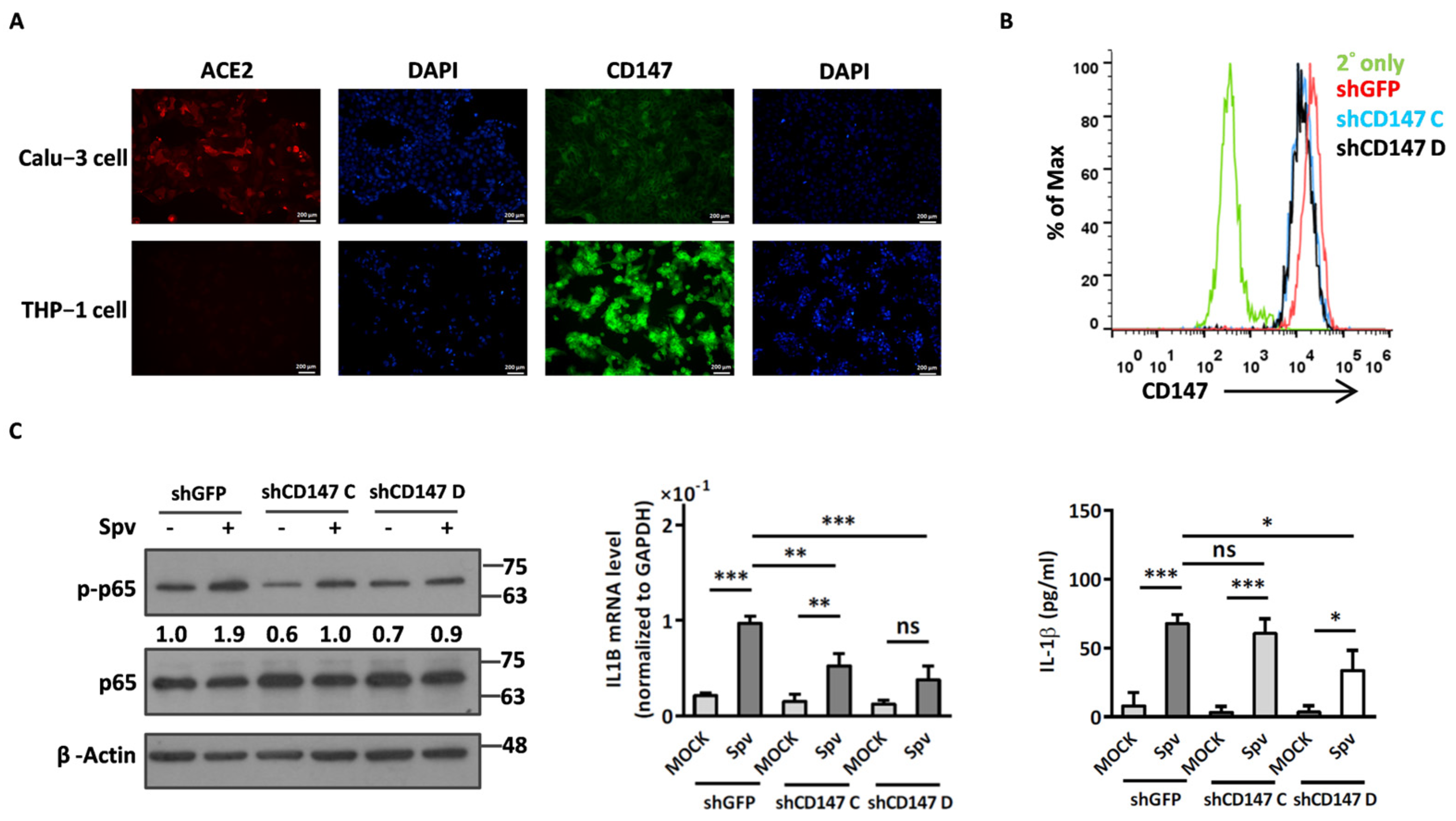
2.1. The SARS-CoV-2 Pseudovirus Activates an Inflammatory Response Independent of Virus Entry
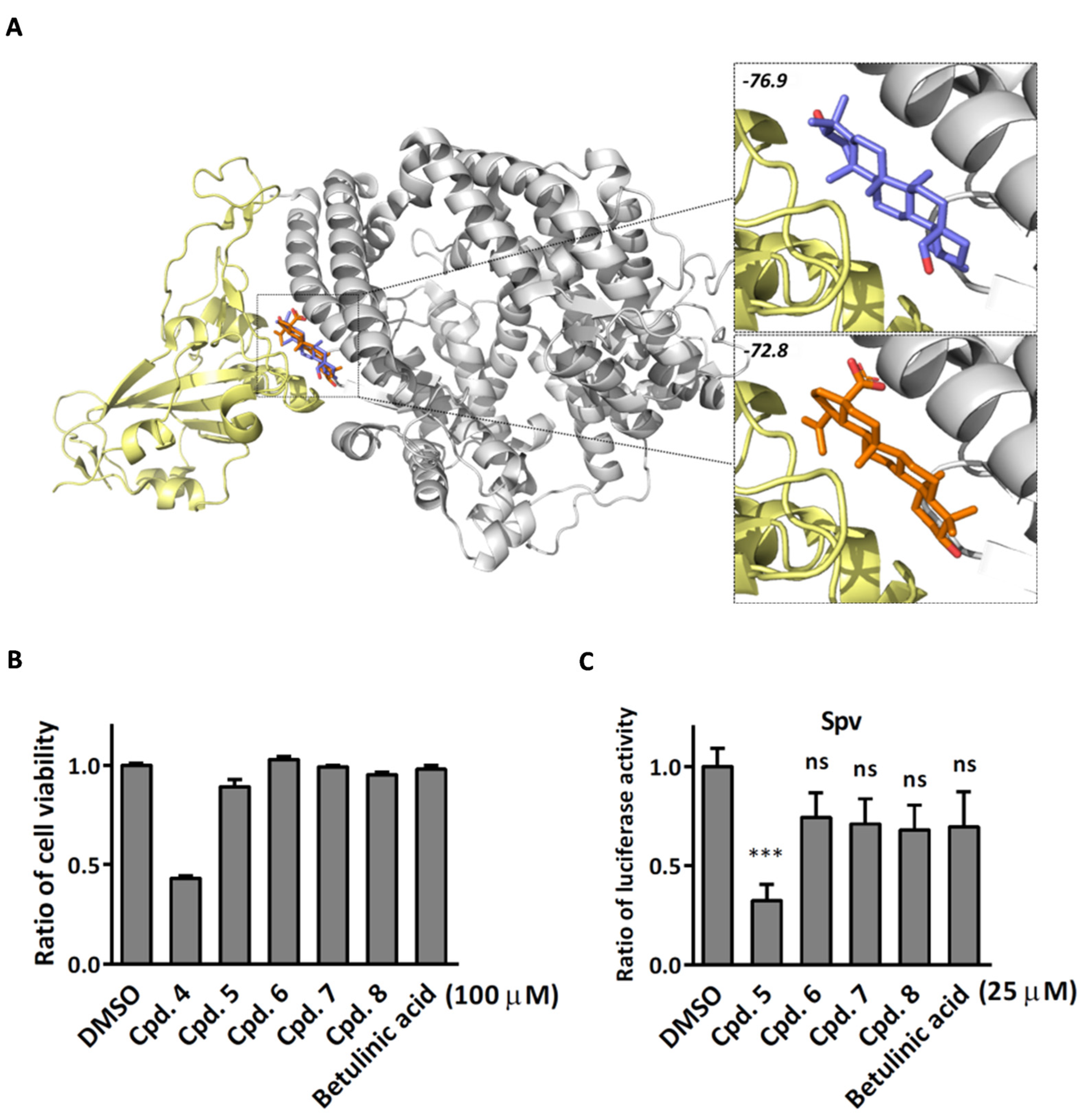
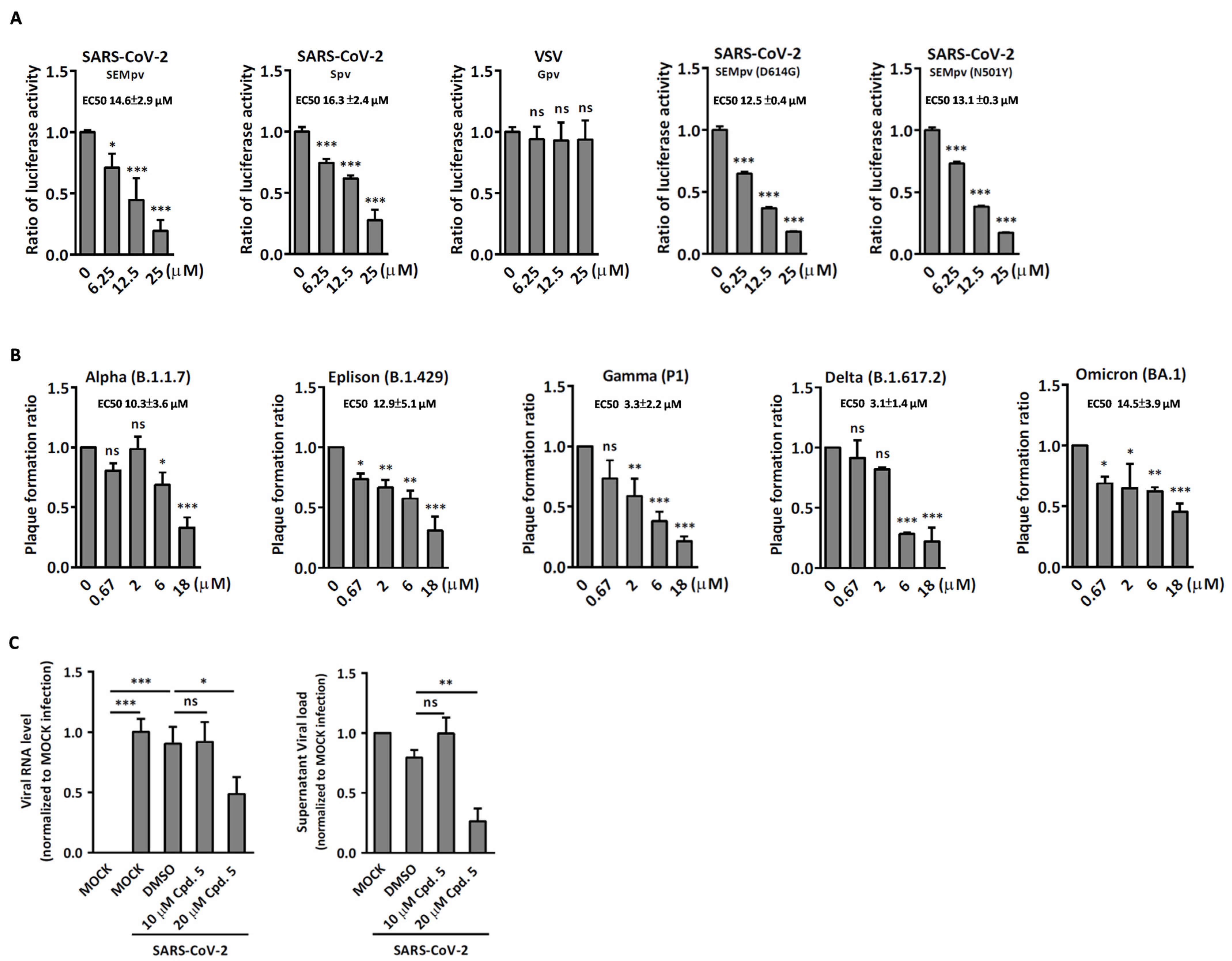
2.2. Compound 5 with Anti-Inflammation Activity Induced by SARS-CoV-2 Pseudoviruses Suppresses Viral Replication
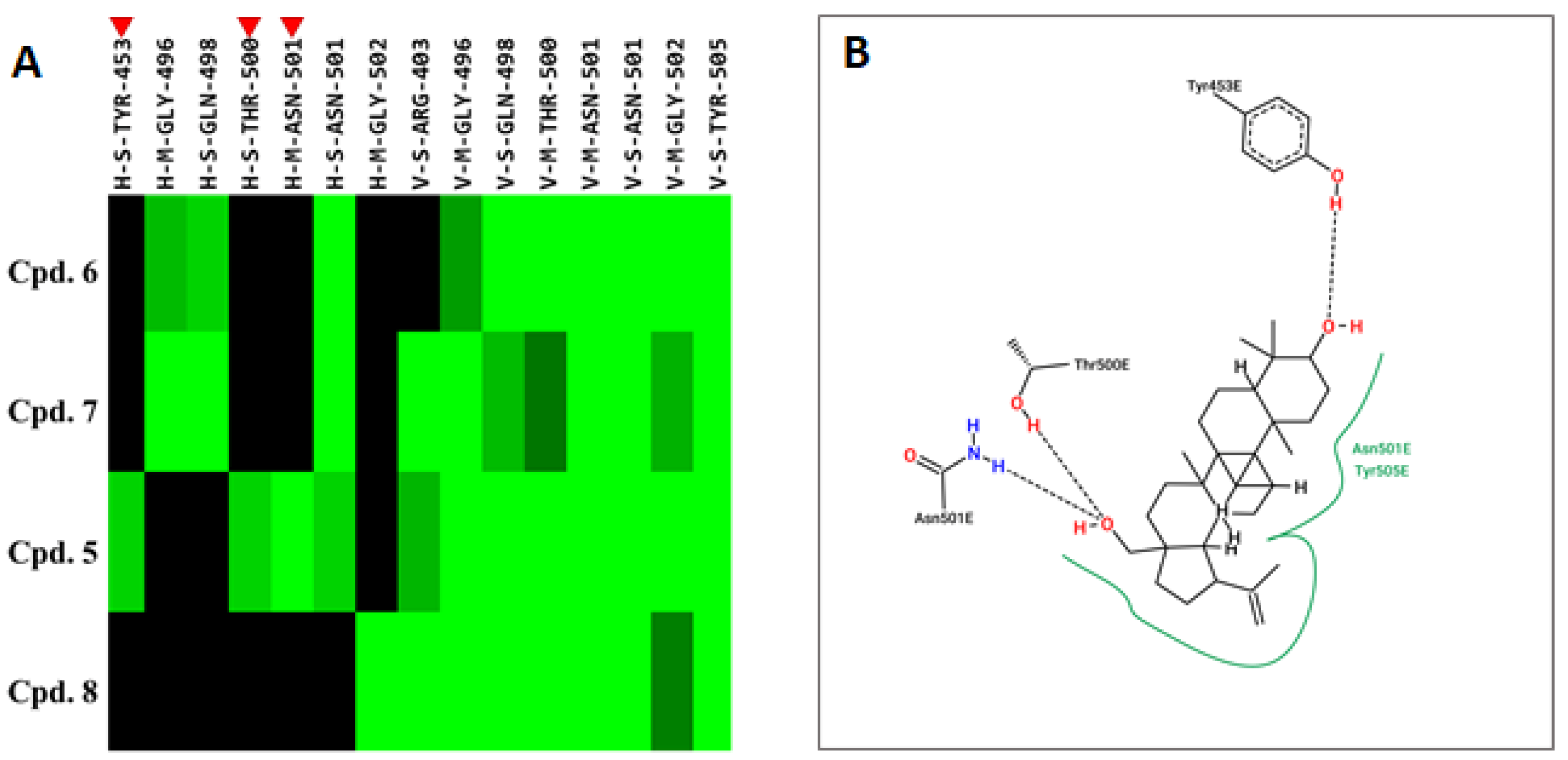
2.3. Compound 5 Docked the RBD Domain of the SARS-CoV-2 Spike Protein and Effectively Inhibited Virus Entry
3. Discussion
4. Materials and Methods
4.1. Purification of Terpenoids from Flueggea virosa and Daphniphyllum glaucescens
4.2. Cells, Viruses, and Compounds
4.3. SARS-CoV-2 Pseudovirus
4.4. Real-Time RT-PCR Assay
4.5. Inflammation Induced by SARS-CoV-2 Pseudovirus
4.6. Flow Cytometric Analysis
4.7. Immunofluorescence Stain
4.8. Cell Viability Assay
4.9. SARS-CoV-2 Pseudovirus Entry Assay
4.10. Plaque Reduction Assay
4.11. Yield Reduction Assay
4.12. Virus Titration
4.13. Molecular Docking and Interaction Analysis
4.14. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, S.; Shafiei, M.S.; Longoria, C.; Schoggins, J.W.; Savani, R.C.; Zaki, H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-kappaB pathway. Elife 2021, 10, e68563. [Google Scholar] [CrossRef]
- Xu, C.; Wang, Y.; Liu, C.; Zhang, C.; Han, W.; Hong, X.; Wang, Y.; Hong, Q.; Wang, S.; Zhao, Q.; et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 2021, 7, eabe5575. [Google Scholar] [CrossRef]
- Ibrahim, I.M.; Abdelmalek, D.H.; Elshahat, M.E.; Elfiky, A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020, 80, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.Q.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Tiwari, S.; Deb, M.K.; Marty, J.L. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): A global pandemic and treatment strategies. Int. J. Antimicrob. Agents 2020, 56, 106054. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022, 20, 270–284. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.; Kalle, A.M.; Reddanna, P. Managing SARS-CoV-2 Infections Through Resolution of Inflammation by Eicosanoids: A Review. J. Inflamm. Res. 2022, 15, 4349–4358. [Google Scholar] [CrossRef]
- Darif, D.; Hammi, I.; Kihel, A.; Saik, I.E.; Guessous, F.; Akarid, K. The pro-inflammatory cytokines in COVID-19 pathogenesis: What goes wrong? Microb. Pathog. 2021, 153, 104799. [Google Scholar] [CrossRef]
- Vollbracht, C.; Kraft, K. Oxidative Stress and Hyper-Inflammation as Major Drivers of Severe COVID-19 and Long COVID: Implications for the Benefit of High-Dose Intravenous Vitamin C. Front. Pharmacol. 2022, 13, 899198. [Google Scholar] [CrossRef]
- Hu, Z.W.; Lin, J.H.; Chen, J.T.; Cai, T.X.; Xia, L.X.; Liu, Y.; Song, X.; He, Z.D. Overview of Viral Pneumonia Associated With Influenza Virus, Respiratory Syncytial Virus, and Coronavirus, and Therapeutics Based on Natural Products of Medicinal Plants. Front. Pharmacol. 2021, 12, 630834. [Google Scholar] [CrossRef]
- Li, T.; Peng, T. Traditional Chinese herbal medicine as a source of molecules with antiviral activity. Antivir. Res. 2013, 97, 1–9. [Google Scholar] [CrossRef]
- Salehi, B.; Kumar, N.V.A.; Sener, B.; Sharifi-Rad, M.; Kilic, M.; Mahady, G.B.; Vlaisavljevic, S.; Iriti, M.; Kobarfard, F.; Setzer, W.N.; et al. Medicinal Plants Used in the Treatment of Human Immunodeficiency Virus. Int. J. Mol. Sci. 2018, 19, 1459. [Google Scholar] [CrossRef]
- Remali, J.; Aizat, W.M. A Review on Plant Bioactive Compounds and Their Modes of Action Against Coronavirus Infection. Front. Pharmacol. 2020, 11, 589044. [Google Scholar] [CrossRef]
- Sruthi, D.; Dhanalakshmi, M.; Rao, H.C.Y.; Parthasarathy, R.; Deepanraj, S.P.; Jayabaskaran, C. Curative Potential of High-Value Phytochemicals on COVID-19 Infection. Biochemistry 2023, 88, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.C.; Chen, Y.J.; Chuang, C.W.; Chao, Y.H.; Huang, H.C.; Lin, C.C.; Chao, C.H. Polyoxygenated Terpenoids and Polyketides from the Roots of and Their Inhibitory Effect against SARS-CoV-2-Induced Inflammation. Molecules 2022, 27, 8548. [Google Scholar] [CrossRef]
- Jeong, G.S.; Bae, J.S. Anti-Inflammatory Effects of Triterpenoids; Naturally Occurring and Synthetic Agents. Mini-Rev. Org. Chem. 2014, 11, 316–329. [Google Scholar] [CrossRef]
- Carino, A.; Moraca, F.; Fiorillo, B.; Marchianò, S.; Sepe, V.; Biagioli, M.; Finamore, C.; Bozza, S.; Francisci, D.; Distrutti, E.; et al. Hijacking SARS-CoV-2/ACE2 Receptor Interaction by Natural and Semi-synthetic Steroidal Agents Acting on Functional Pockets on the Receptor Binding Domain. Front. Chem. 2020, 8, 572885. [Google Scholar] [CrossRef]
- Katrin Stierand, M.R. PoseView—Molecular interaction patterns at a glance. J. Cheminform. 2010, 2, P50. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; HLH Across Speciality Collaboration, UK. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Li, G.; Hilgenfeld, R.; Whitley, R.; De Clercq, E. Therapeutic strategies for COVID-19: Progress and lessons learned. Nat. Rev. Drug Discov. 2023, 22, 449–475. [Google Scholar] [CrossRef] [PubMed]
- Dharra, R.; Sharma, A.K.; Datta, S. Emerging aspects of cytokine storm in COVID-19: The role of proinflammatory cytokines and therapeutic prospects. Cytokine 2023, 169, 156287. [Google Scholar] [CrossRef] [PubMed]
- Barhoumi, T.; Alghanem, B.; Shaibah, H.; Mansour, F.A.; Alamri, H.S.; Akiel, M.A.; Alroqi, F.; Boudjelal, M. SARS-CoV-2 Coronavirus Spike Protein-Induced Apoptosis, Inflammatory, and Oxidative Stress Responses in THP-1-Like-Macrophages: Potential Role of Angiotensin-Converting Enzyme Inhibitor (Perindopril). Front. Immunol. 2021, 12, 728896. [Google Scholar] [CrossRef] [PubMed]
- Avdonin, P.P.; Rybakova, E.Y.; Trufanov, S.K.; Avdonin, P.V. SARS-CoV-2 Receptors and Their Involvement in Cell Infection. Biochem. Mosc. Suppl. Ser. A-Membr. Cell Biol. 2023, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fenizia, C.; Galbiati, S.; Vanetti, C.; Vago, R.; Clerici, M.; Tacchetti, C.; Daniele, T. SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2. Cells 2021, 10, 1434. [Google Scholar] [CrossRef] [PubMed]
- Ragotte, R.J.; Pulido, D.; Donnellan, F.R.; Hill, M.L.; Gorini, G.; Davies, H.; Brun, J.; McHugh, K.; King, L.D.W.; Skinner, K.; et al. Human Basigin (CD147) Does Not Directly Interact with SARS-CoV-2 Spike Glycoprotein. mSphere 2021, 6, e0064721. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Lan, J.; Ju, X.; Gong, M.; Long, Q.; Zhu, Z.; Yu, Y.; Wu, J.; Zhong, J.; Zhang, R.; et al. Mutation Y453F in the spike protein of SARS-CoV-2 enhances interaction with the mink ACE2 receptor for host adaption. PLoS Pathog. 2021, 17, e1010053. [Google Scholar] [CrossRef]
- Burkhanova, T.M.; Krysantieva, A.I.; Babashkina, M.G.; Konyaeva, I.A.; Monina, L.N.; Goncharenko, A.N.; Safin, D.A. In silico analyses of betulin: DFT studies, corrosion inhibition properties, ADMET prediction, and molecular docking with a series of SARS-CoV-2 and monkeypox proteins. Struct. Chem. 2023, 34, 1545–1556. [Google Scholar] [CrossRef]
- Guruprasad, L. Evolutionary relationships and sequence-structure determinants in human SARS coronavirus-2 spike proteins for host receptor recognition. Proteins 2020, 88, 1387–1393. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C.; et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 2022, 602, 294–299. [Google Scholar] [CrossRef]
- Teng, S.; Sobitan, A.; Rhoades, R.; Liu, D.; Tang, Q. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief. Bioinform. 2021, 22, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, M.; Sharma, A.R.; Dhama, K.; Agoramoorthy, G.; Chakraborty, C. Omicron variant (B.1.1.529) of SARS-CoV-2: Understanding mutations in the genome, S-glycoprotein, and antibody-binding regions. Geroscience 2022, 44, 619–637. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.H.; Cheng, J.C.; Shen, D.Y.; Wu, T.S. Anti-Hepatitis C Virus Dinorditerpenes from the Roots of. J. Nat. Prod. 2014, 77, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.H.; Cheng, J.C.; Gonçalves, T.P.; Huang, K.W.; Lin, C.C.; Huang, H.C.; Hwang, S.Y.; Wu, Y.C. Glaulactams A-C, daphniphyllum alkaloids from. Sci. Rep. 2018, 8, 15417. [Google Scholar] [CrossRef]
- Wang, H.I.; Chuang, Z.S.; Kao, Y.T.; Lin, Y.L.; Liang, J.J.; Liao, C.C.; Liao, C.L.; Lai, M.M.C.; Yu, C.Y. Small Structural Proteins E and M Render the SARS-CoV-2 Pseudovirus More Infectious and Reveal the Phenotype of Natural Viral Variants. Int. J. Mol. Sci. 2021, 22, 9087. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. ; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yeh, Y.J.; Tseng, C.P.; Hsu, S.D.; Huang, H.Y.; Lai, M.M.C.; Huang, H.D.; Cheng, J.C. Dual Effects of Let-7b in the Early Stage of Hepatitis C Virus Infection. J. Virol. 2021, 95, e01800-20. [Google Scholar] [CrossRef]
- Yang, J.M.; Chen, C.C. GEMDOCK: A generic evolutionary method for molecular docking. Proteins 2004, 55, 288–304. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Gobeil, S.M.; Janowska, K.; McDowell, S.; Mansouri, K.; Parks, R.; Stalls, V.; Kopp, M.F.; Manne, K.; Li, D.; Wiehe, K.; et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science 2021, 373, eabi6226. [Google Scholar] [CrossRef] [PubMed]
- McCallum, M.; Bassi, J.; De Marco, A.; Chen, A.; Walls, A.C.; Di Iulio, J.; Tortorici, M.A.; Navarro, M.J.; Silacci-Fregni, C.; Saliba, C.; et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 2021, 373, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xiao, T.; Cai, Y.; Lavine, C.L.; Peng, H.; Zhu, H.; Anand, K.; Tong, P.; Gautam, A.; Mayer, M.L.; et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science 2021, 374, 1353–1360. [Google Scholar] [CrossRef] [PubMed]



| No. | Compound Name | Compound Structure |
|---|---|---|
| Cpd. 1 | 12-Hydroxy-13-methyl-ent-podocarp-6,8,11,13-tetraen-3-one | 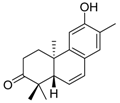 |
| Cpd. 2 | 3β,12-Dihydroxy-13-methylpodocarpa-6,8,11,13-tetraene | 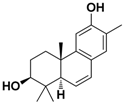 |
| Cpd. 3 | ent-3β,12α-Dihydroxypimara-8(14),15-diene | 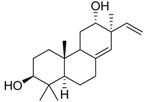 |
| Cpd. 4 | 3-epi-Betulinic acid |  |
| Cpd. 5 | 3-epi-Betulin | 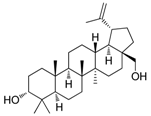 |
| Cpd. 6 | Lup-20(30)-ene-3β,29-diol | 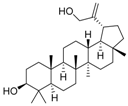 |
| Cpd. 7 | Betulin |  |
| Cpd. 8 | Lup-20(29)-ene-1,3-dione | 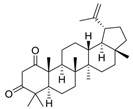 |
| Compound | Energy | Binding Residues |
|---|---|---|
| Cpd. 5 | −76.9 | Y453, Y495, G496, Q498, T500, N501, Y505 |
| Betulinic acid | −72.8 | Y495, G496, Q498, T500, N501, Y505 |
| Target | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| GAPDH | TGGGTGTGAACCATGAGAAG | GCTAAGCAGTTGGTGGTGC |
| FFLuc | ATTACACCCGAGGGGGATGA | CCAGATCCACAACCTTCGCT |
| IL1B | GGACAAGCTGAGGAAGATGC | GATTCTTTTCCTTGAGGCCC |
| IL6 | AATGAGGAGACTTGCCTGGT | GCAGGAACTGGATCAGGACT |
| TNF | CTGCACTTTGGAGTGATCGG | AGGGTTTGCTACAACATGGG |
| RT primer for (+) stranded viral RNA | AGAAGGTTTTACAAGACTCACGTT | |
| RT primer for (−) stranded viral RNA | CGAACTTATGTACTCATTCGTTTCGG | |
| SARS-CoV-2_E | ACAGGTACGTTAATAGTTAATAGCGT | ATATTGCAGCAGTACGCACACA |
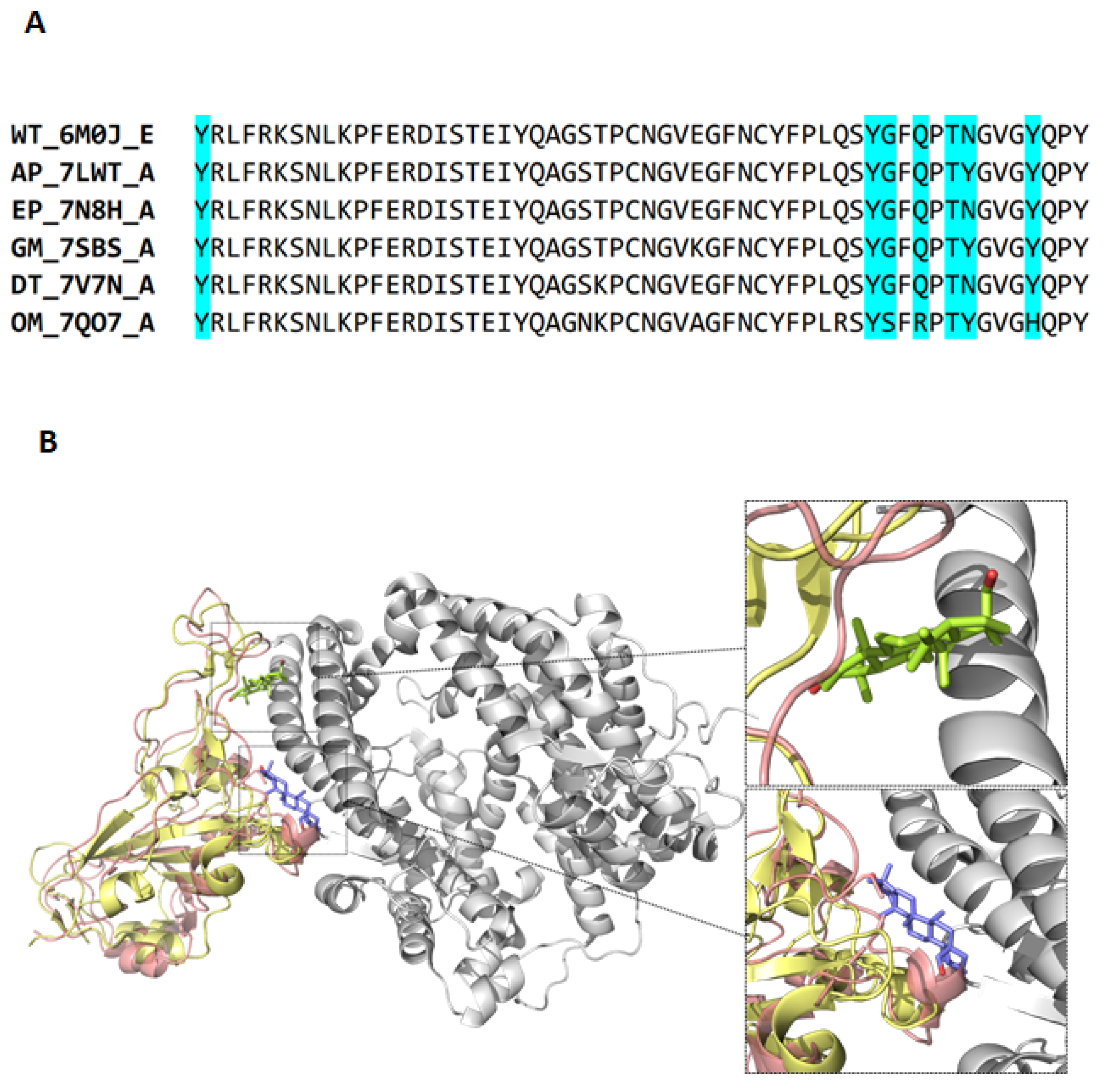
| Strain | Abbreviations | PDB ID | Chain | Reference |
|---|---|---|---|---|
| Wild-type | WT | 6M0J | E | [40] |
| Alpha (B.1.1.7) | AP | 7LWT | A | [41] |
| Epsilon (B.1.429) | EP | 7N8H | A | [42] |
| Gamma (P1) | GM | 7SBS | A | [43] |
| Delta (B.1.617.2) | DT | 7V7N | A | |
| Omicron (BA.1) | OM | 7QO7 | A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeh, Y.-J.; Chao, T.-L.; Chang, Y.-J.; Chang, S.-Y.; Lu, C.-H.; Chao, C.-H.; Su, W.-C.; Tseng, C.-P.; Lai, M.M.C.; Cheng, J.-C. Dual Effects of 3-epi-betulin from Daphniphyllum glaucescens in Suppressing SARS-CoV-2-Induced Inflammation and Inhibiting Virus Entry. Int. J. Mol. Sci. 2023, 24, 17040. https://doi.org/10.3390/ijms242317040
Yeh Y-J, Chao T-L, Chang Y-J, Chang S-Y, Lu C-H, Chao C-H, Su W-C, Tseng C-P, Lai MMC, Cheng J-C. Dual Effects of 3-epi-betulin from Daphniphyllum glaucescens in Suppressing SARS-CoV-2-Induced Inflammation and Inhibiting Virus Entry. International Journal of Molecular Sciences. 2023; 24(23):17040. https://doi.org/10.3390/ijms242317040
Chicago/Turabian StyleYeh, Yung-Ju, Tai-Ling Chao, Yu-Jen Chang, Sui-Yuan Chang, Chih-Hao Lu, Chih-Hua Chao, Wen-Chi Su, Ching-Ping Tseng, Michael M.C. Lai, and Ju-Chien Cheng. 2023. "Dual Effects of 3-epi-betulin from Daphniphyllum glaucescens in Suppressing SARS-CoV-2-Induced Inflammation and Inhibiting Virus Entry" International Journal of Molecular Sciences 24, no. 23: 17040. https://doi.org/10.3390/ijms242317040
APA StyleYeh, Y.-J., Chao, T.-L., Chang, Y.-J., Chang, S.-Y., Lu, C.-H., Chao, C.-H., Su, W.-C., Tseng, C.-P., Lai, M. M. C., & Cheng, J.-C. (2023). Dual Effects of 3-epi-betulin from Daphniphyllum glaucescens in Suppressing SARS-CoV-2-Induced Inflammation and Inhibiting Virus Entry. International Journal of Molecular Sciences, 24(23), 17040. https://doi.org/10.3390/ijms242317040






