Altered Expression of Two Small Secreted Proteins (ssp4 and ssp6) Affects the Degradation of a Natural Lignocellulosic Substrate by Pleurotus ostreatus
Abstract
:1. Introduction
2. Results
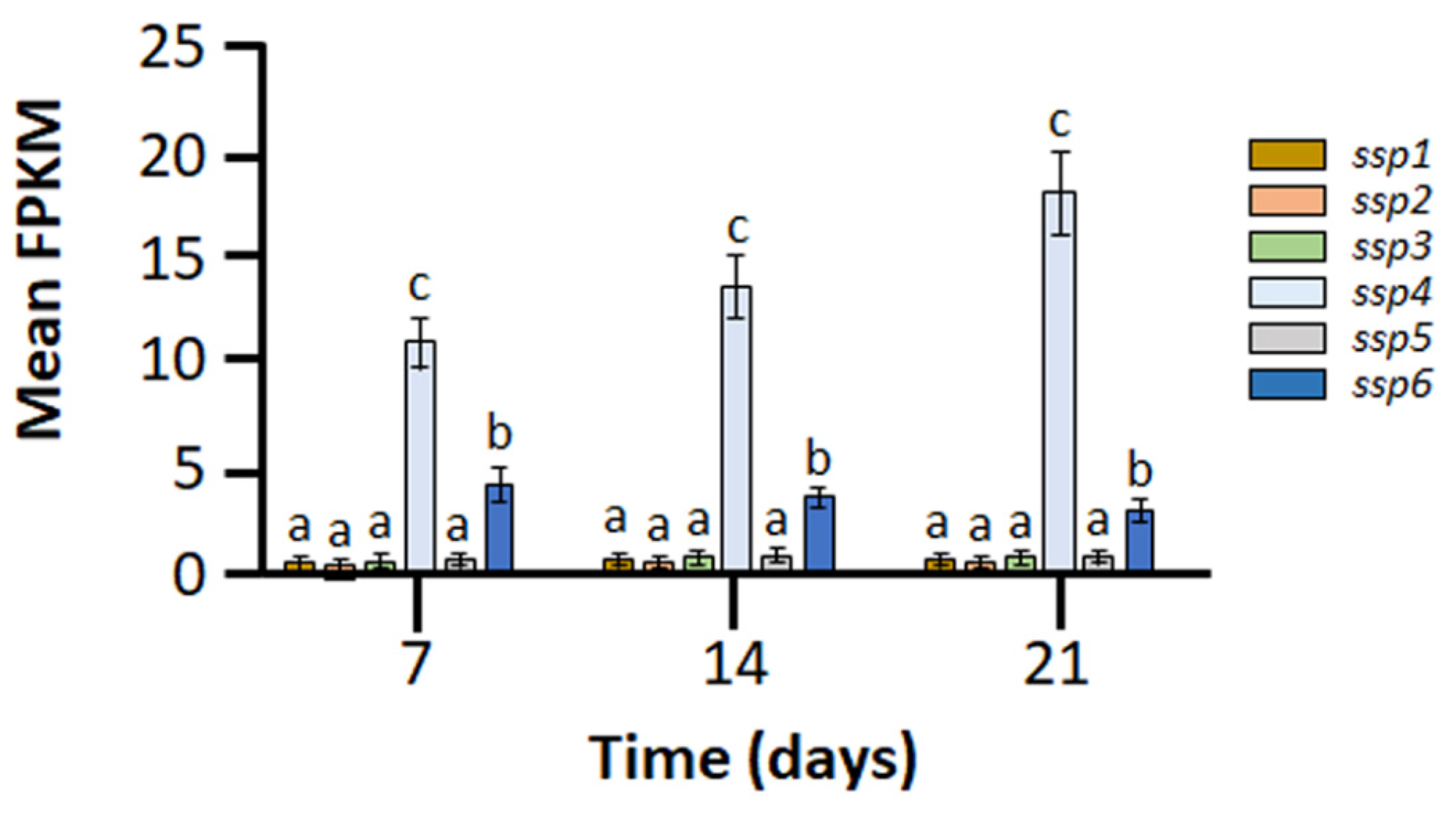
2.1. Members of the Six-Gene ssp Family Exhibit Diverse Expression Patterns during Growth of Pleurotus ostreatus on Wheat Straw
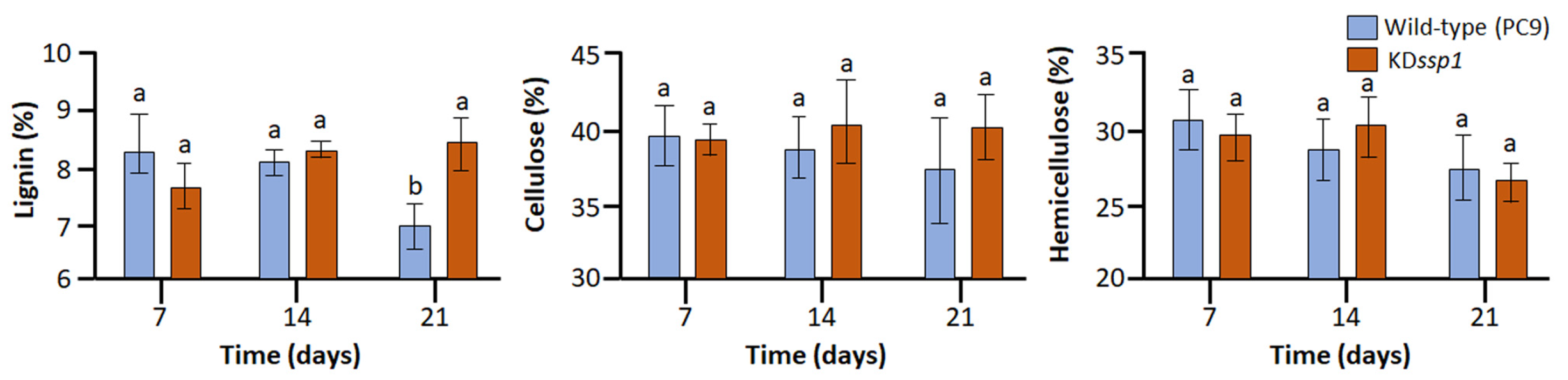
2.2. Changes in the Expression of Two ssp Genes during Growth on Wheat Straw Impose a Change in Lignin Degradation
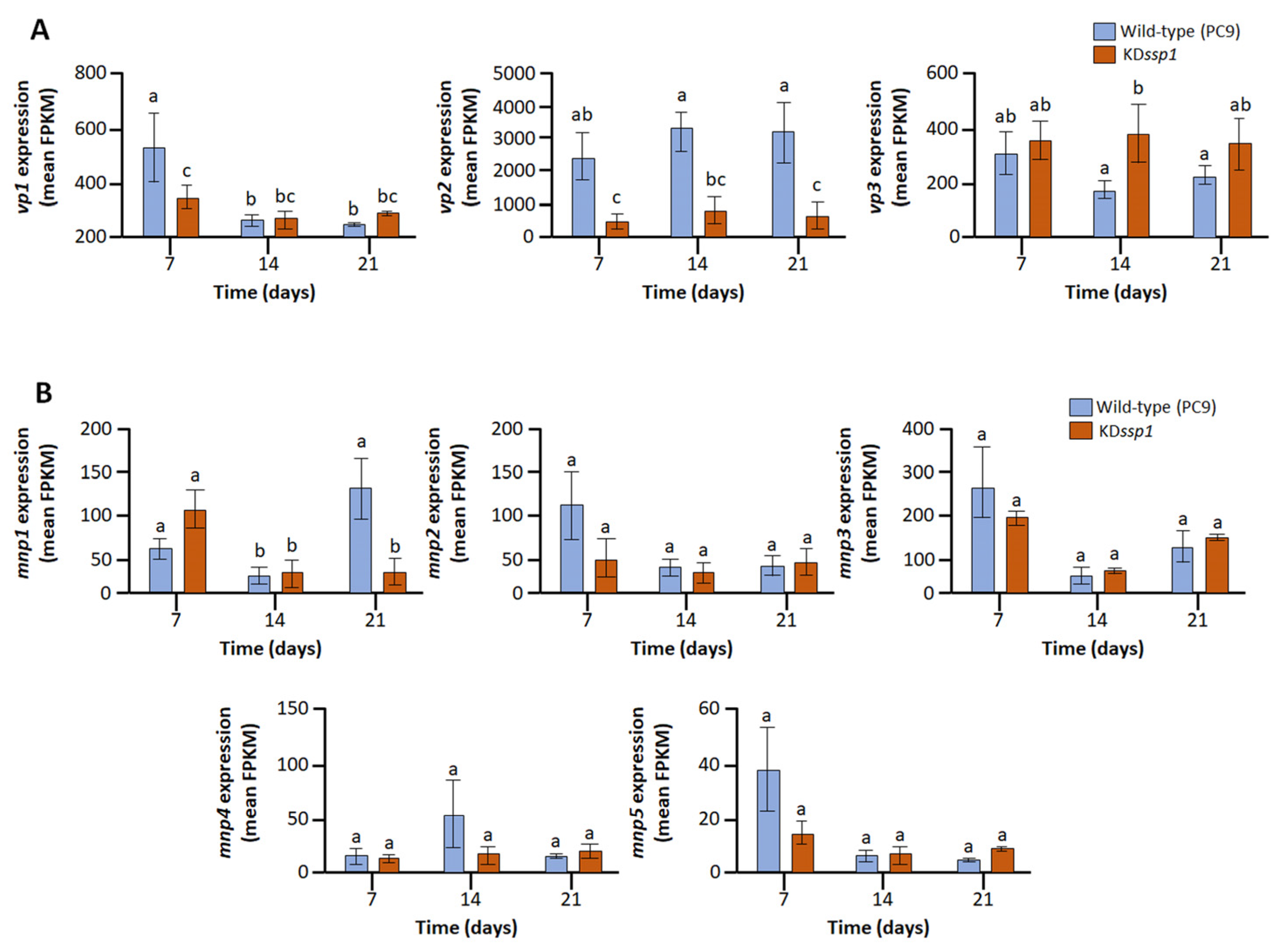
2.3. Expression of vp2 and mnp1, Components of the Lignin Degrading Machinery, Is Compromised in the KDssp1 Strain
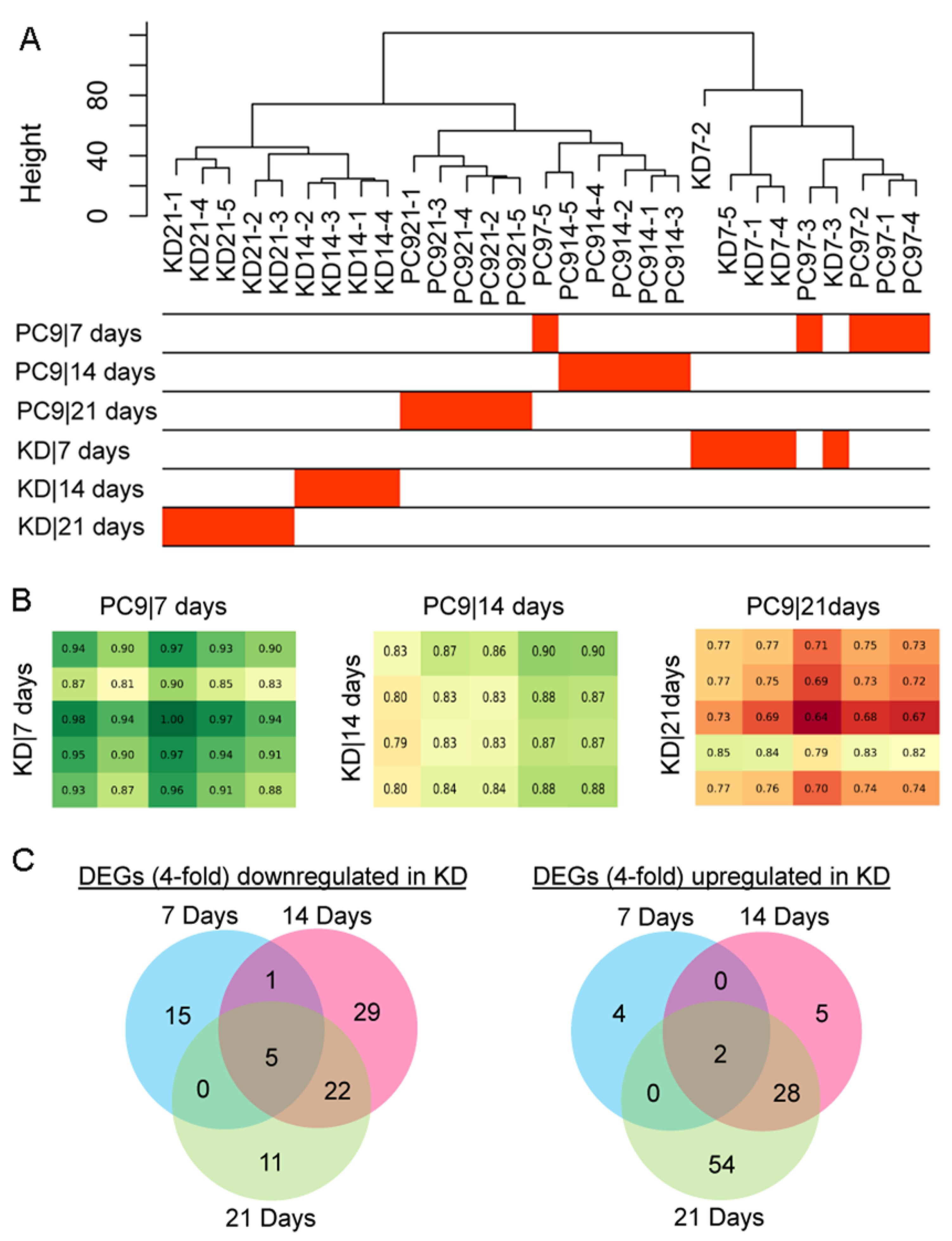
2.4. Identification of Key Differentially Expressed Genes (DEGs) Associated with the Knockdown of ssps
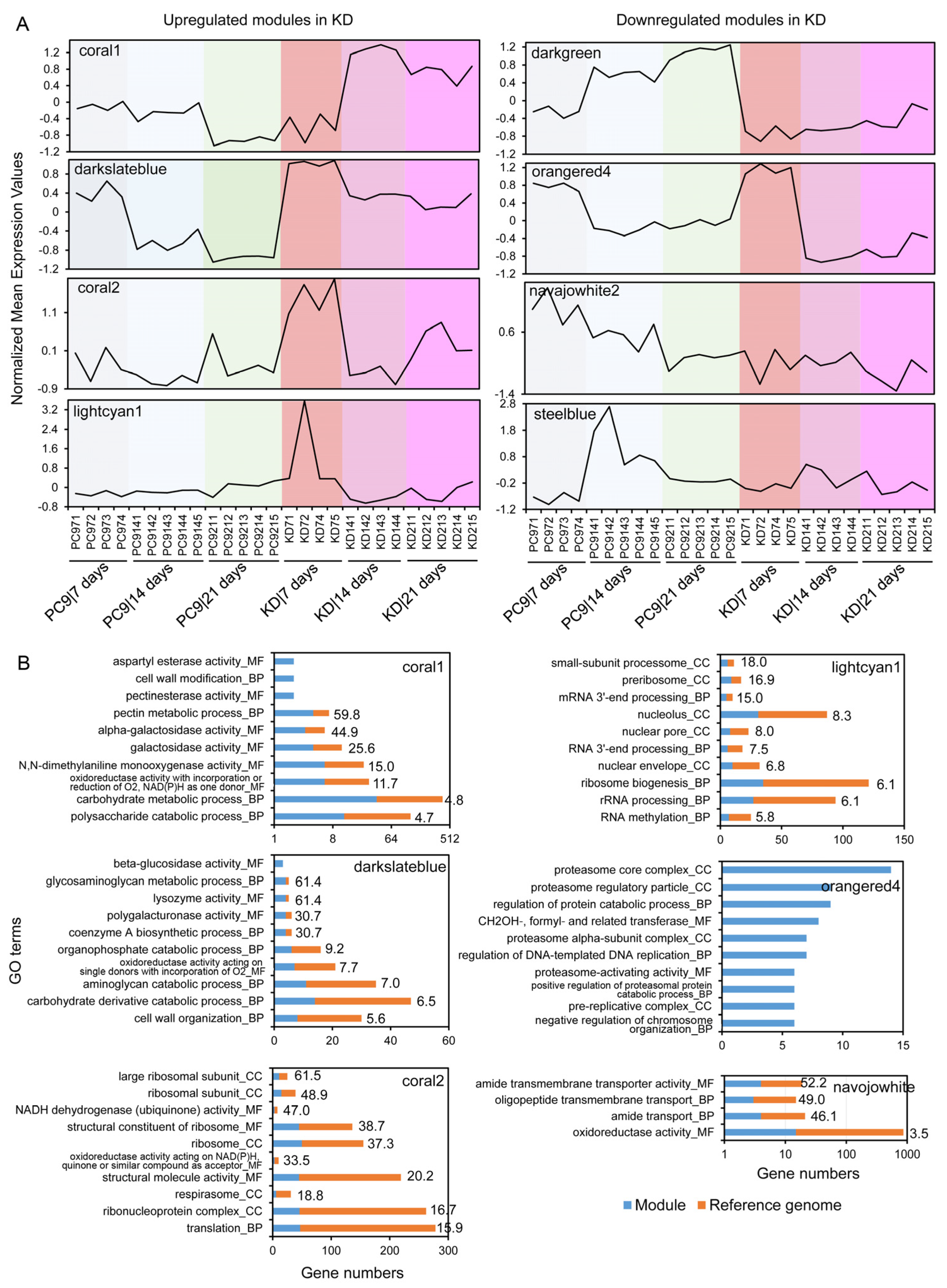
2.5. Co-Expression Network Analysis Suggests the Roles SSPs May Have in Regulating Lignocellulose Degradation and Other Fungal Processes
3. Discussion
4. Materials and Methods
4.1. Fungal Growth and Experimental Conditions
4.2. Nucleic Acid Isolation and Gene Expression Analyses
4.3. RNA-Seq Data Processing and DEGs
4.4. Co-Expression Network Analysis
4.5. Functional Analysis of the Co-Regulated Modules
4.6. Cell Wall Component Analyses
4.7. Pectin Degradation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cohen, L.; Persky, Y.; Hadar, R. Biotechnological applications and potential of wood-degrading mushrooms of the genus Pleurotus. Appl. Microbiol. Biotechnol. 2002, 58, 582–594. [Google Scholar] [CrossRef] [PubMed]
- Kijpornyongpan, T.; Schwartz, A.; Yaguchi, A.; Salvachúa, D. Systems biology-guided understanding of white-rot fungi for biotechnological applications: A review. iScience 2022, 25, 104640. [Google Scholar] [CrossRef] [PubMed]
- Blanchette, R.A. Screening wood decayed by white rot fungi for preferential lignin degradation. Appl. Environ. Microbiol. 1984, 48, 647–653. [Google Scholar] [CrossRef]
- Schilling, J.S.; Kaffenberger, J.T.; Held, B.W.; Ortiz, R.; Blanchette, R.A. Using wood rot phenotypes to illuminate the “gray” among decomposer fungi. Front. Microbiol. 2020, 11, 1288. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Fueyo, E.; Ruiz-Dueñas, F.J.; López-Lucendo, M.F.; Pérez-Boada, M.; Rencoret, J.; Gutiérrez, A.; Pisabarro, A.G.; Ramírez, L.; Martínez, A.T. A secretomic view of woody and nonwoody lignocellulose degradation by Pleurotus ostreatus. Biotechnol. Biofuels 2016, 9, 49. [Google Scholar] [CrossRef]
- Knop, D.; Yarden, O.; Hadar, Y. The ligninolytic peroxidases in the genus Pleurotus: Divergence in activities, expression, and potential applications. Appl. Microbiol. Biotechnol. 2015, 99, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Téllez-Téllez, M.; Díaz-Godínez, G. Omic tools to study enzyme production from fungi in the Pleurotus genus. BioResources 2019, 14, 2420–2457. [Google Scholar] [CrossRef]
- Honda, Y.; Matsuyama, T.; Irie, T.; Watanabe, T.; Kuwahara, M. Carboxin resistance transformation of the homobasidiomycete fungus Pleurotus ostreatus. Curr. Genet. 2000, 37, 209–212. [Google Scholar] [CrossRef]
- Salame, T.M.; Knop, D.; Tal, D.; Levinson, D.; Yarden, O.; Hadar, Y. Predominance of a versatile-peroxidase-encoding gene, mnp4, as demonstrated by gene replacement via a gene targeting system for Pleurotus ostreatus. Appl. Environ. Microbiol. 2012, 78, 5341–5352. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Nakazawa, T.; Zhang, Y.; Oh, M.; Bao, D.; Kawauchi, M.; Sakamoto, M.; Honda, Y. Introducing multiple-gene mutations in Pleurotus ostreatus using a polycistronic tRNA and CRISPR guide RNA strategy. FEMS Microbiol. Lett. 2022, 369, fnac102. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-T.; Jeon, J.; Choi, J.; Cheong, K.; Song, H.; Choi, G.; Kang, S.; Lee, Y.-H. Kingdom-wide analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Front. Plant Sci. 2016, 7, 186. [Google Scholar] [CrossRef]
- Pellegrin, C.; Morin, E.; Martin, F.M.; Veneault-Fourrey, C. Comparative analysis of secretomes from ectomycorrhizal fungi with an emphasis on small-secreted proteins. Front. Microbiol. 2015, 6, 1278. [Google Scholar] [CrossRef] [PubMed]
- Gaderer, R.; Bonazza, K.; Seidl-Seiboth, V. Cerato-platanins: A fungal protein family with intriguing properties and application potential. Appl. Microbiol. Biotechnol. 2014, 98, 4795–4803. [Google Scholar] [CrossRef] [PubMed]
- Selin, C.; de Kievit, T.R.; Belmonte, M.F.; Fernando, W.G.D. Elucidating the role of effectors in plant-fungal interactions: Progress and challenges. Front. Microbiol. 2016, 7, 600. [Google Scholar] [CrossRef] [PubMed]
- Wösten, H.A. Hydrophobins: Multipurpose proteins. Annu. Rev. Microbiol. 2001, 55, 625–646. [Google Scholar] [CrossRef] [PubMed]
- Feldman, D.; Yarden, O.; Hadar, Y. Seeking the roles for fungal small-secreted proteins in affecting saprophytic lifestyles. Front. Microbiol. 2020, 11, 455. [Google Scholar] [CrossRef] [PubMed]
- Feldman, D.; Kowbel, D.J.; Glass, N.L.; Yarden, O.; Hadar, Y. A role for small secreted proteins (SSPs) in a saprophytic fungal lifestyle: Ligninolytic enzyme regulation in Pleurotus ostreatus. Sci. Rep. 2017, 7, 14553. [Google Scholar] [CrossRef]
- Alfaro, M.; Castanera, R.; Lavín, J.L.; Grigoriev, I.V.; Oguiza, J.A.; Ramírez, L.; Pisabarro, A.G. Comparative and transcriptional analysis of the predicted secretome in the lignocellulose-degrading basidiomycete fungus Pleurotus ostreatus. Environ. Microbiol. 2016, 18, 4710–4726. [Google Scholar] [CrossRef]
- Wu, H.; Nakazawa, T.; Xu, H.; Yang, R.; Bao, D.; Kawauchi, M.; Sakamoto, M.; Honda, Y. Comparative Transcriptional analyses of Pleurotus ostreatus mutants on beech wood and rice straw shed light on substrate-biased gene regulation. Appl. Microbiol. Biotechnol. 2021, 105, 1175–1190. [Google Scholar] [CrossRef]
- Nakazawa, T.; Morimoto, R.; Wu, H.; Kodera, R.; Sakamoto, M.; Honda, Y. Dominant effects of gat1 mutations on the ligninolytic activity of the white-rot fngus Pleurotus ostreatus. Fungal Biol. 2019, 123, 209–217. [Google Scholar] [CrossRef]
- Feldman, D.; Amedi, N.; Carmeli, S.; Yarden, O.; Hadar, Y. Manipulating the expression of small secreted protein 1 (Ssp1) alters patterns of development and metabolism in the white-rot fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 2019, 85, e00761-19. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, S.; Kun, R.S.; Peng, M.; Gruben, B.S.; Benoit Gelber, I.; Mäkelä, M.; de Vries, R.P. The cultivation method affects the transcriptomic response of Aspergillus niger to growth on sugar beet pulp. Microbiol. Spectr. 2021, 9, e01064-21. [Google Scholar] [CrossRef]
- Liu, J.; Yang, J.; Wang, R.; Liu, L.; Zhang, Y.; Bao, H.; Jang, J.M.; Wang, E.; Yuan, H. Comparative characterization of extracellular enzymes secreted by Phanerochaete chrysosporium during solid-state and submerged fermentation. Int. J. Biol. Macromol. 2020, 152, 288–294. [Google Scholar] [CrossRef]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Peng, Y.; Zhang, H.; Wang, K.; Zhao, C.; Zhu, G.; Reddy Palli, S.; Han, Z. Off-target effects of RNAi correlate with the mismatch rate between dsRNA and non-target mRNA. RNA Biol. 2021, 18, 1747–1759. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Mandal, M.; Chakraborti, T.; Mandal, A.; Chakraborti, S. Structure and evolutionary aspects of matrix metalloproteinases: A brief overview. Mol. Cell. Biochem. 2003, 253, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Tolosano, E.; Altruda, F. Hemopexin: Structure, function, and regulation. DNA Cell Biol. 2002, 21, 297–306. [Google Scholar] [CrossRef]
- Watkins, J.L.; Li, M.; McQuinn, R.P.; Chan, K.X.; McFarlane, H.E.; Ermakova, M.; Furbank, R.T.; Mares, D.; Dong, C.; Chalmers, K.J.; et al. A GDSL esterase/lipase catalyzes the esterification of lutein in bread wheat. Plant Cell 2019, 31, 3092–3112. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Staszczak, M. The role of the ubiquitin–proteasome system in the response of the ligninolytic fungus Trametes versicolor to nitrogen deprivation. Fungal Genet. Biol. 2008, 45, 328–337. [Google Scholar] [CrossRef]
- Staszczak, M. Fungal secondary metabolites as inhibitors of the ubiquitin–proteasome system. Int. J. Mol. Sci. 2021, 22, 13309. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Nakazawa, T.; Takenaka, A.; Kodera, R.; Morimoto, R.; Sakamoto, M.; Honda, Y. Transcriptional shifts in delignification-defective mutants of the white-rot fungus Pleurotus ostreatus. FEBS Lett. 2020, 594, 3182–3199. [Google Scholar] [CrossRef]
- Yoav, S.; Salame, T.M.; Feldman, D.; Levinson, D.; Ioelovich, M.; Morag, E.; Yarden, O.; Bayer, E.A.; Hadar, Y. Effects of cre1 modification in the white-rot fungus Pleurotus ostreatus PC9: Altering substrate preference during biological pretreatment. Biotechnol. Biofuels 2018, 11, 212. [Google Scholar] [CrossRef]
- Arroyo-Velez, N.; González-Fuente, M.; Peeters, N.; Lauber, E.; Noël, L.D. From effectors to effectomes: Are functional studies of individual effectors enough to decipher plant pathogen infectious strategies? PLoS Pathog. 2020, 16, e1009059. [Google Scholar] [CrossRef]
- Fernández-Fueyo, E.; Castanera, R.; Ruiz-Dueñas, F.J.; López-Lucendo, M.F.; Ramírez, L.; Pisabarro, A.G.; Martínez, A.T. Ligninolytic peroxidase gene expression by Pleurotus ostreatus: Differential regulation in lignocellulose medium and effect of temperature and PH. Fungal Genet. Biol. 2014, 72, 150–161. [Google Scholar] [CrossRef]
- Salame, T.M.; Knop, D.; Levinson, D.; Mabjeesh, S.J.; Yarden, O.; Hadar, Y. Inactivation of a Pleurotus ostreatus versatile peroxidase-encoding gene (mnp2) results in reduced lignin degradation. Environ. Microbiol. 2014, 16, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Salame, T.M.; Knop, D.; Levinson, D.; Yarden, O.; Hadar, Y. Redundancy among manganese peroxidases in Pleurotus ostreatus. Appl. Environ. Microbiol. 2013, 79, 2405–2415. [Google Scholar] [CrossRef]
- Nakazawa, T.; Yamaguchi, I.; Zhang, Y.; Saka, C.; Wu, H.; Kayama, K.; Kawauchi, M.; Sakamoto, M.; Honda, Y. Experimental evidence that lignin-modifying enzymes are essential for degrading plant cell wall lignin by Pleurotus ostreatus using CRISPR/Cas9. Environ. Microbiol. 2023, 25, 1909–1924. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.; Mikelj, M.; Papler, T.; Leonardi, A.; Križaj, I.; Maček, P. Ostreopexin: A hemopexin fold protein from the oyster mushroom, Pleurotus ostreatus. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 1468–1473. [Google Scholar] [CrossRef]
- Hrkal, Z.; Vodrazka, Z.; Kalousek, I. Transfer of heme from ferrihemoglobin and ferrihemoglobin isolated chains to hemopexin. Eur. J. Biochem. 1974, 43, 73–78. [Google Scholar] [CrossRef]
- Lechuga, G.C.; Napoleão-Pêgo, P.; Morel, C.M.; Provance, D.W.; De-Simone, S.G. New insights into hemopexin-binding to hemin and hemoglobin. Int. J. Mol. Sci. 2022, 23, 3789. [Google Scholar] [CrossRef] [PubMed]
- Hamza, R.; Roque, E.; Gómez-Mena, C.; Madueño, F.; Beltrán, J.P.; Cañas, L.A. PsEND1 Is a key player in pea pollen development through the modulation of redox homeostasis. Front. Plant Sci. 2021, 12, 765277. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, S.; Võsa, U.; van der Graaf, A.; Franke, L.; de Magalhães, J.P. Gene co-expression analysis for functional classification and gene–disease predictions. Brief. Bioinform. 2017, 19, bbw139. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Tan, Z.; Wu, X.; Liang, X.; Liu, Y.; Xie, Y.; Li, X.; Xiao, C.; Gao, X.; Chen, S.; et al. Comparative transcriptome analysis of genes and metabolic pathways involved in sporulation in Ganoderma lingzhi. G3 Genes|Genomes|Genet. 2022, 12, jkab448. [Google Scholar] [CrossRef]
- Krizsán, K.; Almási, É.; Merényi, Z.; Sahu, N.; Virágh, M.; Kószó, T.; Mondo, S.; Kiss, B.; Bálint, B.; Kües, U.; et al. Transcriptomic atlas of mushroom development reveals conserved genes behind complex multicellularity in fungi. Proc. Natl. Acad. Sci. USA 2019, 116, 7409–7418. [Google Scholar] [CrossRef] [PubMed]
- Larraya, L.M.; Pérez, G.; Peñas, M.M.; Baars, J.J.P.; Mikosch, T.S.P.; Pisabarro, A.G.; Ramírez, L. Molecular karyotype of the white rot fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 1999, 65, 3413–3417. [Google Scholar] [CrossRef]
- Salame, T.M.; Ziv, C.; Hadar, Y.; Yarden, O. RNAi as a potential tool for biotechnological applications in fungi. Appl. Microbiol. Biotechnol. 2011, 89, 501–512. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Ramírez, F.; Dündar, F.; Diehl, S.; Grüning, B.A.; Manke, T. DeepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014, 42, W187–W191. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Presley, G.N.; Hammel, K.E.; Ryu, J.-S.; Menke, J.R.; Figueroa, M.; Hu, D.; Orr, G.; Schilling, J.S. Localizing gene regulation reveals a staggered wood decay mechanism for the brown rot fungus Postia placenta. Proc. Natl. Acad. Sci. USA 2016, 113, 10968–10973. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Haile, S.; Masi, C.; Tafesse, M. Isolation and characterization of pectinase-producing bacteria (Serratia marcescens) from avocado peel waste for juice clarification. BMC Microbiol. 2022, 22, 145. [Google Scholar] [CrossRef] [PubMed]
- Beg, Q.K.; Bhushan, B.; Kapoor, M.; Hoondal, G.S. Production and characterization of thermostable xylanase and pectinase from Streptomyces sp. QG-11-3. J. Ind. Microbiol. Biotechnol. 2000, 24, 396–402. [Google Scholar] [CrossRef]

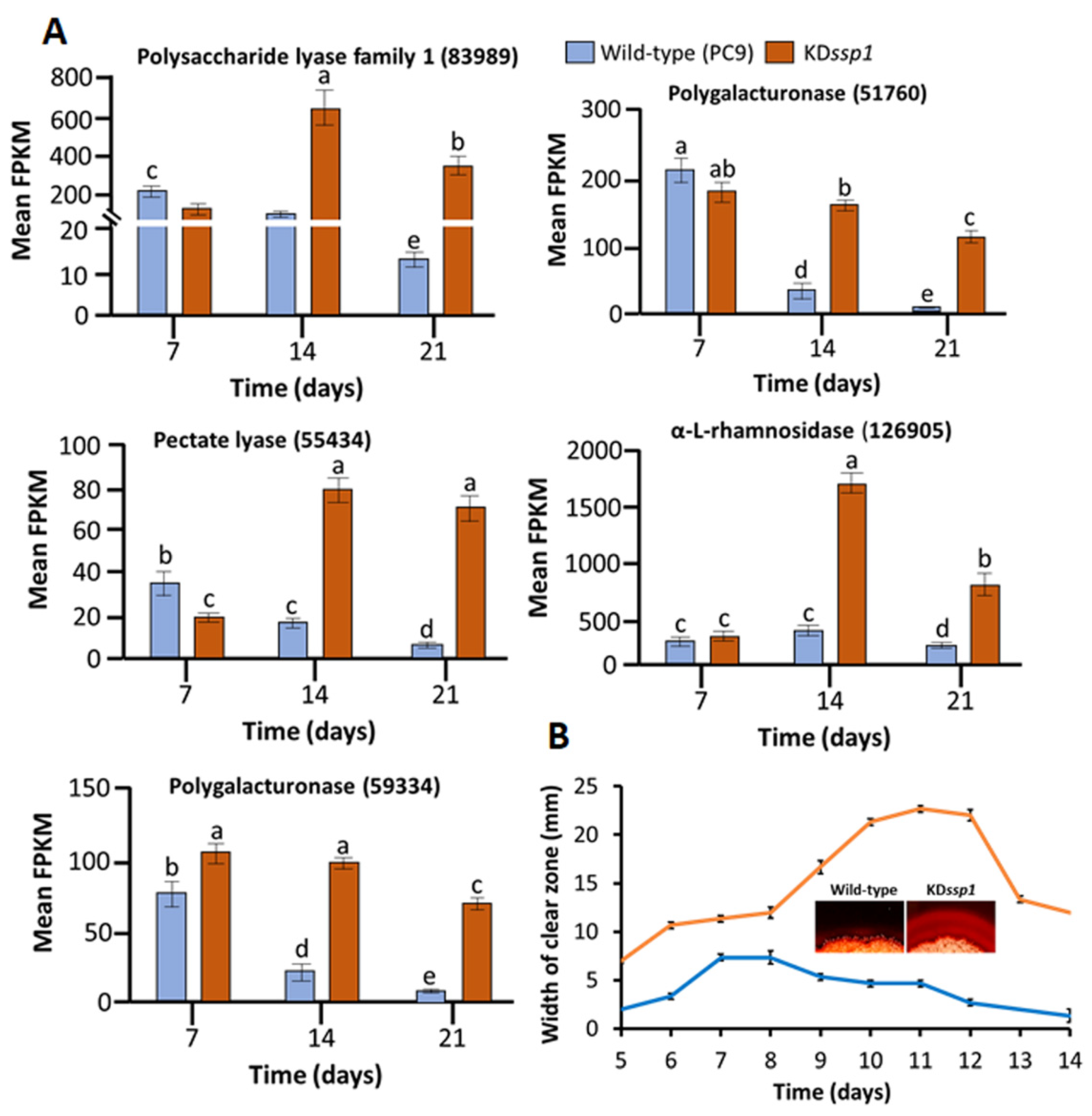

| Protein IDs a | Weighted Gene Co-Expression Network Analysis (WGCNA) Modules b | Biological Description c | SP d | Fold Change of DEGs e | ||
|---|---|---|---|---|---|---|
| 7 d | 14 d | 21 d | ||||
| Genes downregulated in KD mutants | ||||||
| 133592 | darkgreen | Hemopexin-like protein | N | 1.14 | 492.00 | 46.47 |
| 86351 | darkgreen | Hypothetical self-proteolytic membrane RHS protein | N | 1.16 | 14.00 | 22.30 |
| 117678 | darkgreen | Troponin-like | N | 1.01 | 11.43 | 20.20 |
| 108294 | darkgreen | 3′-5′ Exonuclease | N | 1.38 | 10.09 | 15.92 |
| 122257 | darkgreen | Unknown protein | N | 2.45 | 11.17 | 10.01 |
| 89221 | darkgreen | Membrane attack complex component/perforin domain | N | 5.25 | 7.88 | 8.16 |
| 113854 | darkgreen | Unknown protein | N | 16.64 | 13.98 | 8.14 |
| 116044 | darkgreen | O-methyltransferase | N | 3.73 e | 18.79 | 7.92 |
| 87569 | darkgreen | Unknown protein | N | 1.07 f | 7.37 | 7.74 |
| 72169 | darkgreen | Unknown membrane protein | Y | 15.75 | 14.07 | 7.27 |
| 123506 | orangered4 | Unknown protein | N | 0.48 f | 4.20 | 7.16 |
| 117582 | darkgreen | Voltage-gated shaker-like K+ channel, subunit beta/KCNAB | N | 8.02 | 15.53 | 7.11 |
| 122258 | darkgreen | Hypothetical helicase | N | 1.87 | 4.86 | 6.80 |
| 101257 | darkgreen | Aldo/keto reductase | N | 2.70 f | 14.77 | 6.33 |
| 87540 | orangered4 | Unknown protein | N | 2.99 f | 16.94 | 6.15 |
| 88910 | darkgreen | GMC oxidoreductase | N | 1.16 f | 4.48 | 5.91 |
| 39792 | darkgreen | Hypothetical velvet factor | N | 1.24 | 5.26 | 5.71 |
| 60432 | darkgreen | Versatile peroxidase; AA2 | Y | 8.82 | 4.58 | 5.66 |
| 79338 | darkgreen | Nitronate monooxygenase | Y | 1.20 | 5.00 | 5.58 |
| 130698 | darkgreen | Unknown membrane protein | N | 3.92 | 4.82 | 5.39 |
| 81199 | darkgreen | Glucose/ribitol dehydrogenase | Y | 5.73 f | 6.65 | 5.06 |
| 46715 | darkgreen | SKP1/BTB/POZ domain containing | N | 2.93 | 5.59 | 4.98 |
| 126430 | darkgreen | Hypothetical isomerase | N | 3.37 | 6.20 | 4.76 |
| 113642 | darkgreen | Hypothetical zinc finger protein | Y | 1.07 | 5.64 | 4.71 |
| 117317 | darkgreen | Unknown protein | Y | 2.70 | 4.87 | 4.56 |
| 99670 | darkgreen | Galactose oxidase-like, AA5 | Y | 0.82 f | 5.36 | 4.49 |
| 87426 | darkgreen | Asl1-like glycosyl hydrolase | Y | 2.20 | 6.53 | 4.30 |
| Genes upregulated in KD mutants | ||||||
| 89942 | coral1 | alpha-galactosidase, GH27 | Y | 0.74 f | 11.96 | 32.01 |
| 117545 | darkslateblue | Lysozyme, GH25 | Y | 1.98 | 5.93 | 28.01 |
| 83989 | coral1 | Pectin lyase | Y | 0.55 f | 8.57 | 24.72 |
| 44086 | coral1 | Hypothetical cerato-platanin protein | Y | 0.47 f | 9.57 | 19.44 |
| 96680 | darkslateblue | Lysozyme, GH24 | Y | 1.28 f | 5.36 | 12.85 |
| 132186 | black | Hypothetical cerato-platanin protein | N | 2.17 f | 6.01 | 12.68 |
| 110536 | coral1 | Hypothetical short-chain dehydrogenase | Y | 1.22 f | 4.03 | 12.41 |
| 104355 | darkslateblue | Proline-rich protein | Y | 4.16 | 10.74 | 12.03 |
| 97623 | coral1 | alpha-N-arabinofuranosidase, GH43 | Y | 0.57 f | 5.53 | 11.86 |
| 51760 | darkslateblue | Endo-polygalacturonase, GH28 | Y | 0.83 f | 4.33 | 11.84 |
| 87572 | darkslateblue | Hypothetical cupredoxin protein | Y | 1.34 f | 11.25 | 11.60 |
| 56468 | darkslateblue | Sugar transporter | Y | 1.87 | 7.31 | 11.02 |
| 55434 | coral1 | Pectate lyase | Y | 0.54 f | 4.67 | 10.74 |
| 87101 | darkslateblue | Cytochrome P450 CYP2 subfamily | Y | 1.82 | 11.82 | 10.52 |
| 59334 | darkslateblue | Pectin lyase, GH28 | Y | 1.34 f | 5.55 | 10.07 |
| 99620 | coral1 | Protein-tyrosine-phosphatase | N | 0.73 f | 6.09 | 10.01 |
| 98979 | darkslateblue | Unknown protein | N | 2.26 | 4.55 | 8.25 |
| 31260 | darkslateblue | Hypothetical membrane protein | Y | 2.98 | 6.68 | 6.81 |
| 126905 | coral1 | alpha-L-rhamnosidase, GH37 | N | 1.08 f | 6.12 | 6.74 |
| 88830 | black | Hypothetical membrane protein | Y | 6.99 | 10.23 | 6.60 |
| 65815 | black | Carbohydrate-Binding Module Family 13 | Y | 1.48 | 5.12 | 6.58 |
| 61773 | coral1 | Lipase_GDSL, CE16 | Y | 1.14 | 4.37 | 6.56 |
| 116339 | darkslateblue | Carotenoid ester lipase precursor | Y | 2.49 | 4.41 | 5.46 |
| 126566 | darkslateblue | Carotenoid ester lipase precursor | N | 1.96 | 6.52 | 5.46 |
| 91336 | darkslateblue | Unknown protein | N | 3.35 | 5.08 | 4.89 |
| 62766 | darkslateblue | GH32 | Y | 1.87 | 4.24 | 4.86 |
| 95957 | darkslateblue | Hypothetial dienelactone hydrolase | N | 1.38 | 4.58 | 4.83 |
| 117665 | black | Unknown protein | Y | 1.25 | 5.90 | 4.76 |
| 131254 | darkslateblue | Chromo (CHRromatin Organisation MOdifier) domain | N | 2.86 | 4.67 | 4.58 |
| 114119 | darkslateblue | Nitronate monooxygenase | N | 2.39 | 4.47 | 4.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yarden, O.; Zhang, J.; Marcus, D.; Changwal, C.; Mabjeesh, S.J.; Lipzen, A.; Zhang, Y.; Savage, E.; Ng, V.; Grigoriev, I.V.; et al. Altered Expression of Two Small Secreted Proteins (ssp4 and ssp6) Affects the Degradation of a Natural Lignocellulosic Substrate by Pleurotus ostreatus. Int. J. Mol. Sci. 2023, 24, 16828. https://doi.org/10.3390/ijms242316828
Yarden O, Zhang J, Marcus D, Changwal C, Mabjeesh SJ, Lipzen A, Zhang Y, Savage E, Ng V, Grigoriev IV, et al. Altered Expression of Two Small Secreted Proteins (ssp4 and ssp6) Affects the Degradation of a Natural Lignocellulosic Substrate by Pleurotus ostreatus. International Journal of Molecular Sciences. 2023; 24(23):16828. https://doi.org/10.3390/ijms242316828
Chicago/Turabian StyleYarden, Oded, Jiwei Zhang, Dor Marcus, Chunoti Changwal, Sameer J. Mabjeesh, Anna Lipzen, Yu Zhang, Emily Savage, Vivian Ng, Igor V. Grigoriev, and et al. 2023. "Altered Expression of Two Small Secreted Proteins (ssp4 and ssp6) Affects the Degradation of a Natural Lignocellulosic Substrate by Pleurotus ostreatus" International Journal of Molecular Sciences 24, no. 23: 16828. https://doi.org/10.3390/ijms242316828
APA StyleYarden, O., Zhang, J., Marcus, D., Changwal, C., Mabjeesh, S. J., Lipzen, A., Zhang, Y., Savage, E., Ng, V., Grigoriev, I. V., & Hadar, Y. (2023). Altered Expression of Two Small Secreted Proteins (ssp4 and ssp6) Affects the Degradation of a Natural Lignocellulosic Substrate by Pleurotus ostreatus. International Journal of Molecular Sciences, 24(23), 16828. https://doi.org/10.3390/ijms242316828











