Antifungal Effect of Bacillus velezensis ZN-S10 against Plant Pathogen Colletotrichum changpingense and Its Inhibition Mechanism
Abstract
:1. Introduction
2. Results
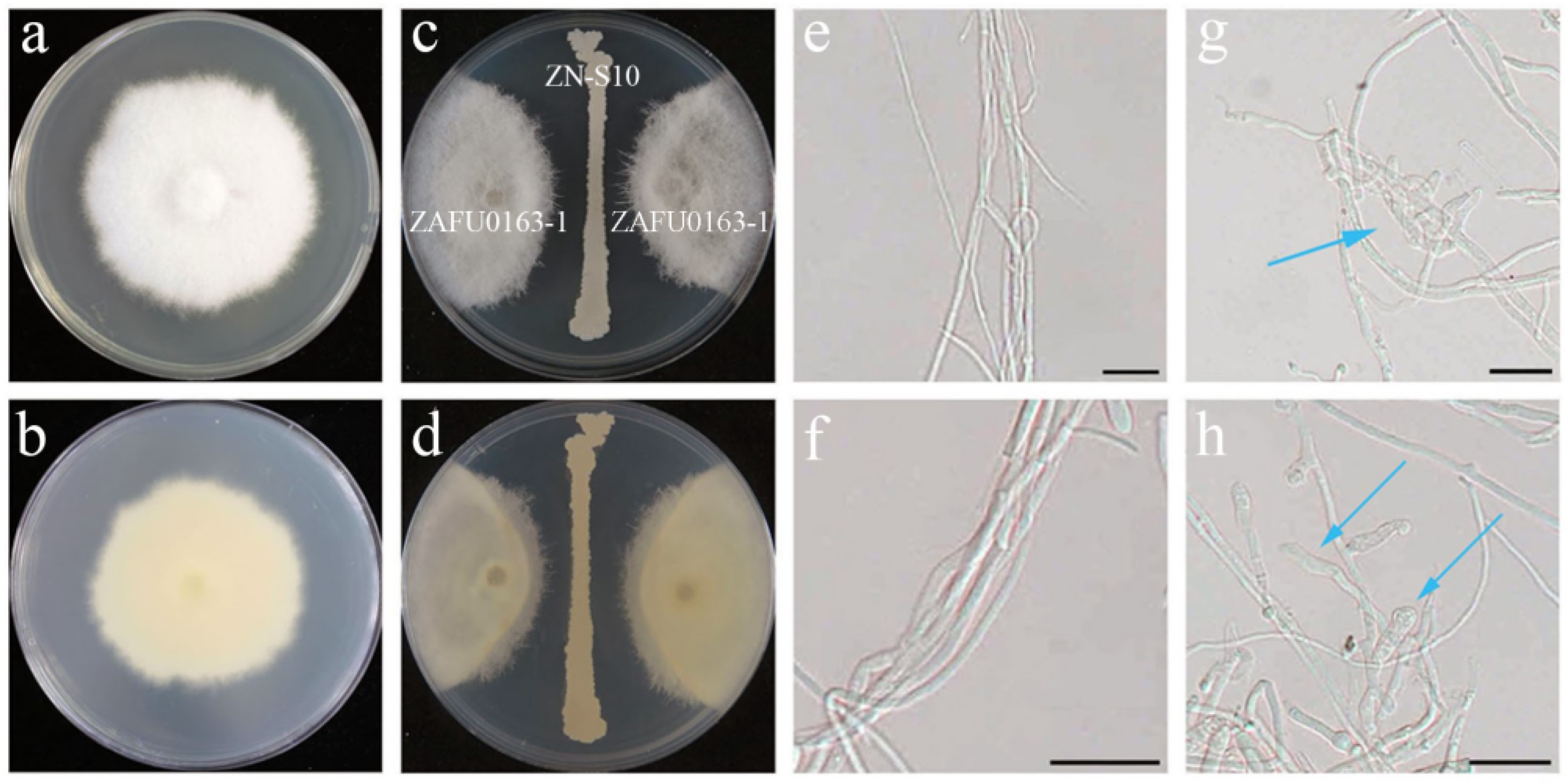
2.1. Antagonistic Activity of B. velezensis ZN-S10 against Phytopathogenic Fungi
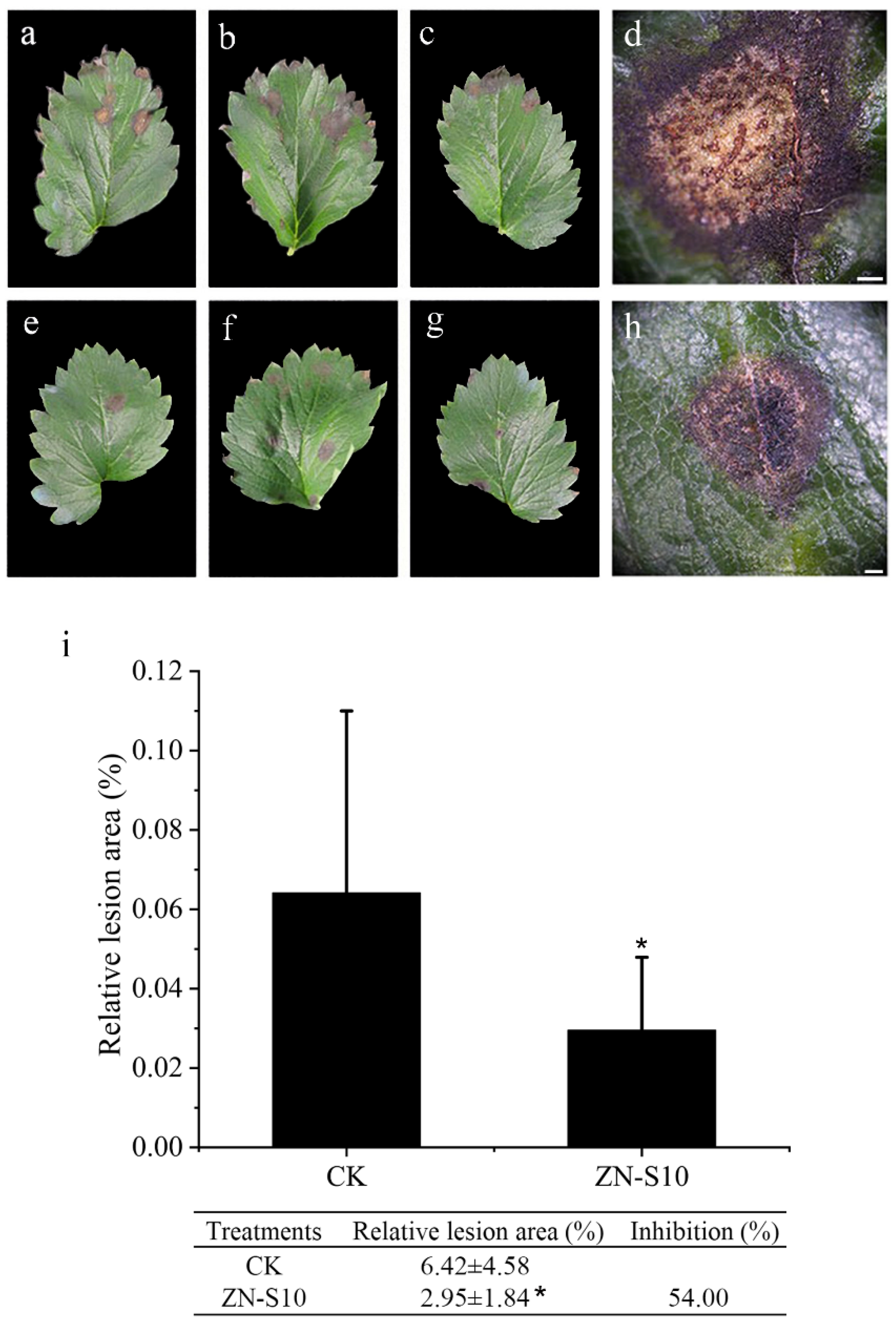
2.2. Detached Leaf Inoculation Test
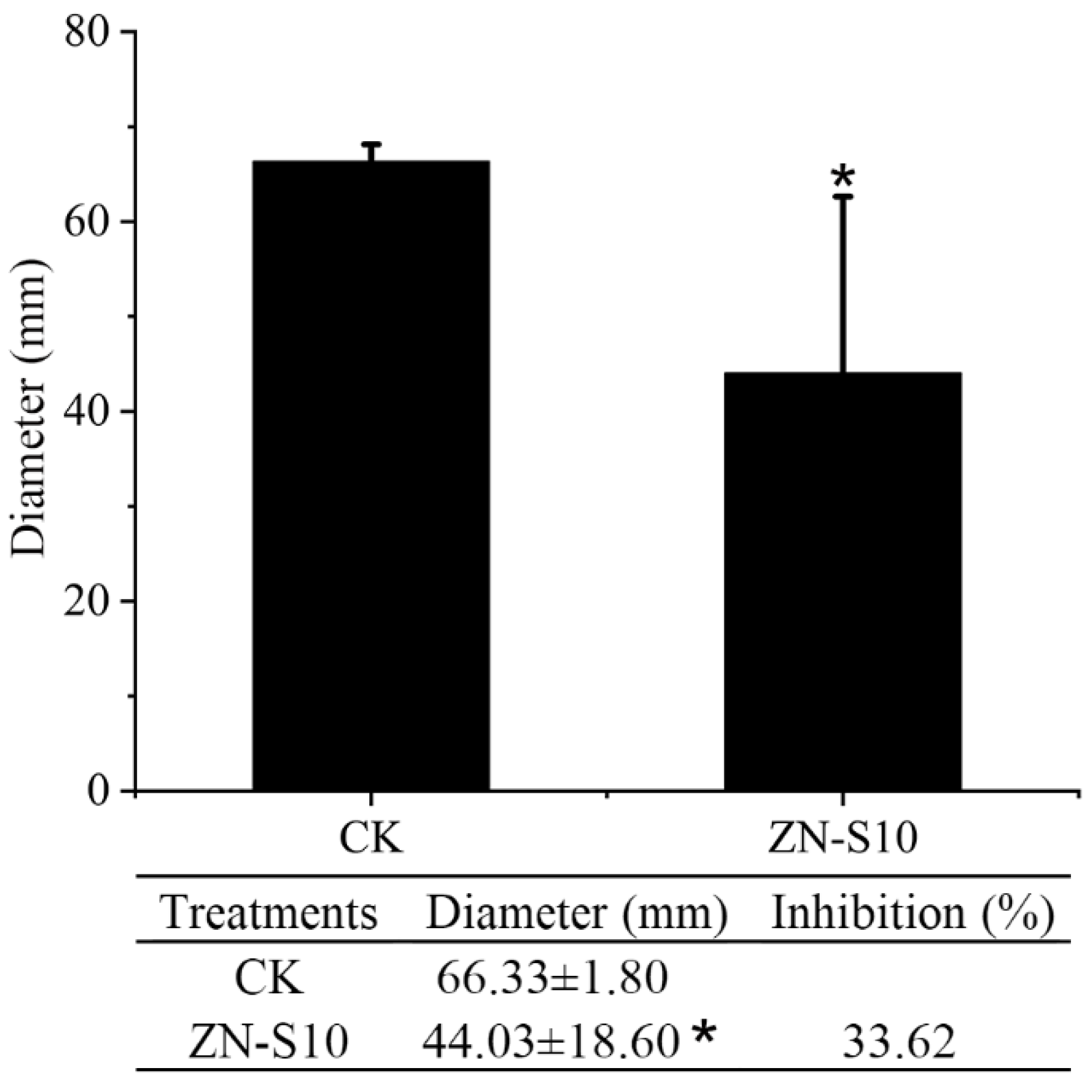
2.3. Inhibitory Effect of Volatile Organic Compounds (VOCs) Released by B. velezensis ZN-S10 on the Growth of C. changpingense ZAFU0163-1
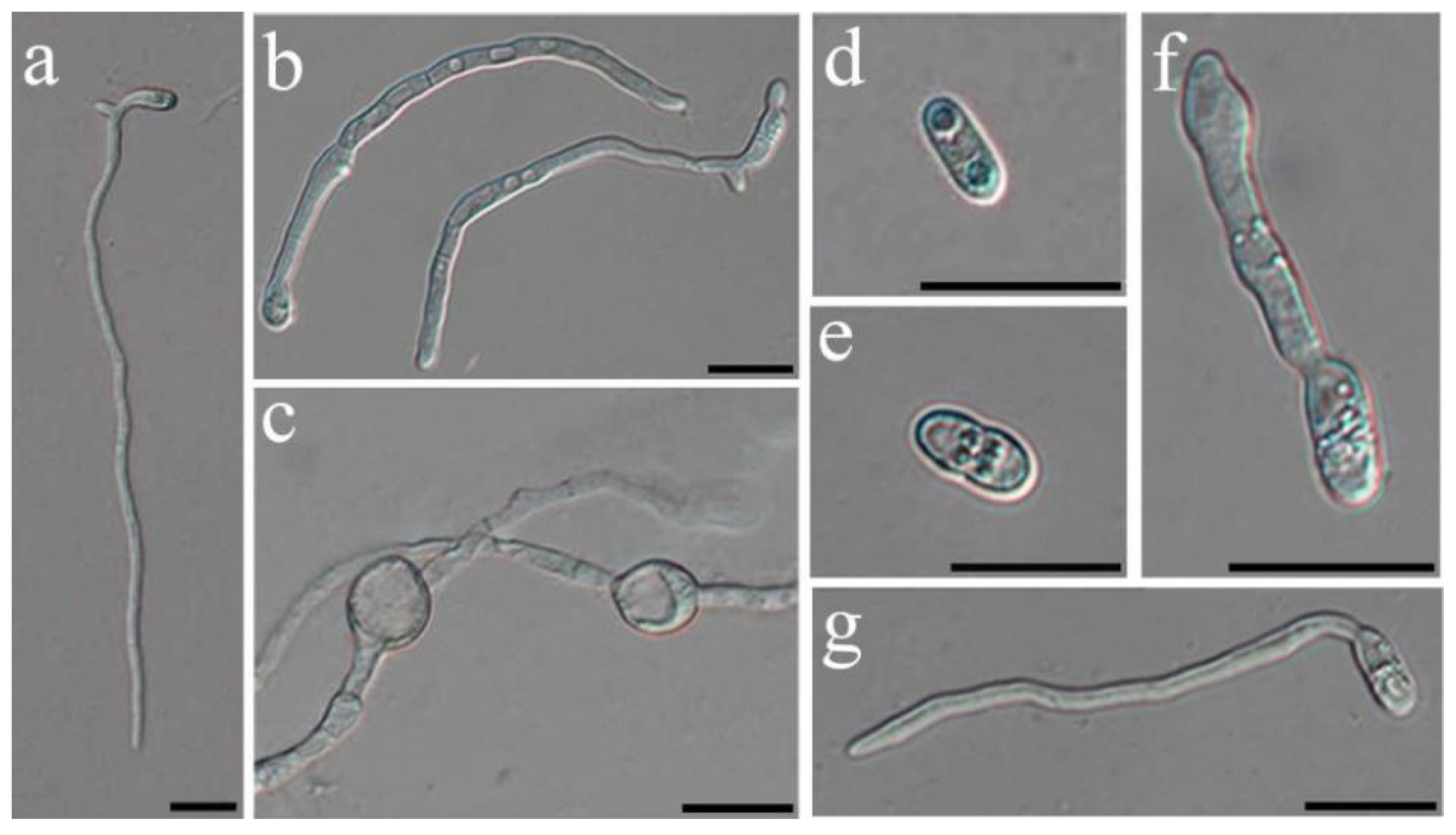
2.4. Nonvolatile Compound Antifungal Test on Hyphal and Conidial Germination
2.5. Metabolic Profiles Analyzed Using Untargeted Metabolomics
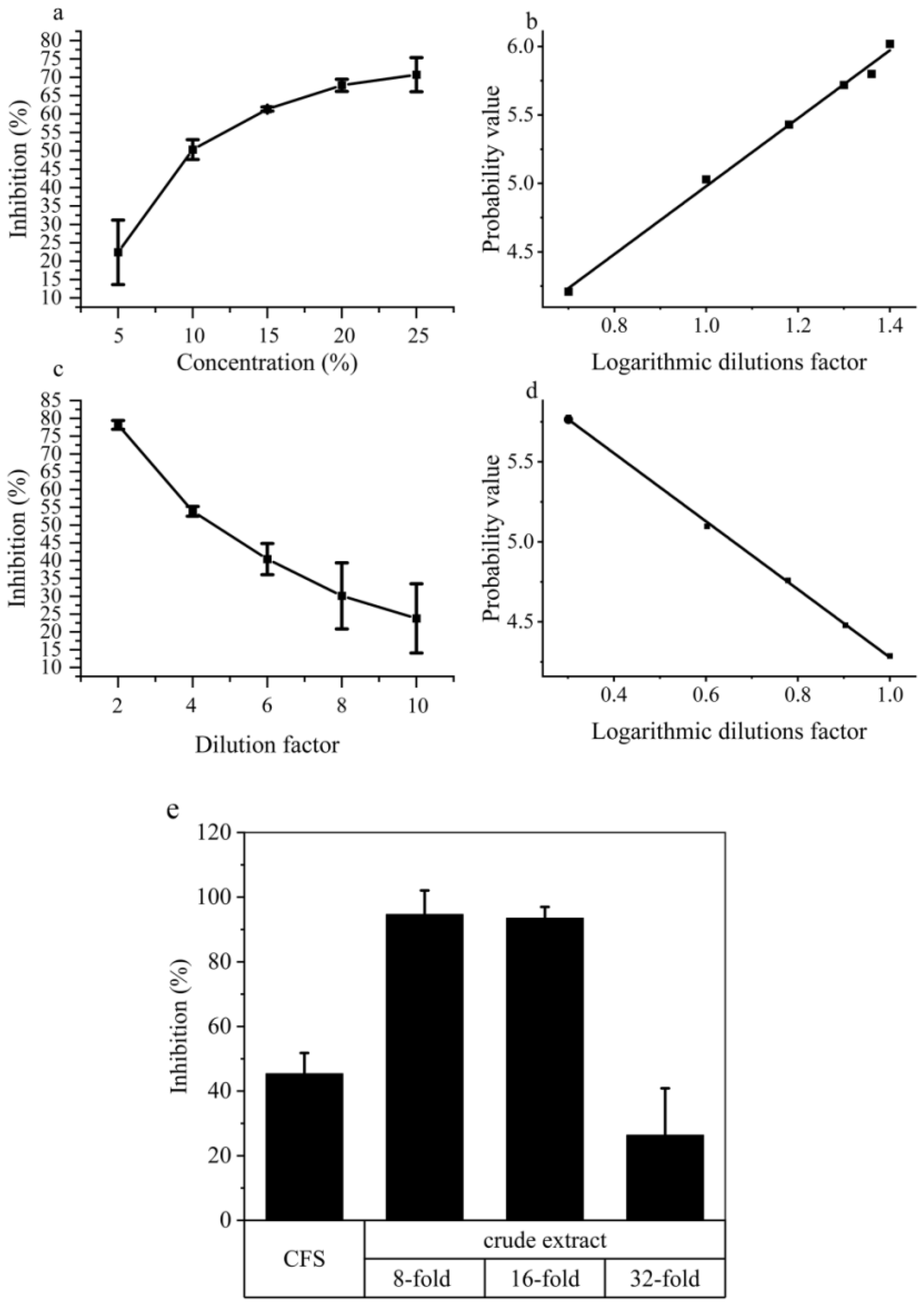
2.6. Differential Metabolites in the Crude Extract Compounds
3. Discussion
4. Materials and Methods
4.1. Culture Conditions for Bacterial and Fungal Strains
4.2. Antagonistic Activity of B. velezensis ZN-S10 against Colletotrichum
4.2.1. Dual-Culture Assay
4.2.2. Detached Leaves Inoculation
4.2.3. Volatile Organic Compound (VOC) Inhibition Assay
4.2.4. Nonvolatile Compound Antifungal Test on Hyphal and Conidial Germination
4.2.5. Extraction of Crude Compounds from the Broth Culture of B. velezensis ZN-10 Using C18 Solid Phase Extraction (SPE) and Inhibition Assay
4.3. LC-MS and GC-MS Analyses
4.4. Differential Metabolites Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 Fungal Pathogens in Molecular Plant Pathology: Top 10 Fungal Pathogens. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Peres, N.A.; Timmer, L.W.; Adaskaveg, J.E.; Correll, J.C. Lifestyles of Colletotrichum acutatum. Plant Dis. 2005, 89, 784–796. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.-C.; Wu, H.-Y.; Wang, Y.-W.; Ariyawansa, H.A.; Hu, H.-P.; Hung, T.-H.; Tzean, S.-S.; Chung, C.-L. Diversity and Pathogenicity of Colletotrichum Species Causing Strawberry Anthracnose in Taiwan and Description of a New Species, Colletotrichum miaoliense Sp. Nov. Sci. Rep. 2020, 10, 14664. [Google Scholar] [CrossRef] [PubMed]
- Alijani, Z.; Amini, J.; Ashengroph, M.; Bahramnejad, B.; Mozafari, A.A. Biocontrol of Strawberry Anthracnose Disease Caused by Colletotrichum Nymphaeae Using Bacillus Atrophaeus Strain DM6120 with Multiple Mechanisms. Trop. Plant Pathol. 2022, 47, 245–259. [Google Scholar] [CrossRef]
- Zubrod, J.P.; Bundschuh, M.; Arts, G.; Brühl, C.A.; Imfeld, G.; Knäbel, A.; Payraudeau, S.; Rasmussen, J.J.; Rohr, J.; Scharmüller, A.; et al. Fungicides: An Overlooked Pesticide Class? Environ. Sci. Technol. 2019, 53, 3347–3365. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, J.A.; Sundh, I.; Becher, P.G.; Björkman, C.; Dubey, M.; Egan, P.A.; Friberg, H.; Gil, J.F.; Jensen, D.F.; Jonsson, M.; et al. When Is It Biological Control? A Framework of Definitions, Mechanisms, and Classifications. J. Pest. Sci. 2021, 94, 665–676. [Google Scholar] [CrossRef]
- Reyes-Perez, J.J.; Hernandez-Montiel, L.G.; Vero, S.; Noa-Carrazana, J.C.; Quiñones-Aguilar, E.E.; Rincón-Enríquez, G. Postharvest Biocontrol of Colletotrichum Gloeosporioides on Mango Using the Marine Bacterium Stenotrophomonas Rhizophila and Its Possible Mechanisms of Action. J. Food Sci. Technol. 2019, 56, 4992–4999. [Google Scholar] [CrossRef] [PubMed]
- Chlebek, D.; Pinski, A.; Żur, J.; Michalska, J.; Hupert-Kocurek, K. Genome Mining and Evaluation of the Biocontrol Potential of Pseudomonas Fluorescens BRZ63, a New Endophyte of Oilseed Rape (Brassica napus L.) against Fungal Pathogens. Int. J. Mol. Sci. 2020, 21, 8740. [Google Scholar] [CrossRef]
- Peeran, M.F.; Krishnan, N.; Thangamani, P.R.; Gandhi, K.; Thiruvengadam, R.; Kuppusamy, P. Development and Evaluation of Water-in-Oil Formulation of Pseudomonas Fluorescens (FP7) against Colletotrichum Musae Incitant of Anthracnose Disease in Banana. Eur. J. Plant Pathol. 2014, 138, 167–180. [Google Scholar] [CrossRef]
- Evangelista-Martínez, Z. Isolation and Characterization of Soil Streptomyces Species as Potential Biological Control Agents against Fungal Plant Pathogens. World J. Microbiol. Biotechnol. 2014, 30, 1639–1647. [Google Scholar] [CrossRef]
- Hu, J.; Zheng, M.; Dang, S.; Shi, M.; Zhang, J.; Li, Y. Biocontrol Potential of Bacillus amyloliquefaciens LYZ69 Against Anthracnose of Alfalfa (Medicago sativa). Phytopathology 2021, 111, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Lee, Y.; Cheon, W.; Park, J.; Kwon, H.-T.; Balaraju, K.; Kim, J.; Yoon, Y.J.; Jeon, Y. Characterization of Bacillus Velezensis AK-0 as a Biocontrol Agent against Apple Bitter Rot Caused by Colletotrichum Gloeosporioides. Sci. Rep. 2021, 11, 626. [Google Scholar] [CrossRef] [PubMed]
- De Silva, N.I.; Brooks, S.; Lumyong, S.; Hyde, K.D. Use of Endophytes as Biocontrol Agents. Fungal Biol. Rev. 2019, 33, 133–148. [Google Scholar] [CrossRef]
- Eljounaidi, K.; Lee, S.K.; Bae, H. Bacterial Endophytes as Potential Biocontrol Agents of Vascular Wilt Diseases—Review and Future Prospects. Biol. Control 2016, 103, 62–68. [Google Scholar] [CrossRef]
- Ryan, R.P.; Germaine, K.; Franks, A.; Ryan, D.J.; Dowling, D.N. Bacterial Endophytes: Recent Developments and Applications. FEMS Microbiol. Lett. 2008, 278, 1–9. [Google Scholar] [CrossRef]
- Piggot, P.J.; Hilbert, D.W. Sporulation of Bacillus Subtilis. Curr. Opin. Microbiol. 2004, 7, 579–586. [Google Scholar] [CrossRef]
- Rabbee, M.; Ali, M.; Choi, J.; Hwang, B.; Jeong, S.; Baek, K. Bacillus Velezensis: A Valuable Member of Bioactive Molecules within Plant Microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef]
- Mahdi, I.; Allaoui, A.; Fahsi, N.; Biskri, L. Bacillus Velezensis QA2 Potentially Induced Salt Stress Tolerance and Enhanced Phosphate Uptake in Quinoa Plants. Microorganisms 2022, 10, 1836. [Google Scholar] [CrossRef]
- Abuhena, M.; Al-Rashid, J.; Azim, M.F.; Khan, M.N.M.; Kabir, M.G.; Barman, N.C.; Rasul, N.M.; Akter, S.; Huq, M.A. Optimization of Industrial (3000 L) Production of Bacillus Subtilis CW-S and Its Novel Application for Minituber and Industrial-Grade Potato Cultivation. Sci. Rep. 2022, 12, 11153. [Google Scholar] [CrossRef]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus Velezensis FZB42 in 2018: The Gram-Positive Model Strain for Plant Growth Promotion and Biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef]
- Wang, J.; Guo, R.; Wang, W.; Ma, G.; Li, S. Insight into the Surfactin Production of Bacillus Velezensis B006 through Metabolomics Analysis. J. Ind. Microbiol. Biotechnol. 2018, 45, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Yang, Y.; Yuan, Q.; Shi, G.; Wu, L.; Lou, Z.; Huo, R.; Wu, H.; Borriss, R.; Gao, X. Bacillomycin D Produced by Bacillus Amyloliquefaciens Is Involved in the Antagonistic Interaction with the Plant-Pathogenic Fungus Fusarium Graminearum. Appl. Env. Microbiol. 2017, 83, e01075-17. [Google Scholar] [CrossRef] [PubMed]
- Hanif, A.; Zhang, F.; Li, P.; Li, C.; Xu, Y.; Zubair, M.; Zhang, M.; Jia, D.; Zhao, X.; Liang, J.; et al. Fengycin Produced by Bacillus Amyloliquefaciens FZB42 Inhibits Fusarium Graminearum Growth and Mycotoxins Biosynthesis. Toxins 2019, 11, 295. [Google Scholar] [CrossRef] [PubMed]
- Cheffi, M.; Bouket, A.C.; Alenezi, F.N.; Luptakova, L.; Belka, M.; Vallat, A.; Rateb, M.E.; Tounsi, S.; Triki, M.A.; Belbahri, L. Olea Europaea L. Root Endophyte Bacillus Velezensis OEE1 Counteracts Oomycete and Fungal Harmful Pathogens and Harbours a Large Repertoire of Secreted and Volatile Metabolites and Beneficial Functional Genes. Microorganisms 2019, 7, 314. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.; Garran, T.A.; Luo, Y.; Cheng, M.; Ren, M.; Zhou, X. Untargeted Metabolomic Analysis and Chemometrics to Identify Potential Marker Compounds for the Chemical Differentiation of Panax Ginseng, P. quinquefolius, P. notoginseng, P. japonicus, and P. japonicus Var. Major. Molecules 2023, 28, 2745. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Hoque, M.A.; Sugimoto, M. Robust Volcano Plot: Identification of Differential Metabolites in the Presence of Outliers. BMC Bioinform. 2018, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Sheflin, A.M.; Broeckling, C.D.; Prenni, J.E. Data Processing for GC-MS- and LC-MS-Based Untargeted Metabolomics. Methods Mol. Biol. 2019, 1978, 287–299. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Wang, Y.; Abozeid, A.; Zu, Y.-G.; Tang, Z.-H. The Integration of GC–MS and LC–MS to Assay the Metabolomics Profiling in Panax Ginseng and Panax Quinquefolius Reveals a Tissue- and Species-Specific Connectivity of Primary Metabolites and Ginsenosides Accumulation. J. Pharm. Biomed. Anal. 2017, 135, 176–185. [Google Scholar] [CrossRef]
- El Sayed, A.M.; Basam, S.M.; El-Naggar, E.-M.B.A.; Marzouk, H.S.; El-Hawary, S. LC–MS/MS and GC–MS Profiling as Well as the Antimicrobial Effect of Leaves of Selected Yucca Species Introduced to Egypt. Sci. Rep. 2020, 10, 17778. [Google Scholar] [CrossRef]
- Naz, S.; Vallejo, M.; García, A.; Barbas, C. Method Validation Strategies Involved in Non-Targeted Metabolomics. J. Chromatogr. A 2014, 1353, 99–105. [Google Scholar] [CrossRef]
- Barnhart, K.; Forman, M.E.; Umile, T.P.; Kueneman, J.; McKenzie, V.; Salinas, I.; Minbiole, K.P.C.; Woodhams, D.C. Identification of Bufadienolides from the Boreal Toad, Anaxyrus Boreas, Active against a Fungal Pathogen. Microb. Ecol. 2017, 74, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Tengattini, S.; Rimaroli, C.; Galmozzi, M.R.; Furlanetto, S.; Massolini, G.; Temporini, C. Effect of Mobile Phase pH on Liquid Chromatography Retention of Mepartricin Related Compounds and Impurities as Support to the Structural Investigation by Liquid Chromatography-Mass Spectrometry. J. Pharm. Biomed. Anal. 2022, 220, 114971. [Google Scholar] [CrossRef] [PubMed]
- Tareq, F.S.; Kim, J.H.; Lee, M.A.; Lee, H.-S.; Lee, J.-S.; Lee, Y.-J.; Shin, H.J. Antimicrobial Gageomacrolactins Characterized from the Fermentation of the Marine-Derived Bacterium Bacillus subtilis under Optimum Growth Conditions. J. Agric. Food Chem. 2013, 61, 3428–3434. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tang, X.-X.; Yan, X.; Wu, Z.; Yi, Z.-W.; Fang, M.-J.; Su, X.; Qiu, Y.-K. A New Macrolactin Antibiotic from Deep Sea-Derived Bacteria Bacillus subtilis B5. Nat. Prod. Res. 2016, 30, 2777–2782. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, D.D.; Zeng, F.; Zhu, Z.H.; Song, P.; Zhao, M.; Duan, J.A. Efficient Biosynthesis, Analysis, Solubility and Anti-Bacterial Activities of Succinylglycosylated Naringenin. Nat. Prod. Res. 2019, 33, 1756–1760. [Google Scholar] [CrossRef]
- Zhang, L.; Jin, M.; Shi, X.; Jin, L.; Hou, X.; Yu, Y.; Liu, B.; Cao, J.; Quan, C. Macrolactin Metabolite Production by Bacillus Sp. ZJ318 Isolated from Marine Sediment. Appl. Biochem. Biotechnol. 2022, 194, 2581–2593. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-C.; Juang, R.-S.; Wei, Y.-H. Applications of a Lipopeptide Biosurfactant, Surfactin, Produced by Microorganisms. Biochem. Eng. J. 2015, 103, 158–169. [Google Scholar] [CrossRef]
- Tiburcio, A.F.; Alcázar, R. Potential Applications of Polyamines in Agriculture and Plant Biotechnology. Methods Mol. Biol. 2018, 1694, 489–508. [Google Scholar] [CrossRef]
- Tyagi, A.; Ali, S.; Ramakrishna, G.; Singh, A.; Park, S.; Mahmoudi, H.; Bae, H. Revisiting the Role of Polyamines in Plant Growth and Abiotic Stress Resilience: Mechanisms, Crosstalk, and Future Perspectives. J. Plant Growth Regul. 2023, 42, 5074–5098. [Google Scholar] [CrossRef]
- Song, Y.; Ren, Y.; Xue, Y.; Lu, D.; Yan, T.; He, J. Putrescine (1,4-Diaminobutane) Enhances Antifungal Activity in Postharvest Mango Fruit against Colletotrichum Gloeosporioides through Direct Fungicidal and Induced Resistance Mechanisms. Pestic. Biochem. Physiol. 2023, 195, 105581. [Google Scholar] [CrossRef]
- Jancewicz, A.L.; Gibbs, N.M.; Masson, P.H. Cadaverine’s Functional Role in Plant Development and Environmental Response. Front. Plant Sci. 2016, 7, 870. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ruan, W.; Li, J.; Xu, H.; Wang, J.; Gao, Y.; Wang, J. Biological Control of Phytopathogenic Fungi by Fatty Acids. Mycopathologia 2008, 166, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Fan, L.; Yang, C.; Cui, L.; Jin, M.; Osei, R. Analysis of Bacillus Mojavensis ZA1 Volatile Anti-Pathogen Substances against Colletotrichum Coccodes. J. Plant Dis. Prot. 2023, 130, 633–642. [Google Scholar] [CrossRef]
- Horne, S.M.; Schroeder, M.; Murphy, J.; Prüβ, B.M. Acetoacetate and Ethyl Acetoacetate as Novel Inhibitors of Bacterial Biofilm. Lett. Appl. Microbiol. 2018, 66, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.-Z.; Cheng, K.; Zhang, X.-M.; Liu, K.; Jiao, Q.-C.; Zhu, H.-L. Synthesis of Some N-Alkyl Substituted Urea Derivatives as Antibacterial and Antifungal Agents. Eur. J. Med. Chem. 2010, 45, 3207–3212. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.F.; Santos Júnior, H.M.D.; Nunes, A.S.; Campos, V.P.; Pinho, R.S.C.D.; Gajo, G.C. Purification and Identification of Metabolites Produced by Bacillus Cereus and B. Subtilis Active against Meloidogyne Exigua, and Their In Silico Interaction with a Putative Phosphoribosyltransferase fromM. Incognita. An. Acad. Bras. Ciênc. 2014, 86, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Mahendran, R.; Thandeeswaran, M.; Kiran, G.; Arulkumar, M.; Ayub Nawaz, K.A.; Jabastin, J.; Janani, B.; Anto Thomas, T.; Angayarkanni, J. Evaluation of Pterin, a Promising Drug Candidate from Cyanide Degrading Bacteria. Curr. Microbiol. 2018, 75, 684–693. [Google Scholar] [CrossRef]
- De Silva, D.D.; Crous, P.W.; Ades, P.K.; Hyde, K.D.; Taylor, P.W.J. Life Styles of Colletotrichum Species and Implications for Plant Biosecurity. Fungal Biol. Rev. 2017, 31, 155–168. [Google Scholar] [CrossRef]
- Perfect, S.E.; Green, J.R. Infection Structures of Biotrophic and Hemibiotrophic Fungal Plant Pathogens: Infection Structures of Biotrophic and Hemibiotrophic Fungal Plant Pathogens. Mol. Plant Pathol. 2001, 2, 101–108. [Google Scholar] [CrossRef]
- Huang, L.; Li, Q.-C.; Hou, Y.; Li, G.-Q.; Yang, J.-Y.; Li, D.-W.; Ye, J.-R. Bacillus velezensis Strain HYEB5-6 as a Potential Biocontrol Agent against Anthracnose on Euonymus japonicus. Biocontrol Sci. Technol. 2017, 27, 636–653. [Google Scholar] [CrossRef]
- Choub, V.; Ajuna, H.B.; Won, S.-J.; Moon, J.-H.; Choi, S.-I.; Maung, C.E.H.; Kim, C.-W.; Ahn, Y.S. Antifungal Activity of Bacillus Velezensis CE 100 against Anthracnose Disease (Colletotrichum gloeosporioides) and Growth Promotion of Walnut (Juglans regia L.) Trees. Int. J. Mol. Sci. 2021, 22, 10438. [Google Scholar] [CrossRef]
- Damasceno, C.L.; Duarte, E.A.A.; Dos Santos, L.B.P.R.; De Oliveira, T.A.S.; De Jesus, F.N.; De Oliveira, L.M.; Góes-Neto, A.; Soares, A.C.F. Postharvest Biocontrol of Anthracnose in Bananas by Endophytic and Soil Rhizosphere Bacteria Associated with Sisal (Agave sisalana) in Brazil. Biol. Control 2019, 137, 104016. [Google Scholar] [CrossRef]
- Ma, Z.; Zhang, S.; Sun, K.; Hu, J. Identification and Characterization of a Cyclic Lipopeptide Iturin A from a Marine-Derived Bacillus Velezensis 11-5 as a Fungicidal Agent to Magnaporthe Oryzae in Rice. J. Plant Dis. Prot. 2020, 127, 15–24. [Google Scholar] [CrossRef]
- Jayawardena, R. An Account of Colletotrichum Species Associated with Strawberry Anthracnose in China Based on Morphology and Molecular Data. Mycosphere 2016, 7, 1147–1163. [Google Scholar] [CrossRef]
- Salazar, F.; Ortiz, A.; Sansinenea, E. A Strong Antifungal Activity of 7-O-Succinyl Macrolactin A vs Macrolactin A from Bacillus Amyloliquefaciens ELI149. Curr. Microbiol. 2020, 77, 3409–3413. [Google Scholar] [CrossRef] [PubMed]
- Sarwar, A.; Hassan, M.N.; Imran, M.; Iqbal, M.; Majeed, S.; Brader, G.; Sessitsch, A.; Hafeez, F.Y. Biocontrol Activity of Surfactin a Purified from Bacillus NH-100 and NH-217 against Rice Bakanae Disease. Microbiol. Res. 2018, 209, 1–13. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Uhl, J.; Grosch, R.; Alquéres, S.; Pittroff, S.; Dietel, K.; Schmitt-Kopplin, P.; Borriss, R.; Hartmann, A. Cyclic Lipopeptides of Bacillus amyloliquefaciens Subsp. Plantarum Colonizing the Lettuce Rhizosphere Enhance Plant Defense Responses Toward the Bottom Rot Pathogen Rhizoctonia solani. MPMI 2015, 28, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Rana, A.; Dhaka, R.K.; Singh, A.P.; Chahar, M.; Singh, S.; Nain, L.; Singh, K.P.; Minz, D. Bacterial Volatile Organic Compounds as Biopesticides, Growth Promoters and Plant-Defense Elicitors: Current Understanding and Future Scope. Biotechnol. Adv. 2023, 63, 108078. [Google Scholar] [CrossRef]
- Veselova, M.A.; Plyuta, V.A.; Khmel, I.A. Volatile Compounds of Bacterial Origin: Structure, Biosynthesis, and Biological Activity. Microbiology 2019, 88, 261–274. [Google Scholar] [CrossRef]
- Ling, L.; Luo, H.; Yang, C.; Wang, Y.; Cheng, W.; Pang, M.; Jiang, K. Volatile Organic Compounds Produced by Bacillus Velezensis L1 as a Potential Biocontrol Agent against Postharvest Diseases of Wolfberry. Front. Microbiol. 2022, 13, 987844. [Google Scholar] [CrossRef]
- Kim, T.Y.; Hwang, S.H.; Noh, J.S.; Cho, J.-Y.; Maung, C.E.H. Antifungal Potential of Bacillus Velezensis CE 100 for the Control of Different Colletotrichum Species through Isolation of Active Dipeptide, Cyclo-(D-Phenylalanyl-D-Prolyl). Int. J. Mol. Sci. 2022, 23, 7786. [Google Scholar] [CrossRef] [PubMed]

| Classification | Metabolites | VIP | q Value | Fold Change | Biocontrol Effects | References |
|---|---|---|---|---|---|---|
| Steroids and steroid derivatives | Telocinobufagin | 16.27 | 2.35 × 10−3 | 3.95 × 103 | Fungi, bacteria | [31] |
| 5beta-cholestan-3alpha,7alpha,12alpha,24(S),27-pentol | 3.48 | 2.25 × 10−6 | 7.22 × 104 | |||
| 17a,21-Dihydroxy-5b-pregnane-3,11,20-trione | 2.22 | 9.29 × 10−6 | 3.68 × 103 | |||
| (22E)-12alpha-Hydroxy-3-oxochola-1,4,22-trien-24-oic Acid | 2.00 | 3.27 × 10−9 | 5.14 × 103 | |||
| Chenodeoxycholylglutamine | 1.92 | 6.88 × 10−5 | 6.68 × 104 | |||
| Fatty acyls | Macrolactin C | 5.02 | 9.05 × 10−4 | 3.41 × 103 | ||
| Mepartricin B | 4.36 | 2.65 × 10−5 | 2.89 × 104 | Fungi | [32] | |
| cis-tetradec-11-enoic acid | 2.78 | 3.83 × 10−4 | 1.01 × 103 | |||
| N-Palmitoyl glutamine | 2.16 | 1.17 × 10−4 | 4.73 × 104 | |||
| MG (18:2(9Z,12Z)/0:0/0:0) | 1.81 | 1.80 × 10−4 | 2.44 × 103 | |||
| Macrolides | 7-O-Succinyl macrolactin A | 17.87 | 3.33 × 10−9 | 1.49 × 104 | Fungi, bacteria | [33,34] |
| Deforolimus | 1.51 | 2.83 × 10−5 | 1.51 × 104 | |||
| Organooxygen compounds | Tunicamycin A | 2.27 | 2.61 × 10−3 | 1.69 × 1010 | Bacteria | [35] |
| Macrolactin B | 1.59 | 4.08 × 10−7 | 3.05 × 103 | Bacteria | [36] | |
| Glycerophospholipids | CDP-DG (16:1(9Z)/22:5(7Z,10Z,13Z,16Z,19Z)) | 2.35 | 1.13 × 10−3 | 3.56 × 104 | ||
| OOV-PS | 2.29 | 2.68 × 10−5 | 4.42 × 103 | |||
| Peptidomimetics | Surfactin A | 14.79 | 1.20 × 10−3 | 3.69 × 103 | Fungi, bacteria, virus, Mycoplasma, biofilm, induction of ISR | [17,37] |
| Prenol lipids | Goshonoside F1 | 2.85 | 2.06 × 10−8 | 4.67 × 103 | ||
| Polyketides | Armochaetoglasin K | 1.94 | 1.22 × 10−3 | 1.61 × 103 |
| Classification | Metabolites | VIP | q Value | Fold Change | Biocontrol Effects | References |
|---|---|---|---|---|---|---|
| Organonitrogen compounds | Putrescine | 2.17 | 1.95 × 10−5 | 1.06 × 102 | Fungi, plant growth, abiotic stress responses | [38,39,40] |
| Spermidine | 1.63 | 4.00 × 10−6 | 1.40 × 101 | Plant growth, abiotic stress responses, biofilm | [38,39] | |
| Ethanolamine | 1.23 | 5.42 × 10−3 | 4.78 × 100 | |||
| Cadaverine | 1.01 | 1.54 × 10−3 | 2.86 × 100 | Plant growth, stress response, insect | [41] | |
| Fatty acyls | Heptadecan-1-Ol | 1.94 | 8.09 × 10−5 | 4.17 × 101 | ||
| 4-Hydroxybutyric acid | 1.60 | 1.90 × 10−3 | 1.19 × 101 | |||
| Margaric acid | 1.57 | 6.23 × 10−4 | 1.04 × 101 | |||
| Caproic acid | 1.19 | 5.92 × 10−4 | 4.15 × 100 | Fungi | [42] | |
| Organooxygen compounds | 2,3-Butanediol | 1.81 | 2.31 × 10−6 | 2.58 × 101 | Induction of ISR, plant growth, bacteria, fungi | [17,43] |
| D-Arabitol | 1.32 | 2.88 × 10−6 | 5.62 × 100 | |||
| Imidazopyrimidines | 2-Hydroxyadenine | 1.34 | 9.87 × 10−3 | 6.43 × 100 | ||
| Hypoxanthine | 1.27 | 2.30 × 10−2 | 5.60 × 100 | |||
| Benzene and substituted derivatives | Phenylboronic acid | 2.43 | 3.29 × 10−4 | 3.39 × 102 | ||
| Keto acids and derivatives | Acetoacetic acid | 1.70 | 1.03 × 10−4 | 1.76 × 101 | Biofilm, bacteria | [44] |
| Organic carbonic acids and derivatives | Urea | 1.54 | 2.72 × 10−2 | 1.13 × 101 | Fungi, bacteria | [45] |
| Purine nucleotides | Inosinic acid | 1.49 | 5.99 × 10−3 | 7.42 × 100 | ||
| Organic phosphoric acids and derivatives | Methylphosphate | 1.49 | 1.56 × 10−5 | 9.00 × 100 | ||
| Diazines | Dihydrouracil | 1.44 | 4.57 × 10−4 | 7.94 × 100 | Nematodes | [46] |
| Pteridines and derivatives | Pterin | 1.36 | 1.07 × 10−3 | 6.36 × 100 | biofilm, bacteria | [47] |
| Purine nucleotides | Adenosine monophosphate | 1.08 | 2.05 × 10−2 | 3.17 × 100 | ||
| Unclassified | 5-Methoxy-1-Pentanol | 2.19 | 5.54 × 10−3 | 1.22 × 102 | ||
| Hydrobenzoin | 1.25 | 3.66 × 10−6 | 4.68 × 100 | |||
| 4-Nitrophenethylamine | 1.24 | 1.19 × 10−4 | 4.66 × 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Q.; Zhong, Z.; Chao, S.; Liu, L.; Chen, M.; Feng, X.; Wu, H. Antifungal Effect of Bacillus velezensis ZN-S10 against Plant Pathogen Colletotrichum changpingense and Its Inhibition Mechanism. Int. J. Mol. Sci. 2023, 24, 16694. https://doi.org/10.3390/ijms242316694
Ye Q, Zhong Z, Chao S, Liu L, Chen M, Feng X, Wu H. Antifungal Effect of Bacillus velezensis ZN-S10 against Plant Pathogen Colletotrichum changpingense and Its Inhibition Mechanism. International Journal of Molecular Sciences. 2023; 24(23):16694. https://doi.org/10.3390/ijms242316694
Chicago/Turabian StyleYe, Qingling, Zhupeiqi Zhong, Shufeng Chao, Lu Liu, Mengli Chen, Xiaoxiao Feng, and Huiming Wu. 2023. "Antifungal Effect of Bacillus velezensis ZN-S10 against Plant Pathogen Colletotrichum changpingense and Its Inhibition Mechanism" International Journal of Molecular Sciences 24, no. 23: 16694. https://doi.org/10.3390/ijms242316694
APA StyleYe, Q., Zhong, Z., Chao, S., Liu, L., Chen, M., Feng, X., & Wu, H. (2023). Antifungal Effect of Bacillus velezensis ZN-S10 against Plant Pathogen Colletotrichum changpingense and Its Inhibition Mechanism. International Journal of Molecular Sciences, 24(23), 16694. https://doi.org/10.3390/ijms242316694







