Alteration of Cadherin 3 Expression and DNA Methylation in Association with Aggressive Renal Cell Carcinoma
Abstract
1. Introduction
2. Results
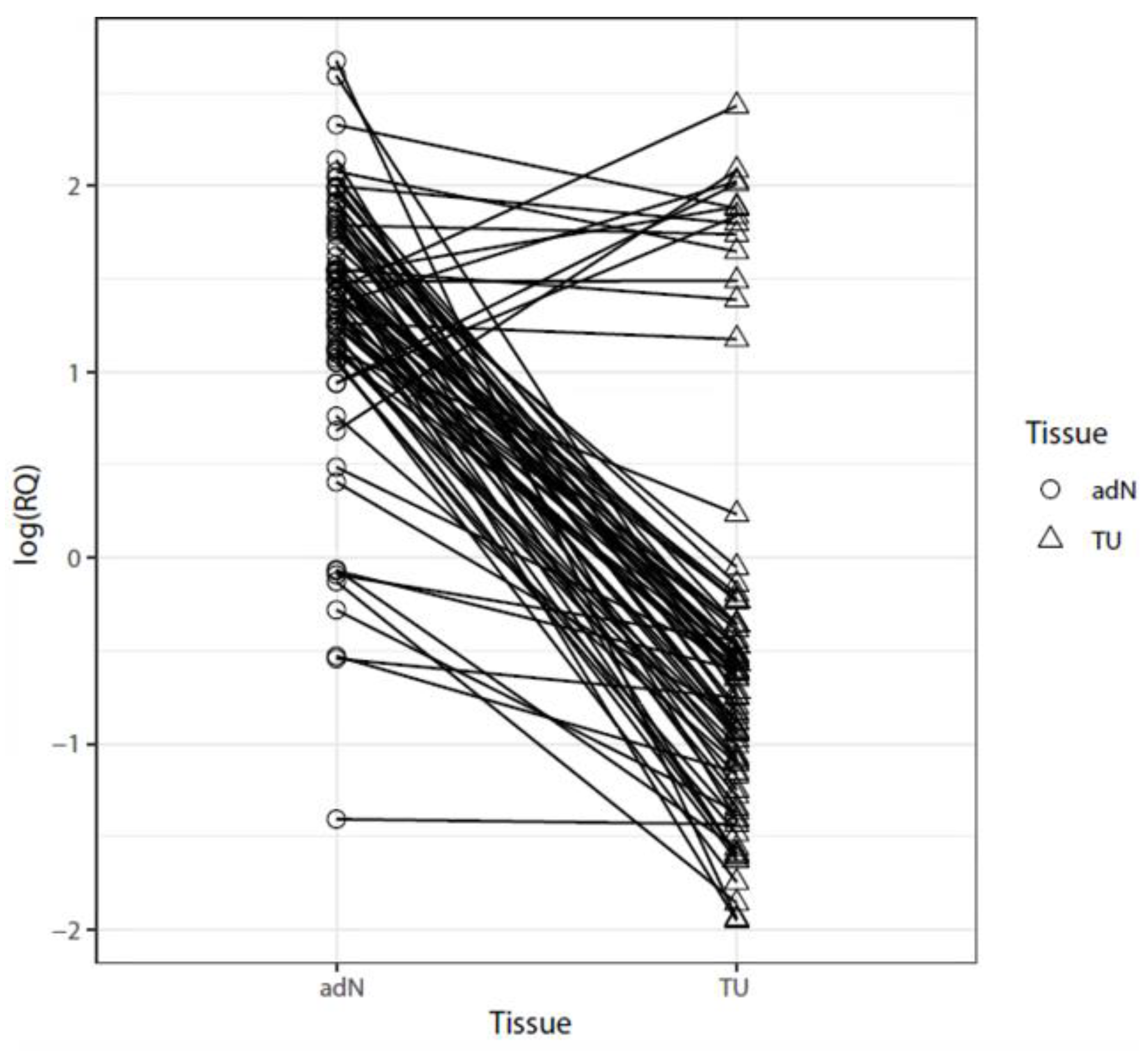
2.1. Analysis of mRNA Expression in Paired Tumor and Normal Adjacent RCC Tissues
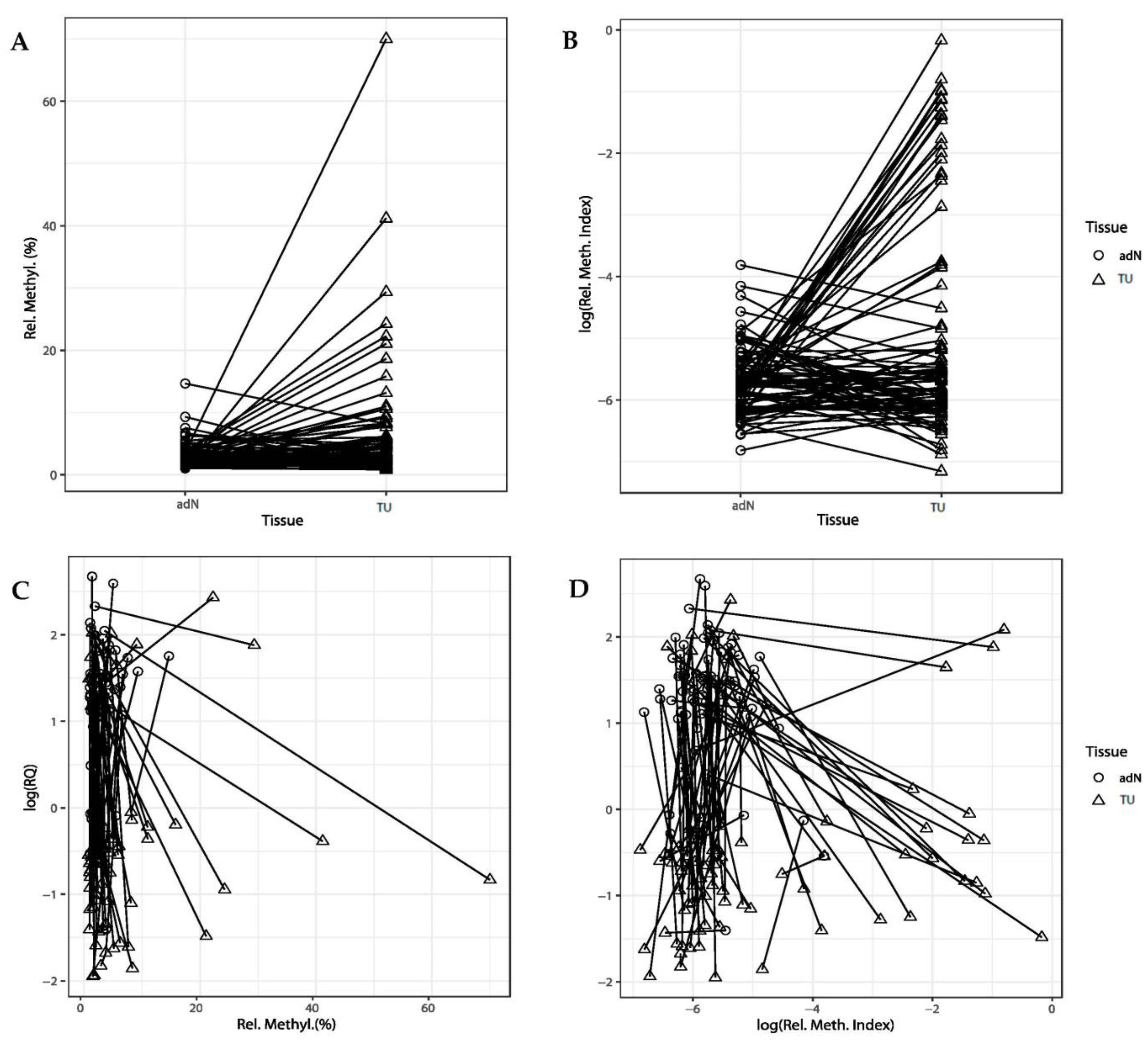
2.2. DNA Methylation of CDH3 in RCC
2.3. Comparison of DNA Methylation and CDH3 mRNA Expression
2.4. Association with Clinicopathological Parameters
2.5. In Silico Analysis of Statistical Associations of CDH3 mRNA Expression and DNA Methylation Using the TCGA-KIRC Data
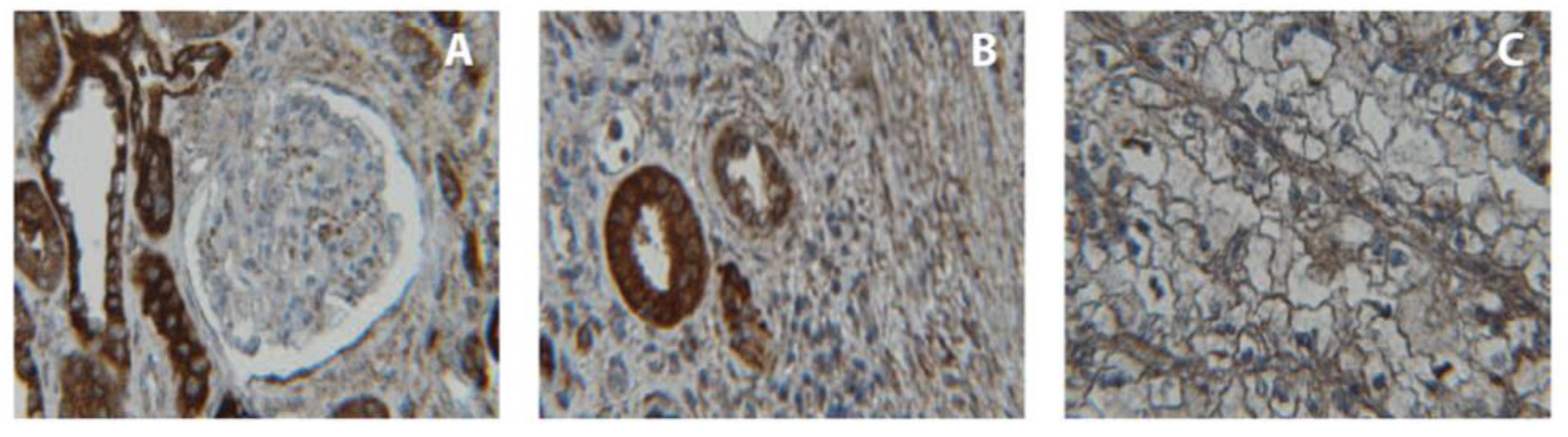
2.6. Immunohistochemical Analysis of CDH3 Protein Expression
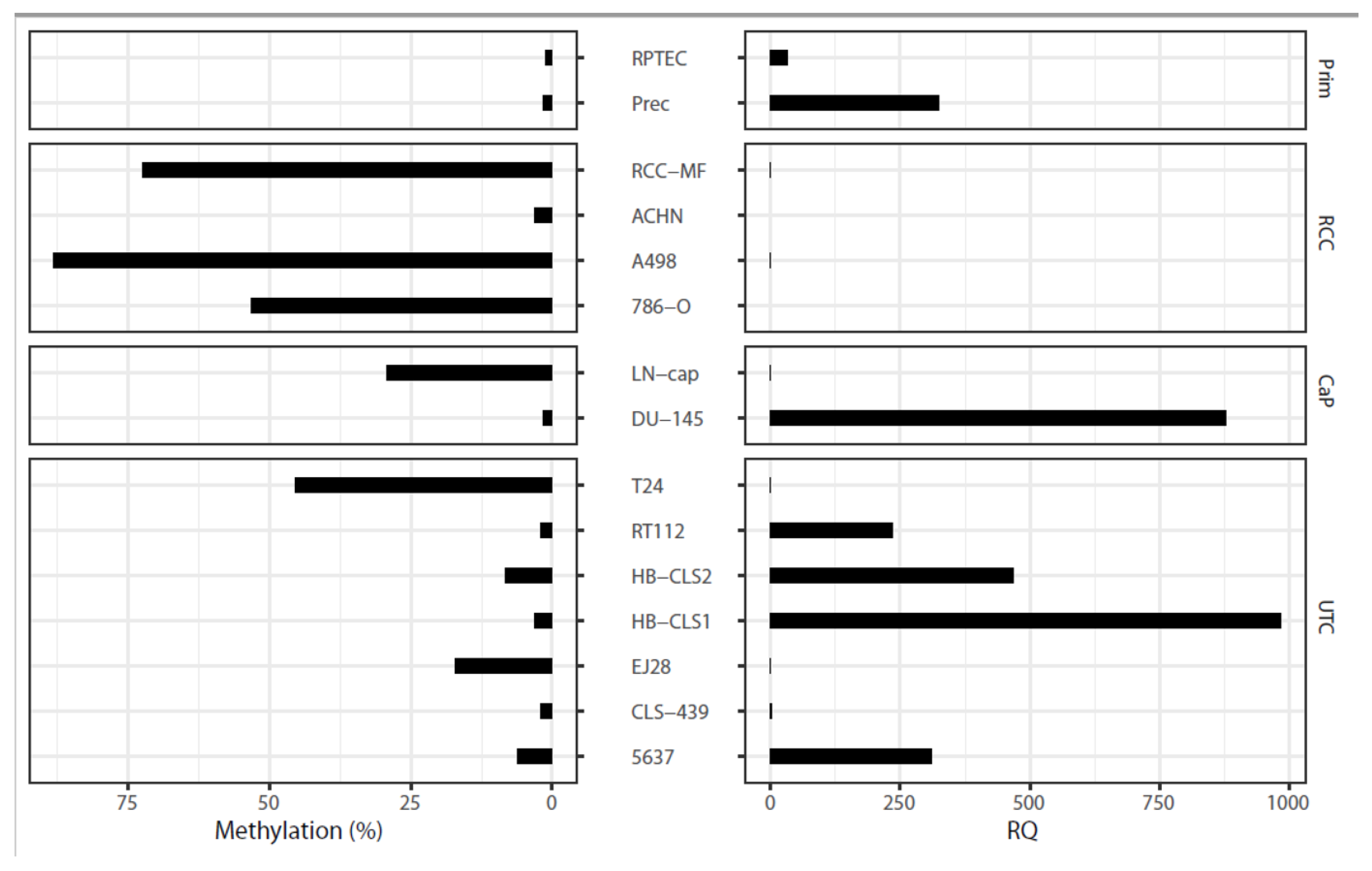
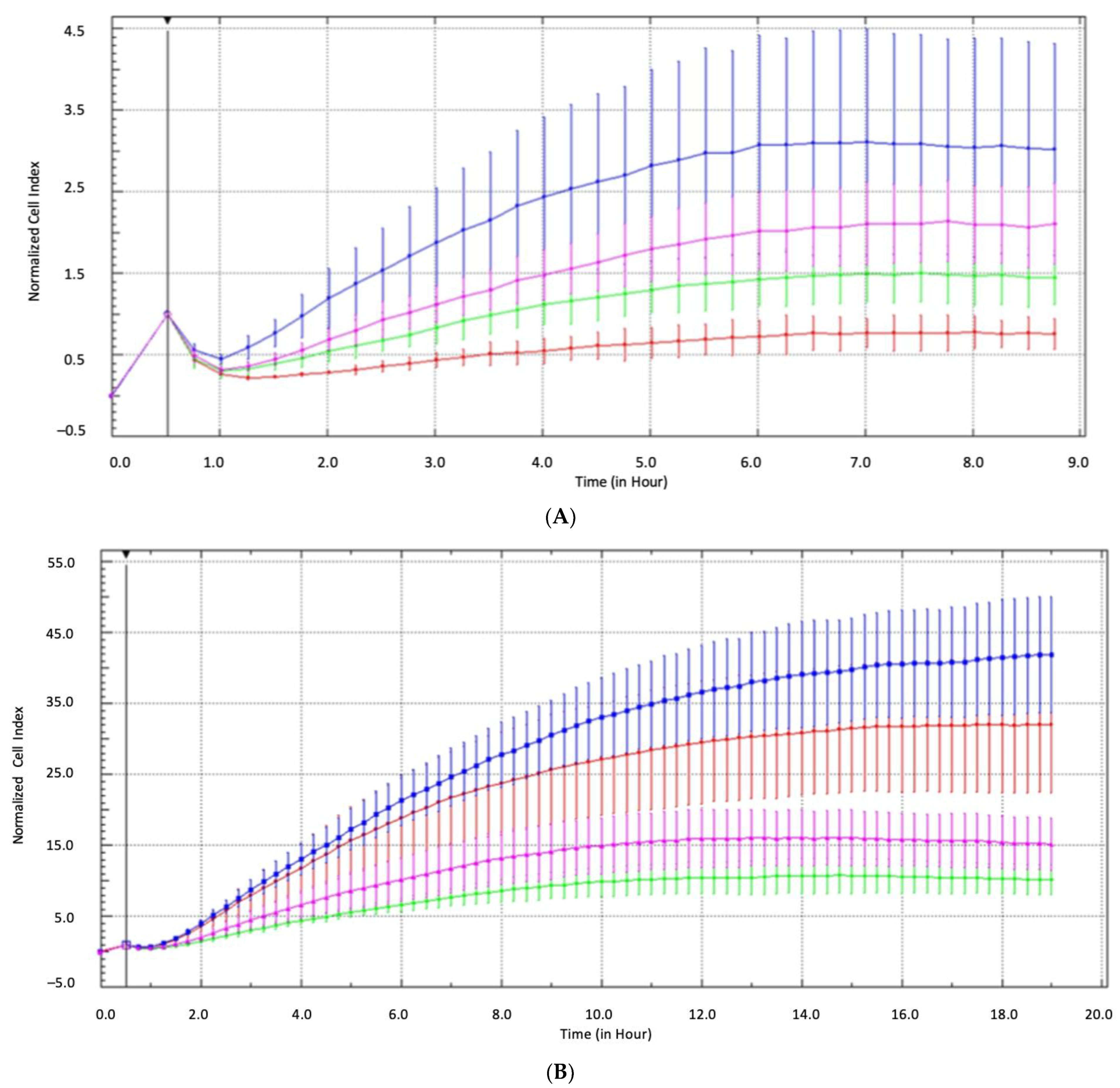
2.7. Epigenetic and Functional Alterations of CDH3 in Cell Line Tumor Models
3. Discussion
4. Material and Methods
4.1. Tumor Cell Lines
4.2. Tissue Specimens
4.3. Immunohistochemical and Western Blotting Analysis of CDH3 Protein Expression
4.4. Isolation of RNA, cDNA Synthesis, and Real-Time PCR Quantitation
4.5. DNA Extraction, Bisulfite Conversion, and Methylation Analyses
4.6. Real-Time Impedance Analysis of Cell Invasiveness
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Rini, B.I.; Campbell, S.C.; Escudier, B. Renal cell carcinoma. Lancet 2009, 373, 1119–1132. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Steliarova-Foucher, E.; Lortet-Tieulent, J.; Rosso, S.; Coebergh, J.W.W.; Comber, H.; Forman, D.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur. J. Cancer 2013, 49, 1374–1403. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.; Heng, D.Y.C.; Brugarolas, J.; Vaishampayan, U. Personalized Management of Advanced Kidney Cancer. Am. Soc. Clin. Oncol. Educ. book. Am. Soc. Clin. Oncol. Annu. Meet. 2018, 38, 330–341. [Google Scholar] [CrossRef]
- Patel, H.D.; Gupta, M.; Joice, G.A.; Srivastava, A.; Alam, R.; Allaf, M.E.; Pierorazio, P.M. Clinical Stage Migration and Survival for Renal Cell Carcinoma in the United States. Eur. Urol. Oncol. 2019, 2, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.H.; Klatte, T.; Usher-Smith, J.; Stewart, G.D. Epidemiology and screening for renal cancer. World J. Urol. 2018, 36, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 313–326.e5. [Google Scholar] [CrossRef]
- Ricketts, C.J.; Hill, V.K.; Linehan, W.M. Tumor-specific hypermethylation of epigenetic biomarkers, including SFRP1, predicts for poorer survival in patients from the TCGA Kidney Renal Clear Cell Carcinoma (KIRC) project. PLoS ONE 2014, 9, e85621. [Google Scholar] [CrossRef]
- Serth, J.; Peters, I.; Katzendorn, O.; Dang, T.N.; Moog, J.; Balli, Z.; Reese, C.; Hennenlotter, J.; Grote, A.; Lafos, M.; et al. Identification of a Novel Renal Metastasis Associated CpG-Based DNA Methylation Signature (RMAMS). Int. J. Mol. Sci. 2022, 23, 11190. [Google Scholar] [CrossRef]
- Peters, I.; Gebauer, K.; Dubrowinskaja, N.; Atschekzei, F.; Kramer, M.W.; Hennenlotter, J.; Tezval, H.; Abbas, M.; Scherer, R.; Merseburger, A.S.; et al. GATA5 CpG island hypermethylation is an independent predictor for poor clinical outcome in renal cell carcinoma. Oncol. Rep. 2014, 31, 1523–1530. [Google Scholar] [CrossRef]
- Tezval, H.; Dubrowinskaja, N.; Peters, I.; Reese, C.; Serth, K.; Atschekzei, F.; Hennenlotter, J.; Stenzl, A.; Kuczyk, M.A.; Serth, J. Tumor Specific Epigenetic Silencing of Corticotropin Releasing Hormone-Binding Protein in Renal Cell Carcinoma: Association of Hypermethylation and Metastasis. PLoS ONE 2016, 11, e0163873. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.L.; Henrique, R.; Ribeiro, F.R.; Pinto, M.; Oliveira, J.; Lobo, F.; Teixeira, M.R.; Jerónimo, C. Quantitative promoter methylation analysis of multiple cancer-related genes in renal cell tumors. BMC Cancer 2007, 7, 133. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Linehan, W.M.; Spellman, P.T.; Ricketts, C.J.; Creighton, C.J.; Fei, S.S.; Davis, C.; Wheeler, D.A.; Murray, B.A.; Schmidt, L.; et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N. Engl. J. Med. 2016, 374, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef]
- Morris, M.R.; Ricketts, C.; Gentle, D.; Abdulrahman, M.; Clarke, N.; Brown, M.; Kishida, T.; Yao, M.; Latif, F.; Maher, E.R. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene 2010, 29, 2104–2117. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.R.; Ricketts, C.J.; Gentle, D.; McRonald, F.; Carli, N.; Khalili, H.; Brown, M.; Kishida, T.; Yao, M.; Banks, R.E.; et al. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene 2011, 30, 1390–1401. [Google Scholar] [CrossRef]
- Henrique, R.; Luís, A.S.; Jerónimo, C. The epigenetics of renal cell tumors: From biology to biomarkers. Front. Genet. 2012, 3, 94. [Google Scholar] [CrossRef]
- Riener, M.-O.; Vogetseder, A.; Pestalozzi, B.C.; Clavien, P.-A.; Probst-Hensch, N.; Kristiansen, G.; Jochum, W. Cell adhesion molecules P-cadherin and CD24 are markers for carcinoma and dysplasia in the biliary tract. Hum. Pathol. 2010, 41, 1558–1565. [Google Scholar] [CrossRef]
- Kourtidis, A.; Lu, R.; Pence, L.J.; Anastasiadis, P.Z. A central role for cadherin signaling in cancer. Exp. Cell Res. 2017, 358, 78–85. [Google Scholar] [CrossRef]
- Ribeiro, A.S.; Albergaria, A.; Sousa, B.; Correia, A.L.; Bracke, M.; Seruca, R.; Schmitt, F.C.; Paredes, J. Extracellular cleavage and shedding of P-cadherin: A mechanism underlying the invasive behaviour of breast cancer cells. Oncogene 2010, 29, 392–402. [Google Scholar] [CrossRef]
- Paredes, J.; Albergaria, A.; Oliveira, J.T.; Jerónimo, C.; Milanezi, F.; Schmitt, F.C. P-cadherin overexpression is an indicator of clinical outcome in invasive breast carcinomas and is associated with CDH3 promoter hypomethylation. Clin. Cancer Res. 2005, 11, 5869–5877. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, K.; Nakagawa, H.; Hosokawa, M.; Nakamura, T.; Eguchi, H.; Ohigashi, H.; Ishikawa, O.; Katagiri, T.; Nakamura, Y. Overexpressed P-cadherin/CDH3 promotes motility of pancreatic cancer cells by interacting with p120ctn and activating rho-family GTPases. Cancer Res. 2005, 65, 3092–3099. [Google Scholar] [CrossRef] [PubMed]
- Han, A.C.; Edelson, M.I.; Peralta Soler, A.; Knudsen, K.A.; Lifschitz-Mercer, B.; Czernobilsky, B.; Rosenblum, N.G.; Salazar, H. Cadherin expression in glandular tumors of the cervix. Cancer 2000, 89, 2053–2058. [Google Scholar] [CrossRef] [PubMed]
- Patel, I.S.; Madan, P.; Getsios, S.; Bertrand, M.A.; MacCalman, C.D. Cadherin switching in ovarian cancer progression. Int. J. Cancer 2003, 106, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Lo Muzio, L.; Campisi, G.; Farina, A.; Rubini, C.; Pannone, G.; Serpico, R.; Laino, G.; De Lillo, A.; Carinci, F. P-cadherin expression and survival rate in oral squamous cell carcinoma: An immunohistochemical study. BMC Cancer 2005, 5, 63. [Google Scholar] [CrossRef][Green Version]
- Imai, K.; Hirata, S.; Irie, A.; Senju, S.; Ikuta, Y.; Yokomine, K.; Harao, M.; Inoue, M.; Tsunoda, T.; Nakatsuru, S.; et al. Identification of a novel tumor-associated antigen, cadherin 3/P-cadherin, as a possible target for immunotherapy of pancreatic, gastric, and colorectal cancers. Clin. Cancer Res. 2008, 14, 6487–6495. [Google Scholar] [CrossRef]
- Yi, S.; Yang, Z.-L.; Miao, X.; Zou, Q.; Li, J.; Liang, L.; Zeng, G.; Chen, S. N-cadherin and P-cadherin are biomarkers for invasion, metastasis, and poor prognosis of gallbladder carcinomas. Pathol. Res. Pract. 2014, 210, 363–368. [Google Scholar] [CrossRef]
- Bauer, R.; Valletta, D.; Bauer, K.; Thasler, W.E.; Hartmann, A.; Müller, M.; Reichert, T.E.; Hellerbrand, C. Downregulation of P-cadherin expression in hepatocellular carcinoma induces tumorigenicity. Int. J. Clin. Exp. Pathol. 2014, 7, 6125–6132. [Google Scholar] [CrossRef]
- Zhu, C.; Feng, X.; Ye, G.; Huang, T. Meta-analysis of possible role of cadherin gene methylation in evolution and prognosis of hepatocellular carcinoma with a PRISMA guideline. Medicine 2017, 96, e6650. [Google Scholar] [CrossRef]
- Mandeville, J.A.; Silva Neto, B.; Vanni, A.J.; Smith, G.L.; Rieger-Christ, K.M.; Zeheb, R.; Loda, M.; Libertino, J.A.; Summerhayes, I.C. P-cadherin as a prognostic indicator and a modulator of migratory behaviour in bladder carcinoma cells. BJU Int. 2008, 102, 1707–1714. [Google Scholar] [CrossRef]
- Jarrard, D.F.; Paul, R.; van Bokhoven, A.; Nguyen, S.H.; Bova, G.S.; Wheelock, M.J.; Johnson, K.R.; Schalken, J.; Bussemakers, M.; Isaacs, W.B. P-Cadherin is a basal cell-specific epithelial marker that is not expressed in prostate cancer. Clin. Cancer Res. 1997, 3, 2121–2128. [Google Scholar] [PubMed]
- Shimazui, T.; Giroldi, L.A.; Bringuier, P.P.; Oosterwijk, E.; Schalken, J.A. Complex cadherin expression in renal cell carcinoma. Cancer Res. 1996, 56, 3234–3237. [Google Scholar] [PubMed]
- Pontén, F.; Gry, M.; Fagerberg, L.; Lundberg, E.; Asplund, A.; Berglund, L.; Oksvold, P.; Björling, E.; Hober, S.; Kampf, C.; et al. A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 2009, 5, 337. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, A.; Watanabe, R.; Tomita, Y. E-cadherin expression in renal cell cancer and its significance in metastasis and survival. Br. J. Cancer 1995, 71, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Nojima, D.; Nakajima, K.; Li, L.C.; Franks, J.; Ribeiro-Filho, L.; Ishii, N.; Dahiya, R. CpG methylation of promoter region inactivates E-cadherin gene in renal cell carcinoma. Mol. Carcinog. 2001, 32, 19–27. [Google Scholar] [CrossRef]
- Li, T.; Pan, H.; Li, R. The dual regulatory role of miR-204 in cancer. Tumour Biol. 2016, 37, 11667–11677. [Google Scholar] [CrossRef]
- Hibi, K.; Goto, T.; Mizukami, H.; Kitamura, Y.-H.; Sakuraba, K.; Sakata, M.; Saito, M.; Ishibashi, K.; Kigawa, G.; Nemoto, H.; et al. Demethylation of the CDH3 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2009, 29, 2215–2217. [Google Scholar]
- Motzer, R.J.; Bander, N.H.; Nanus, D.M. Renal-cell carcinoma. N. Engl. J. Med. 1996, 335, 865–875. [Google Scholar] [CrossRef]
- Liu, X.; Su, L.; Liu, X. Loss of CDH1 up-regulates epidermal growth factor receptor via phosphorylation of YBX1 in non-small cell lung cancer cells. FEBS Lett. 2013, 587, 3995–4000. [Google Scholar] [CrossRef]
- Choueiri, T.K.; Tomczak, P.; Park, S.H.; Venugopal, B.; Ferguson, T.; Chang, Y.-H.; Hajek, J.; Symeonides, S.N.; Lee, J.L.; Sarwar, N.; et al. Adjuvant Pembrolizumab after Nephrectomy in Renal-Cell Carcinoma. N. Engl. J. Med. 2021, 385, 683–694. [Google Scholar] [CrossRef]
- Li, K.K.; Li, F.; Li, Q.S.; Yang, K.; Jin, B. DNA methylation as a target of epigenetic therapeutics in cancer. Anti-Cancer Agents Med. Chem. 2013, 13, 242–247. [Google Scholar] [CrossRef]
- Tezval, H.; Atschekzei, F.; Peters, I.; Waalkes, S.; Hennenlotter, J.; Stenzl, A.; Becker, J.U.; Merseburger, A.S.; Kuczyk, M.A.; Serth, J. Reduced mRNA expression level of corticotropin-releasing hormone-binding protein is associated with aggressive human kidney cancer. BMC Cancer 2013, 13, 199. [Google Scholar] [CrossRef] [PubMed]
- Tezval, H.; Merseburger, A.S.; Matuschek, I.; Machtens, S.; Kuczyk, M.A.; Serth, J. RASSF1A protein expression and correlation with clinicopathological parameters in renal cell carcinoma. BMC Urol. 2008, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Tezval, H.; Jurk, S.; Atschekzei, F.; Serth, J.; Kuczyk, M.A.; Merseburger, A.S. The involvement of altered corticotropin releasing factor receptor 2 expression in prostate cancer due to alteration of anti-angiogenic signaling pathways. Prostate 2009, 69, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Colella, S.; Shen, L.; Baggerly, K.A.; Issa, J.P.; Krahe, R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques 2003, 35, 146–150. [Google Scholar] [CrossRef]
- Gebauer, K.; Peters, I.; Dubrowinskaja, N.; Hennenlotter, J.; Abbas, M.; Scherer, R.; Tezval, H.; Merseburger, A.S.; Stenzl, A.; Kuczyk, M.A.; et al. Hsa-mir-124-3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br. J. Cancer 2013, 108, 131–138. [Google Scholar] [CrossRef]
- Weisenberger, D.J.; Campan, M.; Long, T.I.; Kim, M.; Woods, C.; Fiala, E.; Ehrlich, M.; Laird, P.W. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005, 33, 6823–6836. [Google Scholar] [CrossRef]


| CDH3 Methylation | OR (95% CI) | p Value |
|---|---|---|
| ccRCC vs. papRCC | 11.64 (2.47–72.30) | 0.0041 |
| Tumor stage (low vs. high T *) | ||
| All RCC | 6.16 (1.99–21.60) | 0.0026 |
| ccRCC | 3.49 (0.98–13.90) | 0.061 |
| Lymph node status (N0 vs. N) | ||
| All RCC | 1.12 (0.19–5.60) | 0.893 |
| ccRCC | 0.98 (0.09–8.14) | 0.983 |
| Metastasis (M0 vs. M) | ||
| All RCC | 1.90 (0.56–6.43) | 0.297 |
| ccRCC | 1.00 (0.24–4.04) | 0.995 |
| Differentiation (low vs. high G **) | ||
| All RCC | 5.59 (1.41–24.86) | 0.017 |
| ccRCC | 4.67 (0.98–25.68) | 0.059 |
| State of disease (loc. vs. adv. ***) | ||
| All RCC | 6.94 (2.26–24.41) | 0.0013 |
| ccRCC | 4.31 (1.20–17.94) | 0.032 |
| Total Cases, n (%) | 107 (100%) |
|---|---|
| Histology | |
| ccRCC | 75 (70.1%) |
| papRCC | 22 (20.6%) |
| Chromophobe | 3 (2.8%) |
| Mixed histology | 3 (2.8%) |
| Other | 4 (3.7%) |
| Sex | |
| Female | 40 (37.4%) |
| Male | 67 (62.6%) |
| Age, median (range) | 64 years (35–91) |
| Synchronous lymph node metastasis | |
| N0 | 95 (88.8%) |
| N+ | 12 (11.2%) |
| Synchronous distant metastasis | |
| M0 | 85 (79.4%) |
| M+ | 22 (20.6%) |
| T-classification | |
| pT1 | 59 (55.14%) |
| pT2 | 7 (6.5) |
| pT3 | 39 (36.45%) |
| pT4 | 1 (0.93%) |
| na | 1 (0.93%) |
| Differentiation | |
| G1 | 20 (18.7%) |
| G1–G2 | 14 (13.1%) |
| G2 | 56 (52.3%) |
| G2–G3 | 6 (5.6%) |
| G3 | 11 (10.3%) |
| State of disease | |
| Localized (≤pT2, N0, M0, G1 or G1–2) | 56 (52.3%) |
| Advanced (≥pT3 and/or N+, M+ or G2–3 or G3) | 50 (46.73%) |
| NA | 1 (0.9%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faraj Tabrizi, P.; Peters, I.; Schimansky, I.; Dubrowinskaja, N.; Reese, C.; Tezval, H.; Kuczyk, M.A.; Serth, J. Alteration of Cadherin 3 Expression and DNA Methylation in Association with Aggressive Renal Cell Carcinoma. Int. J. Mol. Sci. 2023, 24, 16476. https://doi.org/10.3390/ijms242216476
Faraj Tabrizi P, Peters I, Schimansky I, Dubrowinskaja N, Reese C, Tezval H, Kuczyk MA, Serth J. Alteration of Cadherin 3 Expression and DNA Methylation in Association with Aggressive Renal Cell Carcinoma. International Journal of Molecular Sciences. 2023; 24(22):16476. https://doi.org/10.3390/ijms242216476
Chicago/Turabian StyleFaraj Tabrizi, Pouriya, Inga Peters, Inga Schimansky, Natalia Dubrowinskaja, Christel Reese, Hossein Tezval, Markus Antonius Kuczyk, and Jürgen Serth. 2023. "Alteration of Cadherin 3 Expression and DNA Methylation in Association with Aggressive Renal Cell Carcinoma" International Journal of Molecular Sciences 24, no. 22: 16476. https://doi.org/10.3390/ijms242216476
APA StyleFaraj Tabrizi, P., Peters, I., Schimansky, I., Dubrowinskaja, N., Reese, C., Tezval, H., Kuczyk, M. A., & Serth, J. (2023). Alteration of Cadherin 3 Expression and DNA Methylation in Association with Aggressive Renal Cell Carcinoma. International Journal of Molecular Sciences, 24(22), 16476. https://doi.org/10.3390/ijms242216476





