Microalga Broths Synthesize Antibacterial and Non-Cytotoxic Silver Nanoparticles Showing Synergy with Antibiotics and Bacterial ROS Induction and Can Be Reused for Successive AgNP Batches
Abstract
:1. Introduction
2. Results
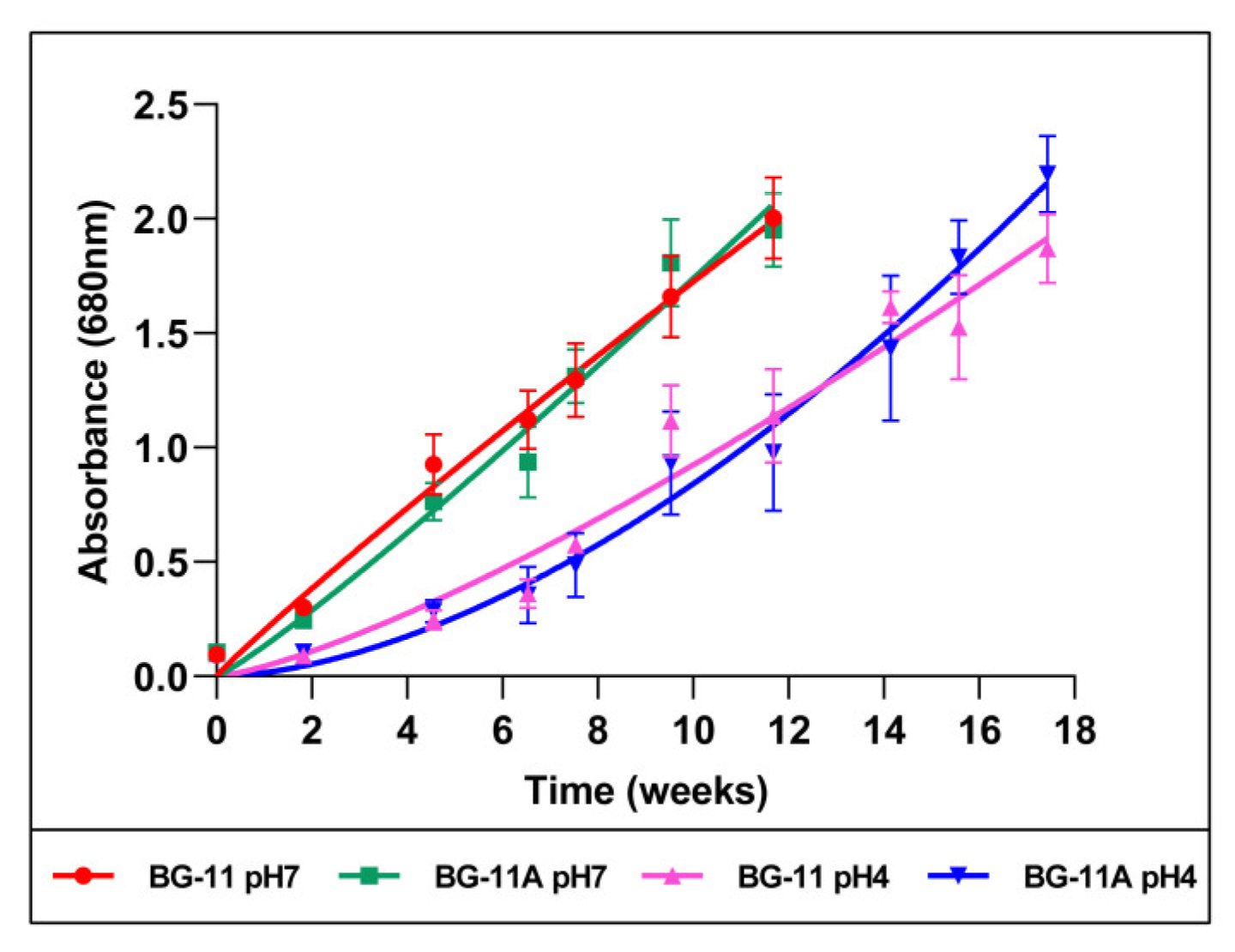
2.1. Microalga Cultures and Growth Curves
2.2. Biosynthesis of Silver Nanoparticles
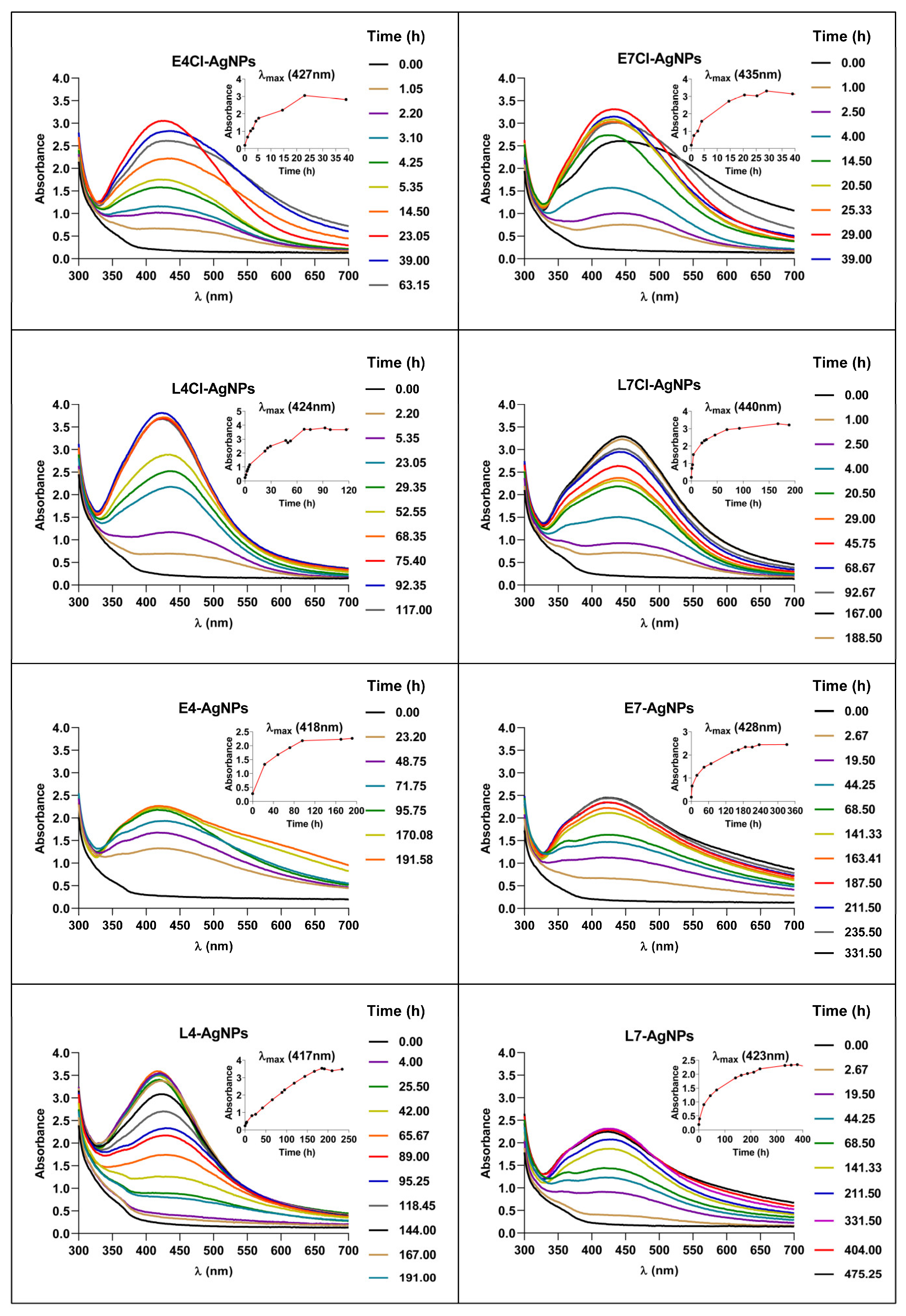
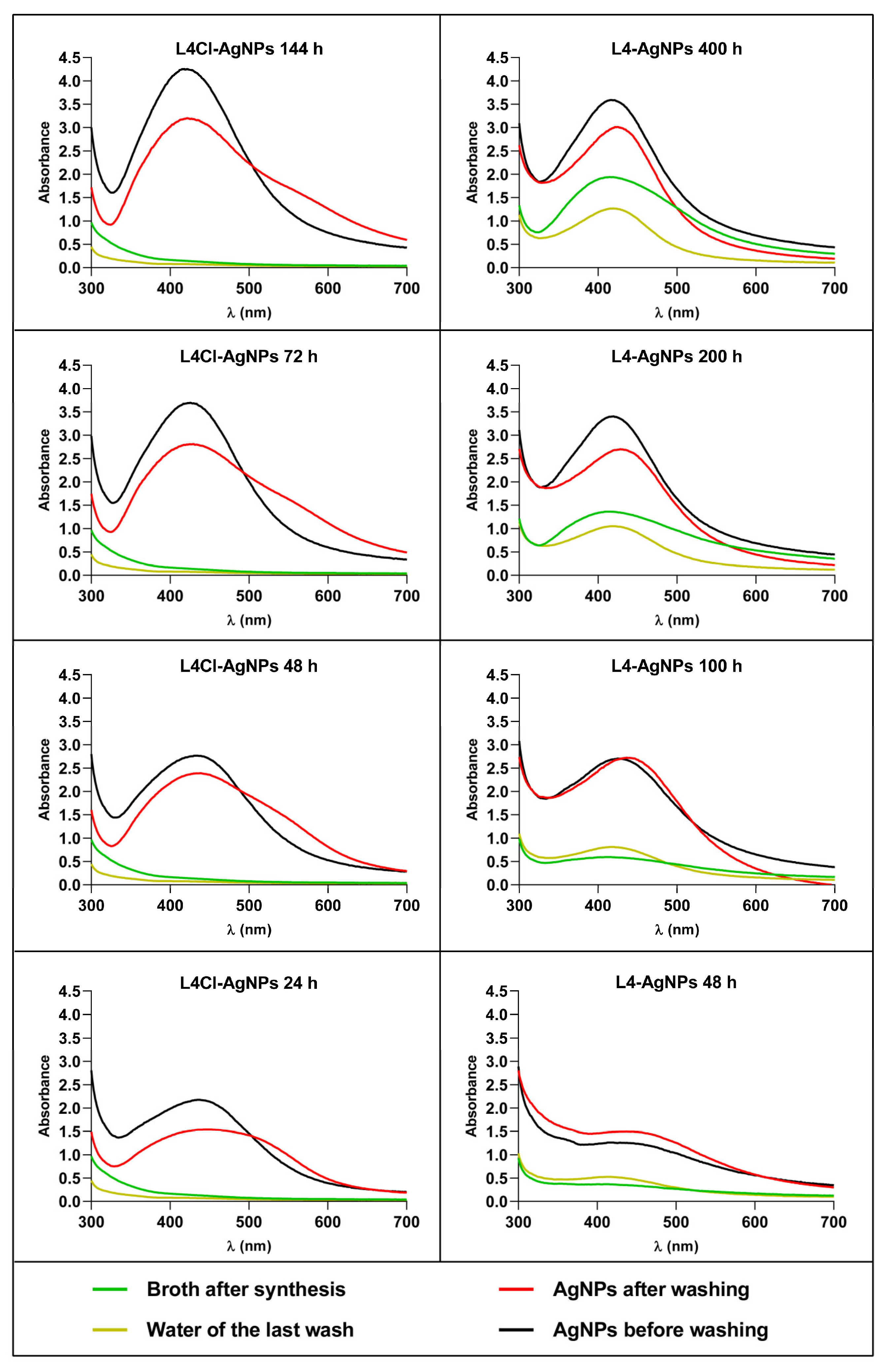
2.2.1. Synthesis Kinetics and Stability after Washing of AgNPs Produced at Different Times of the Synthesis Reaction
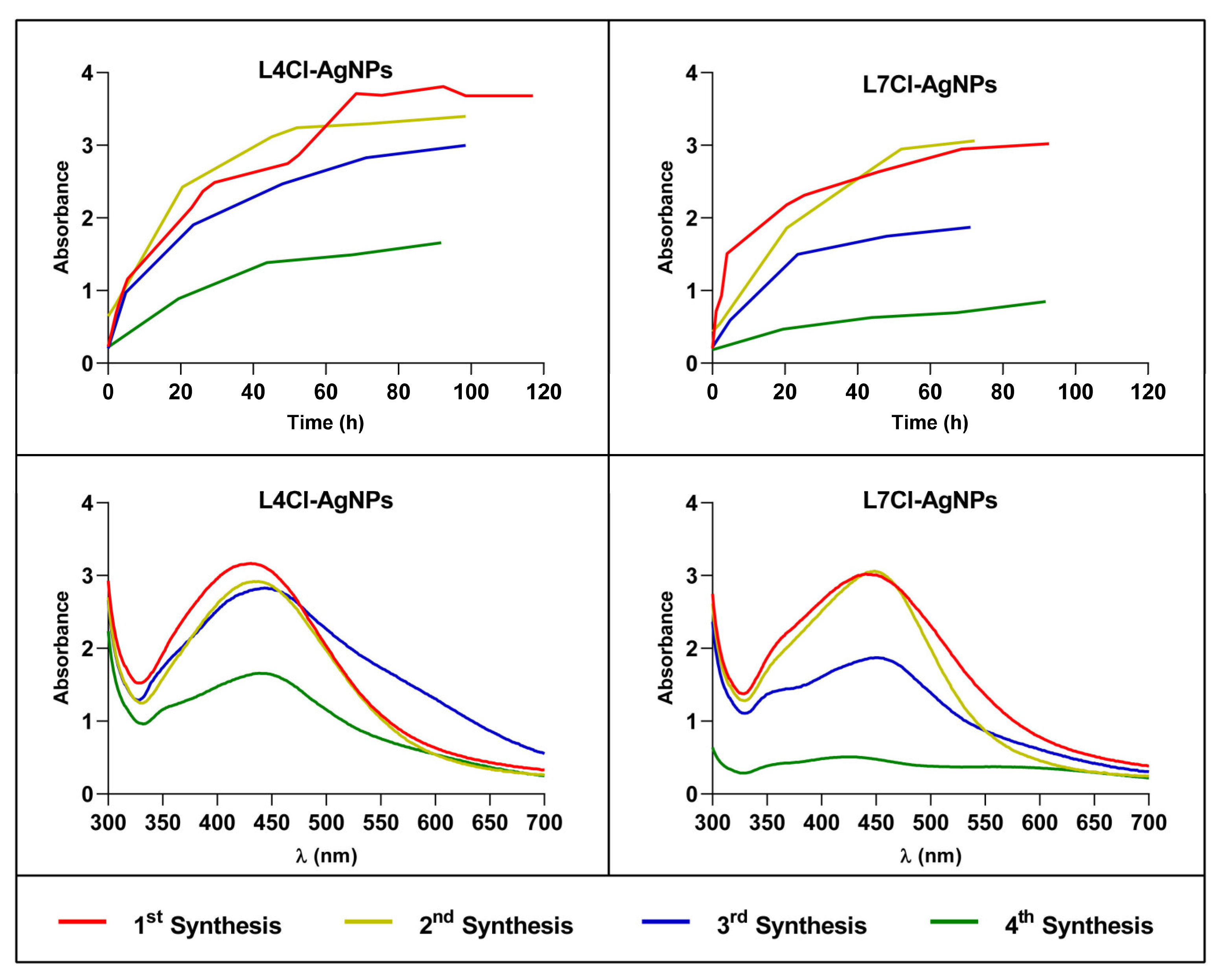
2.2.2. Kinetics of Successive AgNP Syntheses from the Same Broth
2.3. Characterization of Biosynthesized AgNPs
2.3.1. Physicochemical Properties
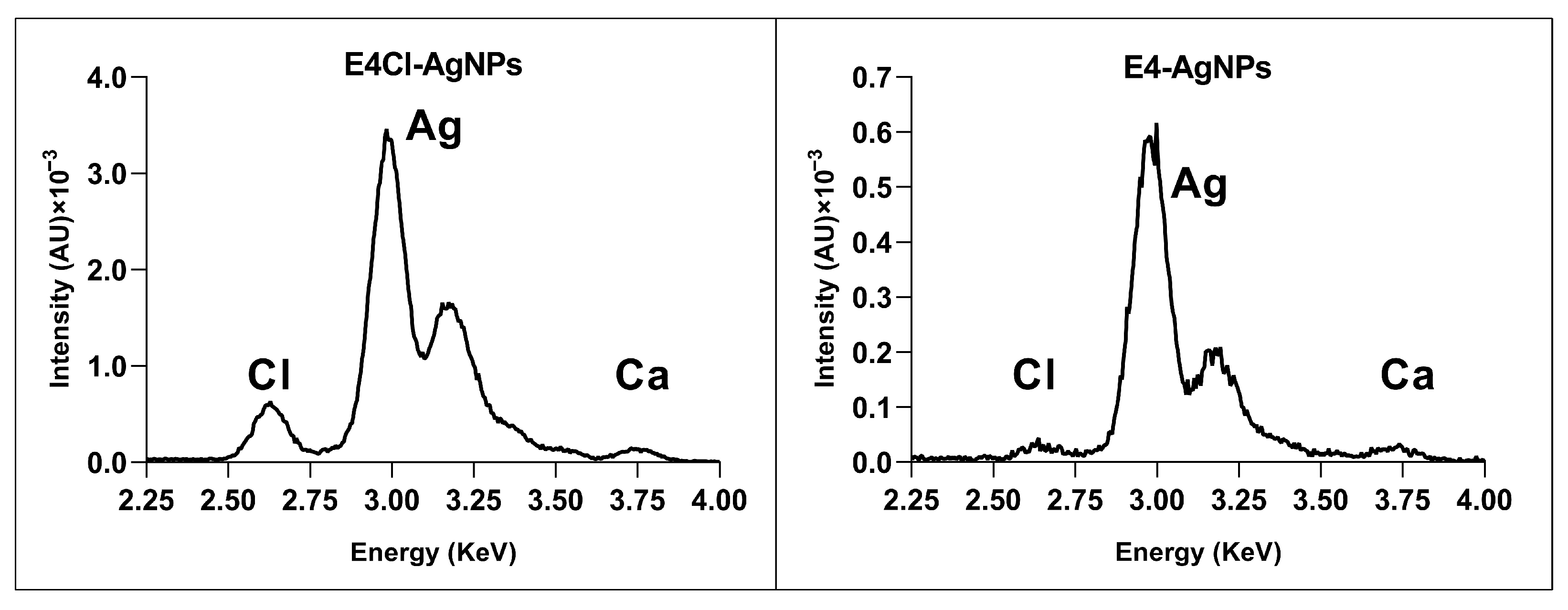
Elemental Composition of AgNPs by Total Reflection X-ray Fluorescence (TXRF)
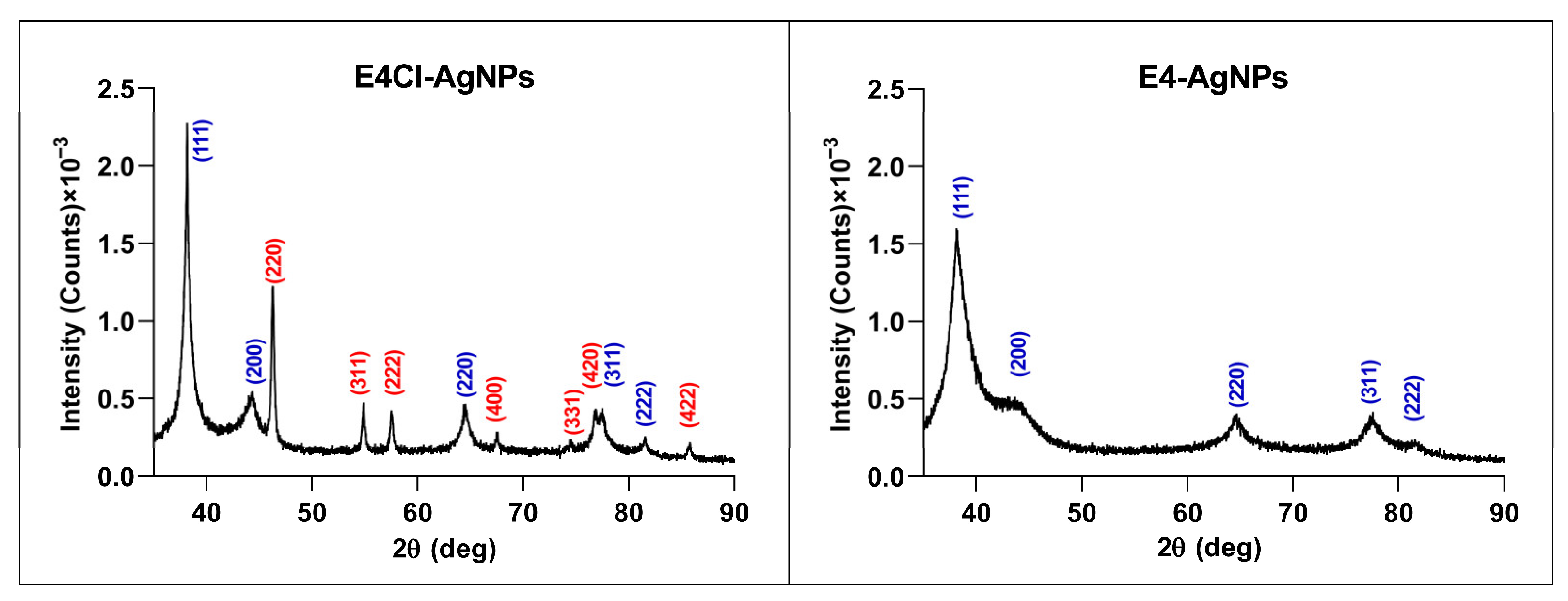
Crystallinity by Powder X-ray Diffraction (XRD)
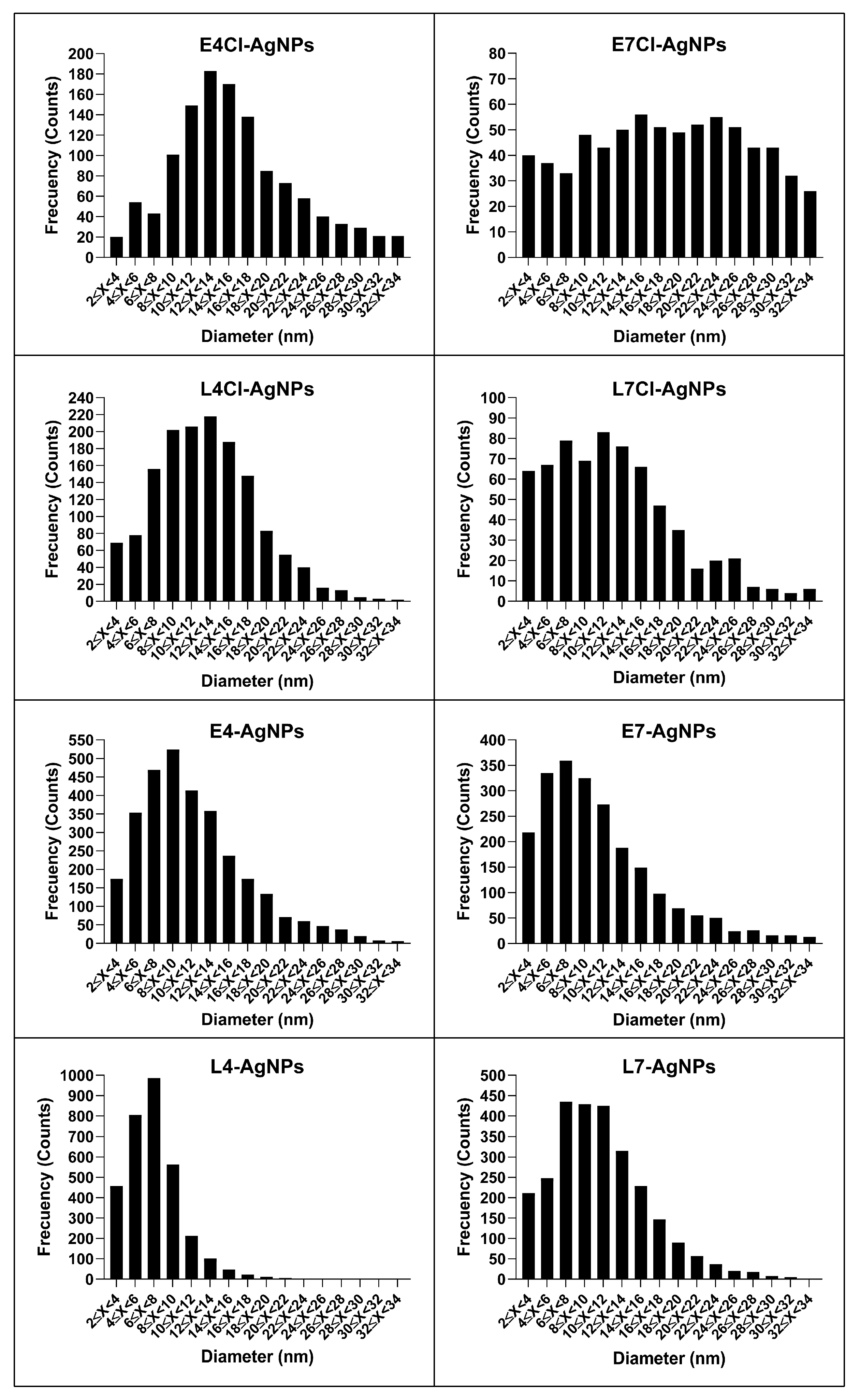
AgNP Core Size and Shape by Transmission Electron Microscopy (TEM)
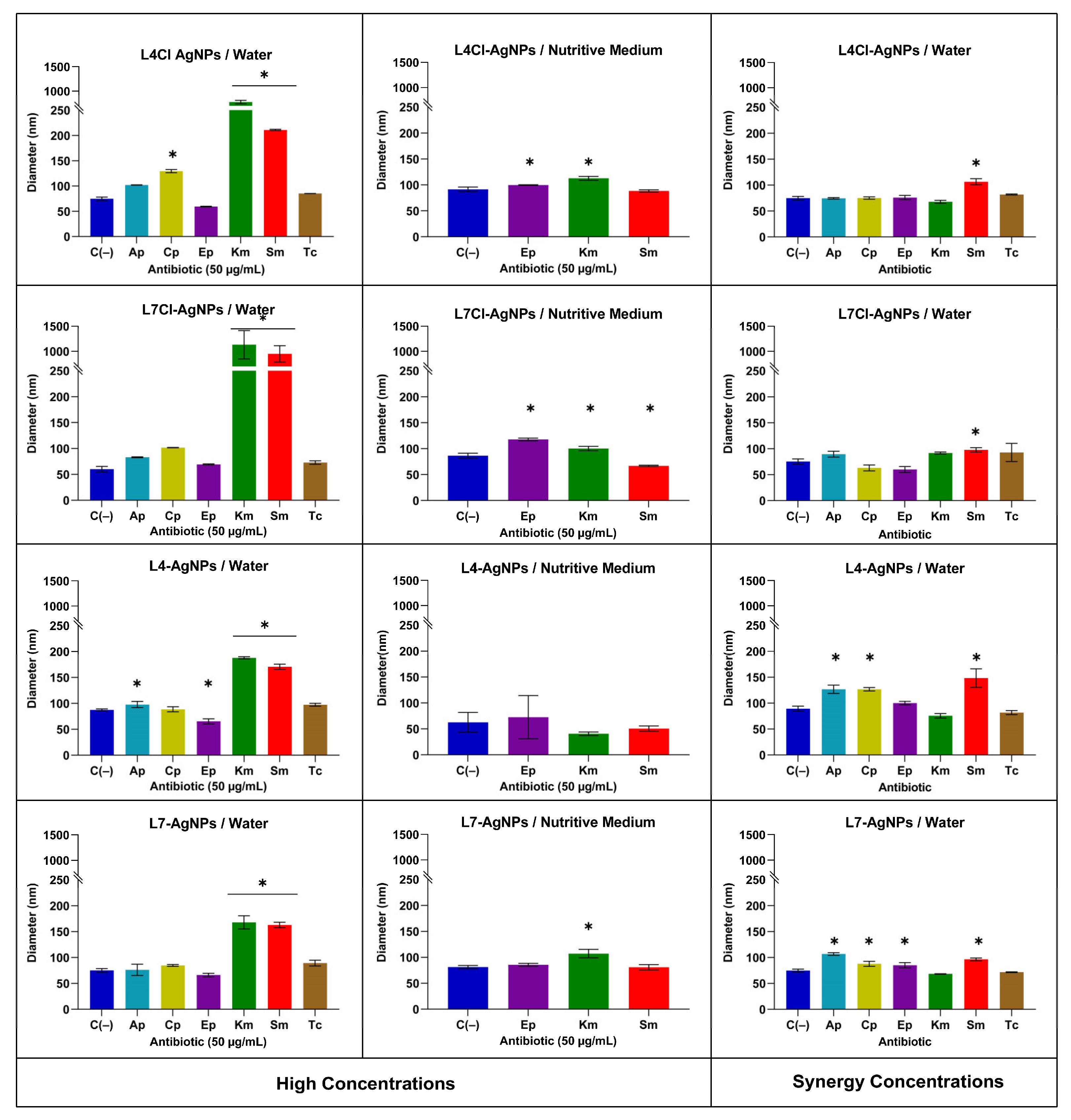
The Z-Potential and the Hydrodynamic Diameter of the AgNPs by Dynamic Light Scattering (DLS)
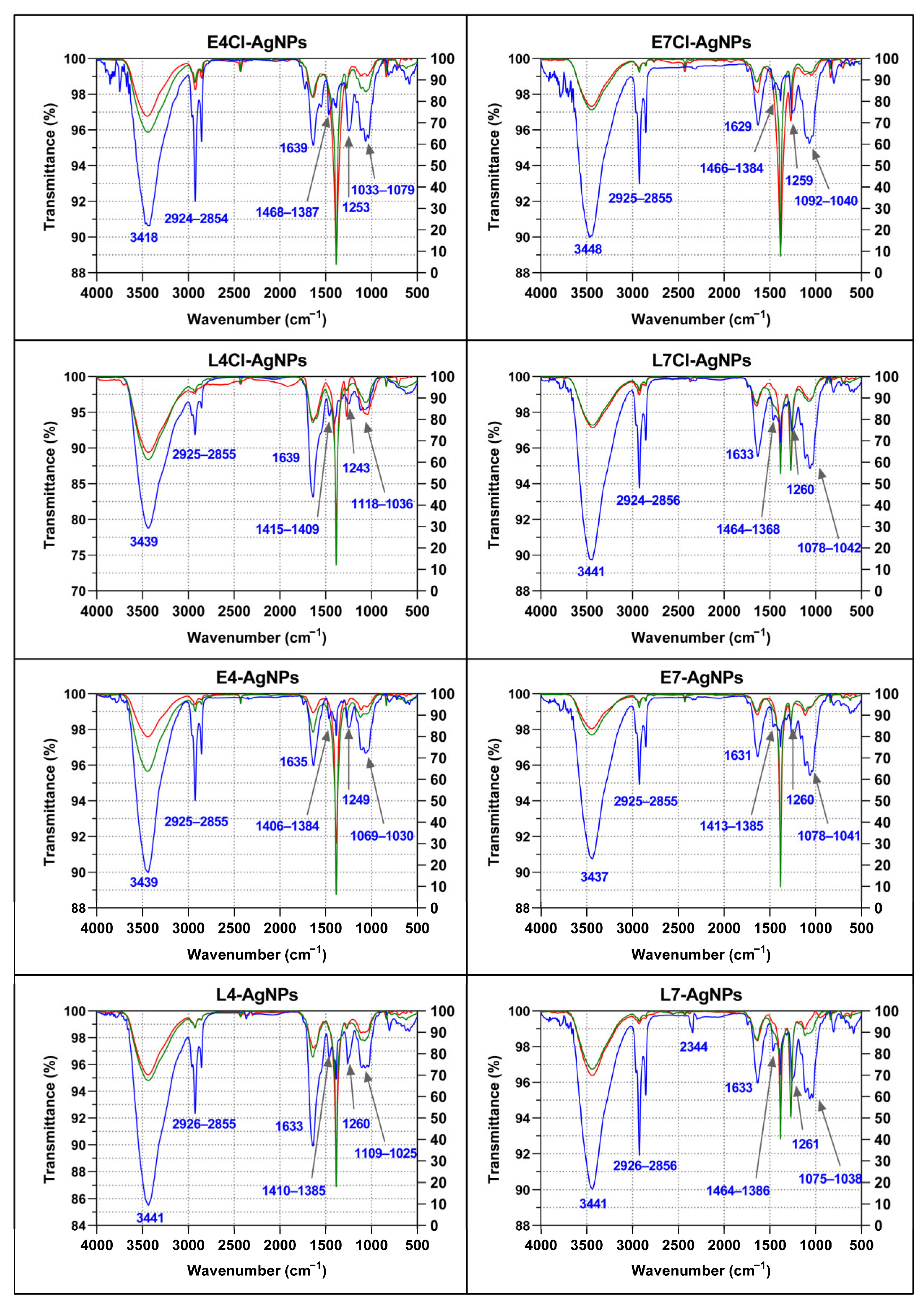
Corona Composition by Fourier Transform Infrared Spectroscopy (FTIR)
2.3.2. Biological Activity of the AgNPs
Antibacterial and Antibiofilm Activity
Antibacterial Activity of AgNPs Harvested at Different Times in the Synthesis Reaction
Antibacterial Activity of Successive Batches of AgNPs Using the Same Broth
AgNPs’ Cytotoxicity on Cultured Intestinal Human Cells
2.3.3. Antibacterial Synergy of the AgNPs with Classic Antibiotics by the Checkboard Assay
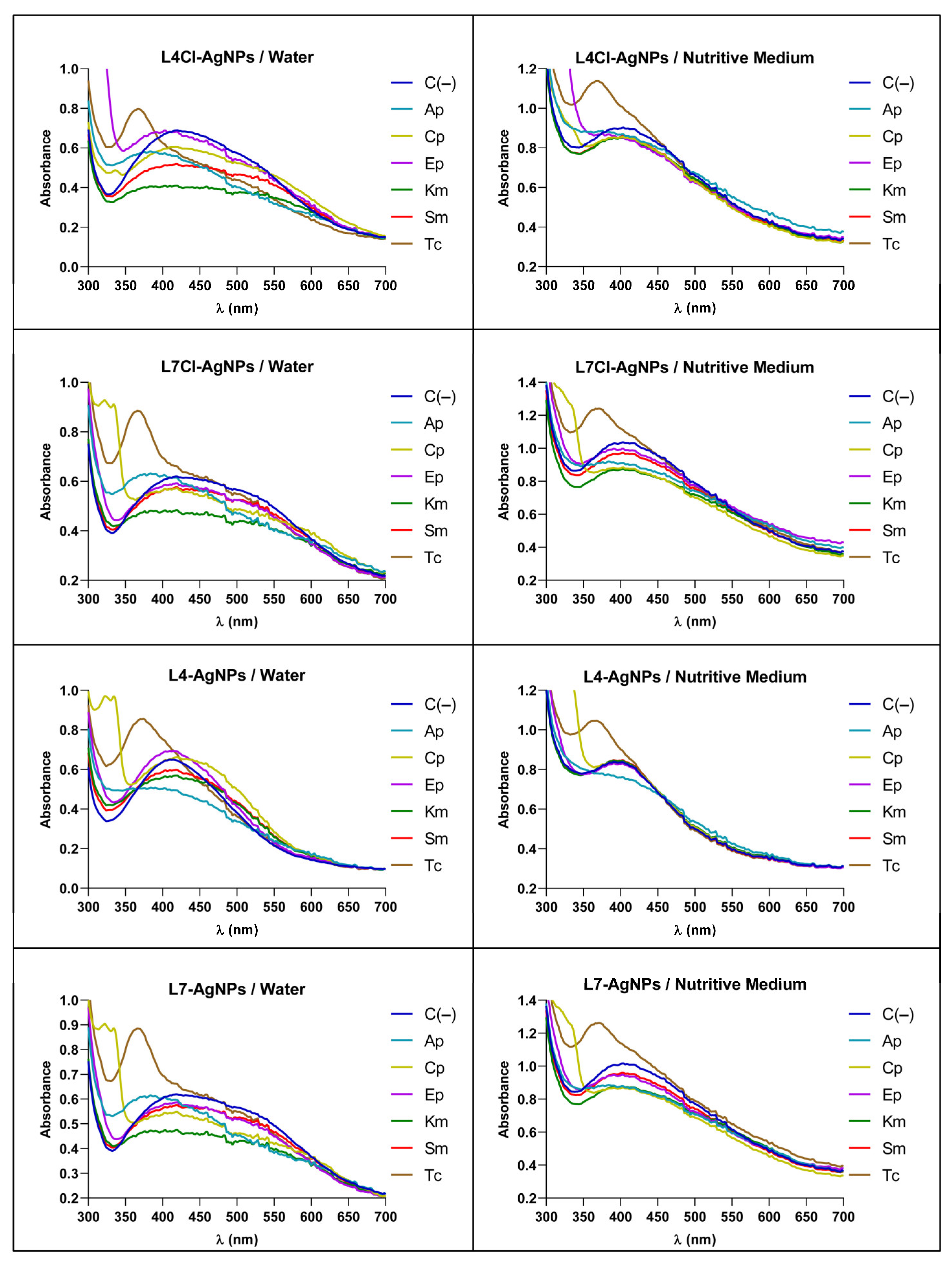
Insights into the Mechanism of the AgNPs’ Antibiotic Synergy
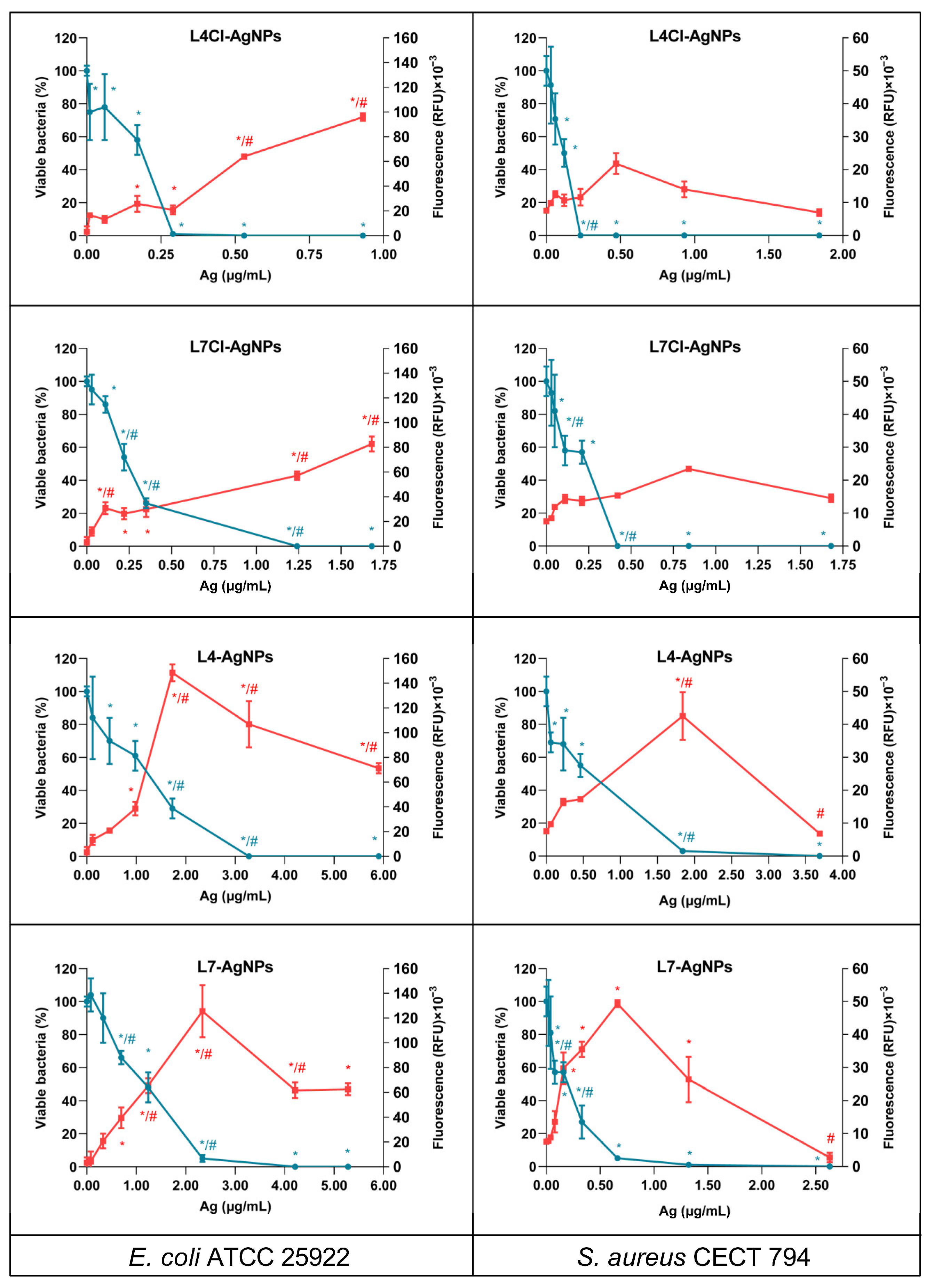
2.4. ROS Production and Bacterial Killing by the AgNPs
3. Discussion
3.1. Microalga Growth and Tested Conditions
3.2. Kinetics of AgNP Synthesis
3.3. Insight into the AgNPs’ Synthesis over Time and Reuse of Broths
3.4. Physicochemical Characteristics of the AgNPs
3.5. Antibacterial Activity
3.6. Cytotoxicity
3.7. Synergy
3.8. ROS and Bacterial Killing by the AgNPs
4. Materials and Methods
4.1. Microorganisms and Culture Media Used
4.2. Cell-Free Broth Preparation
4.3. Green Synthesis of AgNPs
4.4. Characterization of the AgNPs
4.4.1. Determination of the AgNPs’ Physicochemical Properties
4.4.2. Evaluation of the AgNPs’ Antibacterial Activity
4.4.3. Assessment of the AgNPs’ Cytotoxicity on Cultured Human Cells
4.4.4. Assessment of the Synergy between AgNPs and Classic Antibiotics
Analysis of the Interaction of AgNPs and Antibiotics
4.5. Reactive Oxygen Species (ROS) Production by the AgNPs and Their Bacterial Killing Rate
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; The Review on Antimicrobial Resistance; HM Government and the Wellcome Trust: London, UK, 2016. [Google Scholar]
- Rudramurthy, G.; Swamy, M.K.; Sinniah, U.R.; Ghasemzadeh, A. Nanoparticles: Alternatives Against Drug-Resistant Pathogenic Microbes. Molecules 2016, 21, 836. [Google Scholar] [CrossRef] [PubMed]
- Mba, I.E.; Nweze, E.I. Nanoparticles as Therapeutic Options for Treating Multidrug-Resistant Bacteria: Research Progress, Challenges, and Prospects. World J. Microbiol. Biotechnol. 2021, 37, 108. [Google Scholar] [CrossRef] [PubMed]
- Crisan, C.M.; Mocan, T.; Manolea, M.; Lasca, L.I.; Tăbăran, F.-A.; Mocan, L. Review on Silver Nanoparticles as a Novel Class of Antibacterial Solutions. Appl. Sci. 2021, 11, 1120. [Google Scholar] [CrossRef]
- Mateo, E.M.; Jiménez, M. Silver Nanoparticle-Based Therapy: Can It Be Useful to Combat Multi-Drug Resistant Bacteria? Antibiotics 2022, 11, 1205. [Google Scholar] [CrossRef] [PubMed]
- Plotniece, A.; Sobolev, A.; Supuran, C.T.; Carta, F.; Björkling, F.; Franzyk, H.; Yli-Kauhaluoma, J.; Augustyns, K.; Cos, P.; De Vooght, L.; et al. Selected Strategies to Fight Pathogenic Bacteria. J. Enzyme Inhib. Med. Chem. 2023, 38, 2155816. [Google Scholar] [CrossRef] [PubMed]
- Gordienko, M.G.; Palchikova, V.V.; Kalenov, S.V.; Belov, A.A.; Lyasnikova, V.N.; Poberezhniy, D.Y.; Chibisova, A.V.; Sorokin, V.V.; Skladnev, D.A. Antimicrobial Activity of Silver Salt and Silver Nanoparticles in Different Forms against Microorganisms of Different Taxonomic Groups. J. Hazard. Mater. 2019, 378, 120754. [Google Scholar] [CrossRef]
- Kędziora, A.; Speruda, M.; Krzyżewska, E.; Rybka, J.; Łukowiak, A.; Bugla-Płoskońska, G. Similarities and Differences between Silver Ions and Silver in Nanoforms as Antibacterial Agents. Int. J. Mol. Sci. 2018, 19, 444. [Google Scholar] [CrossRef]
- Kodaparthi, A.; Hameeda, B.; Bastipati, S.B.; Golla, S. Bactericidal Effects: Microbial Nanoparticles as Next-Generation Antimicrobials. In Microbial Processes for Synthesizing Nanomaterials; Environmental and Microbial Biotechnology; Maddela, N.R., Rodríguez Díaz, J.M., Branco da Silva Montenegro, M.C., Prasad, R., Eds.; Springer: Singapore, 2023; pp. 261–283. [Google Scholar] [CrossRef]
- Chapa González, C.; González García, L.I.; Burciaga Jurado, L.G.; Carrillo Castillo, A. Bactericidal Activity of Silver Nanoparticles in Drug-Resistant Bacteria. Braz. J. Microbiol. 2023, 54, 691–701. [Google Scholar] [CrossRef]
- De Azeredo, H.M.C. Antimicrobial Nanostructures in Food Packaging. Trends Food Sci. Technol. 2013, 30, 56–69. [Google Scholar] [CrossRef]
- Xi, J.; Kan, W.; Zhu, Y.; Huang, S.; Wu, L.; Wang, J. Synthesis of Silver Nanoparticles Using Eucommia ulmoides Extract and Their Potential Biological Function in Cosmetics. Heliyon 2022, 8, e10021. [Google Scholar] [CrossRef]
- Rujido-Santos, I.; del Castillo Busto, M.E.; Abad-Alvaro, I.; Herbello-Hermelo, P.; Bermejo-Barrera, P.; Barciela-Alonso, M.C.; Goneaga-Infante, H.; Moreda-Piñeiro, A. SpICP-MS Characterisation of Released Silver Nanoparticles from (Nano)Textile Products. Spectrochim. Acta B At. Spectrosc. 2022, 195, 106505. [Google Scholar] [CrossRef]
- Amaro, F.; Morón, Á.; Díaz, S.; Martín-González, A.; Gutiérrez, J.C. Metallic Nanoparticles—Friends or Foes in the Battle against Antibiotic-Resistant Bacteria? Microorganisms 2021, 9, 364. [Google Scholar] [CrossRef] [PubMed]
- Kawata, K.; Osawa, M.; Okabe, S. In Vitro Toxicity of Silver Nanoparticles at Noncytotoxic Doses to HepG2 Human Hepatoma Cells. Environ. Sci. Technol. 2009, 43, 6046–6051. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, M.; Karns, M.; Goodson, M.; Rowe, J.; Hussain, S.M.; Schlager, J.J.; Hong, Y. DNA Damage Response to Different Surface Chemistry of Silver Nanoparticles in Mammalian Cells. Toxicol. Appl. Pharmacol. 2008, 233, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Garrido, I.; Pérez, S.; Blasco, J. Toxicity of Silver and Gold Nanoparticles on Marine Microalgae. Mar. Environ. Res. 2015, 111, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Daphedar, A.; Taranath, T.C. Characterization and Cytotoxic Effect of Biogenic Silver Nanoparticles on Mitotic Chromosomes of Drimia polyantha (Blatt. & McCann) Stearn. Toxicol. Rep. 2018, 5, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Zannatul, F.; Nemmar, A. Health Impact of Silver Nanoparticles: A Review of the Biodistribution and Toxicity Following Various Routes of Exposure. Int. J. Mol. Sci. 2020, 2, 2375. [Google Scholar] [CrossRef]
- Mussin, J.; Robles-Botero, V.; Casañas-Pimentel, R.; Rojas, F.; Angiolella, L.; San Martín-Martínez, E.; Giusiano, G. Antimicrobial and Cytotoxic Activity of Green Synthesis Silver Nanoparticles Targeting Skin and Soft Tissue Infectious Agents. Sci. Rep. 2021, 11, 14566. [Google Scholar] [CrossRef]
- Skóra, B.; Krajewska, U.; Nowak, A.; Dziedzic, A.; Barylyak, A.; Kus-Liśkiewicz, M. Noncytotoxic Silver Nanoparticles as a New Antimicrobial Strategy. Sci. Rep. 2021, 11, 13451. [Google Scholar] [CrossRef]
- Samuel, M.S.; Jose, S.; Selvarajan, E.; Mathimani, T.; Pugazhendhi, A. Biosynthesized Silver Nanoparticles Using Bacillus amyloliquefaciens; Application for Cytotoxicity Effect on A549 Cell Line and Photocatalytic Degradation of p-Nitrophenol. J. Photochem. Photobiol. B Biol. 2020, 202, 111642. [Google Scholar] [CrossRef] [PubMed]
- Vishwanath, R.; Negi, B. Conventional and Green Methods of Synthesis of Silver Nanoparticles and Their Antimicrobial Properties. Curr. Res. Green Sustain. Chem. 2021, 4, 100205. [Google Scholar] [CrossRef]
- Miu, B.A.; Dinischiotu, A. New Green Approaches in Nanoparticles Synthesis: An Overview. Molecules 2022, 27, 6472. [Google Scholar] [CrossRef] [PubMed]
- Babaniyi, B.R.; Ogundele, O.D.; Thompson, S.O.; Aransiola, S.A. Microbial Nanomaterial Synthesis: Types and Applications. In Microbial Processes for Synthesizing Nanomaterials; Environmental and Microbial, Biotechnology; Maddela, N.R., Rodríguez Díaz, J.M., Branco da Silva Montenegro, M.C., Prasad, R., Eds.; Springer: Singapore, 2023; pp. 3–28. [Google Scholar] [CrossRef]
- Huq, M.A.; Ashrafudoulla, M.; Rahman, M.M.; Balusamy, S.R.; Akter, S. Green Synthesis and Potential Antibacterial Applications of Bioactive Silver Nanoparticles: A Review. Polymers 2022, 14, 742. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Usman, M.; Ji, Z.; Rafiq, M.; Yu, B.; Shen, Y.; Cong, H. Nature-Inspired Biogenic Synthesis of Silver Nanoparticles for Antibacterial Applications. Mater. Today Chem. 2023, 27, 101339. [Google Scholar] [CrossRef]
- Clifford, C.B. What are Algae? EGEE 439: Alternative Fuels from Biomass Sources. The John A. Dutton Institute for Teaching and Learning Excellence. College of Earth and Mineral Sciences at The Pennsylvania State University. Available online: https://www.e-education.psu.edu/egee439/node/693 (accessed on 9 August 2023).
- Khan, M.I.; Shin, J.H.; Kim, J.D. The Promising Future of Microalgae: Current Status, Challenges, and Optimization of a Sustainable and Renewable Industry for Biofuels, Feed, and Other Products. Microb. Cell Factories 2018, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Lan, C.Q. Advances in Biosynthesis of Noble Metal Nanoparticles Mediated by Photosynthetic Organisms—A Review. Colloids Surf. B Biointerfaces 2019, 184, 110519. [Google Scholar] [CrossRef] [PubMed]
- Akintelu, S.A.; Bo, Y.; Folorunso, A.S. A Review on Synthesis, Optimization, Mechanism, Characterization, and Antibacterial Application of Silver Nanoparticles Synthesized from Plants. J. Chem. 2020, 2020, 3189043. [Google Scholar] [CrossRef]
- Rizwan, M.; Mujtaba, G.; Memon, S.; Lee, K.; Rashid, N. Exploring the Potential of Microalgae for New Biotechnology Applications and Beyond: A Review. Renew. Sustain. Energy Rev. 2018, 92, 394–404. [Google Scholar] [CrossRef]
- Jacob, J.M.; Ravindran, R.; Narayanan, M.; Samuel, S.M.; Pugazhendhi, A.; Kumar, G. Microalgae: A Prospective Low Cost Green Alternative for Nanoparticle Synthesis. Curr. Opin. Environ. Sci. Health 2021, 20, 100163. [Google Scholar] [CrossRef]
- Moraes, L.C.; Figueiredo, R.C.; Ribeiro-Andrade, R.; Pontes-Silva, A.V.; Arantes, M.L.; Giani, A.; Figueredo, C.C. High Diversity of Microalgae as a Tool for the Synthesis of Different Silver Nanoparticles: A Species-Specific Green Synthesis. Colloid Interface Sci. Commun. 2021, 42, 100420. [Google Scholar] [CrossRef]
- Mukherjee, A.; Sarkar, D.; Sasmal, S. A Review of Green Synthesis of Metal Nanoparticles Using Algae. Front. Microbiol. 2021, 12, 693899. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.S.; Low, S.S.; Chew, K.W.; Ling, T.C.; Rinklebe, J.; Juan, J.C.; Ng, E.P.; Show, P.L. Prospects and Environmental Sustainability of Phyconanotechnology: A review on Algae-Mediated Metal Nanoparticles Synthesis and Mechanism. Environ. Res. 2022, 212, 113140. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.; Berthold, D.; Puranik, P.; Gantar, M. Screening of Cyanobacteria and Microalgae for Their Ability to Synthesize Silver Nanoparticles with Antibacterial Activity. Biotechnol. Rep. 2015, 5, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Shankar, P.D.; Shobana, S.; Karuppusamy, I.; Pugazhendhi, A.; Ramkumar, V.S.; Arvindnarayan, S.; Kumar, G. A Review on the Biosynthesis of Metallic Nanoparticles (Gold and Silver) Using Bio-Components of Microalgae: Formation Mechanism and Applications. Enzyme Microb. Technol. 2016, 95, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Hamouda, R.A.; Hussein, M.H.; Abo-Elmagd, R.A.; Bawazir, S.S. Synthesis and Biological Characterization of Silver Nanoparticles Derived from the Cyanobacterium Oscillatoria limnetica. Sci. Rep. 2019, 9, 13071. [Google Scholar] [CrossRef] [PubMed]
- Kholssi, R.; Ramos, P.V.; Marks, E.A.N.; Montero, O.; Rad, C. Biotechnological Uses of Microalgae: A Review on The State of The Art and Challenges for The Circular Economy. Biocatal. Agric. Biotechnol. 2021, 36, 102114. [Google Scholar] [CrossRef]
- Olabi, A.G.; Shehata, N.; Sayed, E.T.; Rodriguez, C.; Anyanwu, R.C.; Russell, C.; Abdelkareem, M.A. Role of Microalgae in Achieving Sustainable Development Goals and Circular Economy. Sci. Total Environ. 2023, 854, 158689. [Google Scholar] [CrossRef]
- Barwal, I.; Ranjan, P.; Kateriya, S.; Yadav, S.C. Cellular Oxido-Reductive Proteins of Chlamydomonas reinhardtii Control the Biosynthesis of Silver Nanoparticles. J. Nanobiotechnol. 2011, 9, 56. [Google Scholar] [CrossRef]
- Mora-Godínez, S.; Abril-Martínez, F.; Pacheco, A. Green Synthesis of Silver Nanoparticles Using Microalgae Acclimated to High CO2. Mater. Today Proc. 2022, 48, 5–9. [Google Scholar] [CrossRef]
- El Ouardy, K.; Lbouhmadi, R.; Attaoui, H.; Mouzaki, M.; Mouine, H.; Lemkhente, Z.; Mir, Y. Biosynthesis of Silver Nanoparticles Using Parachlorella kessleri and Cyclotella spp. and Evaluation of Its Antibacterial Activity. SSRN J. 2022, 24, 10599. [Google Scholar] [CrossRef]
- Bruna, T.; Maldonado-Bravo, F.; Jara, P.; Caro, N. Silver Nanoparticles and Their Antibacterial Applications. Int. J. Mol. Sci. 2021, 22, 7202. [Google Scholar] [CrossRef] [PubMed]
- Feizi, S.; Cooksley, C.M.; Nepal, R.; Psaltis, A.J.; Wormald, P.-J.; Vreugde, S. Silver Nanoparticles as a Bioadjuvant of Antibiotics against Biofilm-Mediated Infections with Methicillin-Resistant Staphylococcus aureus and Pseudomonas aeruginosa in Chronic Rhinosinusitis Patients. Pathology 2022, 54, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Carrizales, M.; Velasco, K.; Castillo, C.; Flores, A.; Magaña, M.; Martinez-Castanon, G.; Martinez-Gutierrez, F. In Vitro Synergism of Silver Nanoparticles with Antibiotics as an Alternative Treatment in Multiresistant Uropathogens. Antibiotics 2018, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Allend, S.O.; Garcia, M.O.; da Cunha, K.F.; de Albernaz, D.T.F.; da Silva, M.E.; Ishikame, R.Y.; Panagio, L.A.; Nakazaro, G.; Reis, G.F.; Pereira, D.B.; et al. Biogenic Silver Nanoparticle (Bio-AgNP) Has an Antibacterial Effect against Carbapenem-resistant Acinetobacter baumannii with Synergism and Additivity When Combined with Polymyxin B. J. Appl. Microbiol. 2022, 132, 1036–1047. [Google Scholar] [CrossRef] [PubMed]
- Garibo Ruiz, D.; Nefedova, E.; Shkil, N.N.; Shkil, N.A.; Vazquez-Gomez, R.L.; Pestryakov, A.; Bogdanchikova, N. Silver Nanoparticles Targeting the Drug Resistance Problem of Streptococcus dysgalactiae: Susceptibility to Antibiotics and Efflux Effect. Int. J. Mol. Sci. 2022, 23, 6024. [Google Scholar] [CrossRef] [PubMed]
- Malawong, S.; Thammawithan, S.; Sirithongsuk, P.; Daduang, S.; Klaynongsruang, S.; Wong, P.T.; Patramanon, R. Silver Nanoparticles Enhance Antimicrobial Efficacy of Antibiotics and Restore That Efficacy against the Melioidosis Pathogen. Antibiotics 2021, 10, 839. [Google Scholar] [CrossRef] [PubMed]
- McShan, D.; Zhang, Y.; Deng, H.; Ray, P.C.; Yu, H. Synergistic Antibacterial Effect of Silver Nanoparticles Combined with Ineffective Antibiotics on Drug Resistant Salmonella typhimurium DT104. J. Environ. Sci. Health C Toxicol. 2015, 33, 369–384. [Google Scholar] [CrossRef]
- Vazquez-Muñoz, R.; Meza-Villezcas, A.; Fournier, P.G.J.; Soria-Castro, E.; Juarez-Moreno, K.; Gallego-Hernández, A.L.; Bogdanchikova, N.; Vazquez-Duhalt, R.; Huerta-Saquero, A. Enhancement of Antibiotics Antimicrobial Activity Due to the Silver Nanoparticles Impact on the Cell Membrane. PLoS ONE 2019, 14, e0224904. [Google Scholar] [CrossRef]
- Panáček, A.; Smékalová, M.; Večeřová, R.; Bogdanová, K.; Röderová, M.; Kolář, M.; Kilianová, M.; Hradilová, Š.; Froning, J.P.; Havrdová, M.; et al. Silver Nanoparticles Strongly Enhance and Restore Bactericidal Activity of Inactive Antibiotics against Multiresistant Enterobacteriaceae. Colloids Surf. B Biointerfaces 2016, 142, 392–399. [Google Scholar] [CrossRef]
- Ahmad, S.A.; Das, S.S.; Khatoon, A.; Ansari, M.T.; Afzal, M.; Hasnain, M.S.; Nayak, A.K. Bactericidal Activity of Silver Nanoparticles: A Mechanistic Review. Mater. Sci. Energy Technol. 2020, 3, 756–769. [Google Scholar] [CrossRef]
- Dakal, T.C.; Kumar, A.; Majumdar, R.S.; Yadav, V. Mechanistic Basis of Antibacterial Actions of Silver Nanoparticles. Front. Microbiol. 2016, 7, 1831. [Google Scholar] [CrossRef] [PubMed]
- Navarro Gallón, S.M.; Alpaslan, E.; Wang, M.; Larese-Casanova, P.; Londoño, M.E.; Atehortúa, L.; Pavón, J.J.; Webster, T.J. Characterization and Study of the Antibacterial Mechanisms of Silver Nanoparticles Prepared with Microalgal Exopolysaccharides. Mater. Sci. Eng. C 2019, 99, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kaur, K. Chapter 8—Biological and Physical Applications of Silver Nanoparticles with Emerging Trends of Green Synthesis. In Engineered Nanomaterials—Health and Safety; Marius Avramescu, S., Akhtar, K., Fierascu, I., Bahadar Khan, S., Ali, F., Asiri, M.A., Eds.; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Khan, F.; Shahid, A.; Zhu, H.; Wang, N.; Javed, M.R.; Ahmad, N.; Xu, J.; Alam, A.; Mehmood, M.A. Prospects of Algae-Based Green Synthesis of Nanoparticles for Environmental Applications. Chemosphere 2022, 293, 133571. [Google Scholar] [CrossRef]
- Kareem, M.A.; Bello, I.T.; Shittu, H.A.; Awodele, M.K.; Adedokun, O.; Sanusi, Y.K. Green Synthesis of Silver Nanoparticles (AgNPs) for Optical and Photocatalytic Applications: A Review. IOP Conf. Ser. Mater. Sci. Eng. 2020, 805, 012020. [Google Scholar] [CrossRef]
- Gebauer, J.S.; Treuel, L. Influence of Individual Ionic Components on the Agglomeration Kinetics of Silver Nano-particles. J. Colloid Interface Sci. 2011, 354, 546–554. [Google Scholar] [CrossRef]
- Javani, S.; Marín, I.; Amils, R.; Abad, J.P. Four Psychrophilic Bacteria from Antarctica Extracellularly Biosynthesize at Low Temperature Highly Stable Silver Nanoparticles with Outstanding Antimicrobial Activity. Colloids Surf. A Physicochem. Eng. Asp. 2015, 483, 60–69. [Google Scholar] [CrossRef]
- Pernas-Pleite, C.; Conejo-Martínez, A.M.; Marín, I.; Abad, J.P. Green Extracellular Synthesis of Silver Nanoparticles by Pseudomonas alloputida, Their Growth and Biofilm-Formation Inhibitory Activities and Synergic Behavior with Three Classical Antibiotics. Molecules 2022, 27, 7589. [Google Scholar] [CrossRef]
- Nobbmann, U.L. Polydispersity—What Does It Mean for DLS and Chromatography? Available online: https://www.materials-talks.com/polydispersity-what-does-it-mean-for-dls-and-chromatography/ (accessed on 1 September 2023).
- Ferro, L.; Gojkovic, Z.; Gorzsás, A.; Funk, C. Statistical Methods for Rapid Quantification of Proteins, Lipids, and Carbohydrates in Nordic Microalgal Species Using ATR-FTIR Spectroscopy. Molecules 2019, 24, 3237. [Google Scholar] [CrossRef]
- Levison, M.E. Pharmacodynamics of Antimicrobial Drugs. Infect. Dis. Clin. N. Am. 2004, 18, 451–465. [Google Scholar] [CrossRef]
- Odds, F.C. Synergy, Antagonism, and What the Chequerboard Puts Between Them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.K.; Moellering, R.C., Jr.; Eliopoulos, G.M. Antimicrobial Combinations. In Antibiotics in Laboratory Medicine, 5th ed.; Lorian, V., Ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2005; pp. 365–440. [Google Scholar]
- Gómara, M.; Ramón-García, S. The FICI Paradigm: Correcting Flaws in Antimicrobial in Vitro Synergy Screens at Their Inception. Biochem. Pharmacol. 2019, 163, 299–307. [Google Scholar] [CrossRef] [PubMed]
- López-Archilla, A.I.; Marín, I.; Amils, R. Microbial Community Composition and Ecology of An Acidic Aquatic Environment: The Tinto River, Spain. Microb. Ecol. 2001, 41, 20–35. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Contreras, L.M.; Keitz, B.K.; Liu, S.-J. Imposed Environmental Stresses Facilitate Cell-Free Nanoparticle Formation by Deinococcus radiodurans. Appl. Environ. Microbiol. 2017, 83, e00798-17. [Google Scholar] [CrossRef] [PubMed]
- Darwesh, O.; Matter, I.; Eida, M.; Moawad, H.; Oh, Y.-K. Influence of Nitrogen Source and Growth Phase on Extracellular Biosynthesis of Silver Nanoparticles Using Cultural Filtrates of Scenedesmus obliquus. Appl. Sci. 2019, 9, 1465. [Google Scholar] [CrossRef]
- Fernández-Llamosas, H.; Castro, L.; Blázquez, M.L.; Díaz, E.; Carmona, M. Biosynthesis of Selenium Nanoparticles by Azoarcus sp. CIB. Microb. Cell Factories 2016, 15, 109. [Google Scholar] [CrossRef] [PubMed]
- Kaler, A.; Jain, S.; Banerjee, U.C. Green and Rapid Synthesis of Anticancerous Silver Nanoparticles by Saccharomyces boulardii and Insight into Mechanism of Nanoparticle Synthesis. Biomed. Res. Int. 2013, 2013, 872940. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, R.Y.; Mao, C.; Gao, X.; Burt, J.L.; Belcher, A.M.; Georgiou, G.; Iverson, B.L. Bacterial Biosynthesis of Cadmium Sulfide Nanocrystals. Chem. Biol. 2004, 11, 1553–1559. [Google Scholar] [CrossRef]
- Tikariha, S.; Banerjee, S.; Dev, A.; Singh, S. Growth Phase-Dependent Synthesis of Gold Nanoparticles Using Bacillus licheniformis. In Applications of Biotechnology for Sustainable Development; Mukhopadhyay, K., Sachan, A., Kumar, M., Eds.; Springer: Singapore, 2017; pp. 121–128. [Google Scholar] [CrossRef]
- Jalab, J.; Abdelwahed, W.; Kitaz, A.; Al-Kayali, R. Green synthesis of Silver Nanoparticles Using Aqueous Extract of Acacia cyanophylla and Its Antibacterial Activity. Heliyon 2021, 7, e08033. [Google Scholar] [CrossRef]
- Singh, H.; Du, J.; Singh, P.; Yi, T.H. Extracellular Synthesis of Silver Nanoparticles by Pseudomonas sp. THG-LS1.4 and Their Antimicrobial Application. J. Pharm. Anal. 2018, 8, 258–264. [Google Scholar] [CrossRef]
- Abd Alamer, I.S.; Tomah, A.A.; Ahmed, T.; Li, B.; Zhang, J. Biosynthesis of Silver Chloride Nanoparticles by Rhizospheric Bacteria and Their Antibacterial Activity against Phytopathogenic Bacterium Ralstonia solanacearum. Molecules 2021, 27, 224. [Google Scholar] [CrossRef] [PubMed]
- Ghiuta, I.; Croitoru, C.; Kost, J.; Wenkert, R.; Munteanu, D. Bacteria-Mediated Synthesis of Silver and Silver Chloride Nanoparticles and Their Antimicrobial Activity. Appl. Sci. 2021, 11, 3134. [Google Scholar] [CrossRef]
- Alves, M.F.; Murray, P.G. Biological Synthesis of Monodisperse Uniform-Size Silver Nanoparticles (AgNPs) by Fungal Cell-Free Extracts at Elevated Temperature and pH. J. Fungi 2022, 8, 439. [Google Scholar] [CrossRef] [PubMed]
- Velgosová, O.; Mražíková, A.; Marcinčáková, R. Influence of pH on Green Synthesis of Ag Nanoparticles. Mater. Lett. 2016, 180, 336–339. [Google Scholar] [CrossRef]
- Prasad, S.R.; Teli, S.B.; Ghosh, J.; Prasad, N.R.; Shaikh, V.S.; Nazeruddin, G.M.; Al-Sehemi, A.G.; Patel, I.; Shaikh, Y.I. A Review on Bio-Inspired Synthesis of Silver Nanoparticles: Their Antimicrobial Efficacy and Toxicity. Eng. Sci. 2021, 16, 90–128. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and Zeta Potential—What They Are and What They Are Not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Badawy, A.M.E.; Silva, R.G.; Morris, B.; Scheckel, K.G.; Suidan, M.T.; Tolaymat, T.M. Surface Charge-Dependent Toxicity of Silver Nanoparticles. Environ. Sci. Technol. 2011, 45, 283–287. [Google Scholar] [CrossRef]
- Babapour, E.; Haddadi, A.; Mirnejad, R.; Angaji, S.-A.; Amirmozafari, N. Biofilm Formation in Clinical Isolates of Nosocomial Acinetobacter baumannii and its Relationship with Multidrug Resistance. Asian Pac. J. Trop. Biomed. 2016, 6, 528–533. [Google Scholar] [CrossRef]
- Nourbakhsh, F.; Nasrollahzadeh, M.S.; Tajani, A.S.; Soheili, V.; Hadizadeh, F. Bacterial Biofilms and Their Resistance Mechanisms: A Brief Look at Treatment with Natural Agents. Folia Microbiol. 2022, 67, 535–554. [Google Scholar] [CrossRef]
- Musini, A.; Pravalika, E.; Preethi, M.G.; Sri, I.J. Microbiologically Synthesized Nanoparticles and Their Role in Biofilm Inhibition. In Microbial Processes for Synthesizing Nanomaterials; Environmental and Microbial Biotechnology; Maddela, N.R., Rodríguez Díaz, J.M., Branco da Silva Montenegro, M.C., Prasad, R., Eds.; Springer: Singapore, 2023; pp. 285–315. [Google Scholar] [CrossRef]
- Lok, C.-N.; Ho, C.-M.; Chen, R.; He, Q.-Y.; Yu, W.-Y.; Sun, H.; Tam, P.K.-H.; Chiu, J.-F.; Che, C.-M. Proteomic Analysis of the Mode of Antibacterial Action of Silver Nanoparticles. J. Proteome Res. 2006, 5, 916–924. [Google Scholar] [CrossRef]
- Salvioni, L.; Galbiati, E.; Collico, V.; Alessio, G.; Avvakumova, S.; Corsi, F.; Tortora, P.; Prosperi, D.; Colombo, M. Negatively Charged Silver Nanoparticles with Potent Antibacterial Activity and Reduced Toxicity for Pharmaceutical Preparations. Int. J. Nanomed. 2017, 12, 2517–2530. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-R.; Sun, T.-L.; Zhou, S.-L.; Ma, Y.-K.; Shi, Q.-S.; Xie, X.-B.; Huang, X.-M. A Comparative Analysis of Antibacterial Activity, Dynamics and Effects of Silver Ions and Silver Nanoparticles against Four Bacterial Strains. Int. Biodeterior. Biodegrad. 2017, 123, 304–310. [Google Scholar] [CrossRef]
- Hachicho, N.; Hoffmann, P.; Ahlert, K.; Heipieper, H.J. Effect of Silver Nanoparticles and Silver Ions on Growth and Adaptive Response Mechanisms of Pseudomonas putida mt-2. FEMS Microbiol. Lett. 2014, 355, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Abeer Mohammed, A.B.; Abd Elhamid, M.M.; Khalil, M.K.M.; Ali, A.S.; Abbas, R.N. The potential Activity of Biosynthesized Silver Nanoparticles of Pseudomonas aeruginosa as an Antibacterial Agent against Multidrug-Resistant Isolates from Intensive Care Unit and Anticancer Agent. Environ. Sci. Eur. 2022, 34, 109. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Exner, M.; Bhattacharya, S.; Christiansen, B.; Gebel, J.; Goroncy-Bermes, P.; Hartemann, P.; Heeg, P.; Ilschner, C.; Kramer, A.; Larson, E.; et al. Antibiotic Resistance: What Is so Special about Multidrug-Resistant Gram-negative Bacteria? GMS Hyg. Infect. Control 2017, 12, Doc05. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Ghoshal, G.; Jain, A.; Goyal, M. Rapid Green Synthesis of Silver Nanoparticles (AgNPs) Using (Prunus persica) Plants Extract: Exploring its Antimicrobial and Catalytic Activities. J. Nanomed. Nanotechnol. 2017, 8, 452. [Google Scholar] [CrossRef]
- Fathil, M.; Aqil, M.; Taufeq, F.Y.F.; Abdalla, S.S.I.; Katas, H. Roles of Chitosan in Synthesis, Antibacterial and Anti-Biofilm Properties of Bionano Silver and Gold. RSC Adv. 2022, 12, 19297–19312. [Google Scholar] [CrossRef]
- Li, J.; Rong, K.; Zhao, H.; Li, F.; Lu, Z.; Chen, R. Highly Selective Antibacterial Activities of Silver Nanoparticles against Bacillus subtilis. J. Nanosci. Nanotechnol. 2013, 13, 6806–6813. [Google Scholar] [CrossRef]
- Panáček, A.; Smékalová, M.; Kilianová, M.; Prucek, R.; Bogdanová, K.; Večeřová, R.; Kolář, M.; Havrdová, M.; Płaza, G.A.; Chojniak, J.; et al. Strong and Nonspecific Synergistic Antibacterial Efficiency of Antibiotics Combined with Silver Nanoparticles at Very Low Concentrations Showing No Cytotoxic Effect. Molecules 2015, 21, 26. [Google Scholar] [CrossRef]
- Matzke, M.; Jurkschat, K.; Backhaus, T. Toxicity of Differently Sized and Coated Silver Nanoparticles to The Bacterium Pseudomonas putida: Risks for The Aquatic Environment? Ecotoxicology 2014, 23, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Longhin, E.M.; Yamani, N.E.; Rundén-Pran, E.; Dusinska, M. The Alamar Blue Assay in The Context of Safety Testing of Nanomaterials. Front. Toxicol. 2022, 4, 981701. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, K.; Chan, A.; Greenberg, B.; Dixon, D.; Bols, N. Methodology for Demonstrating and Measuring the Photocytotoxicity of Fluoranthene to Fish Cells in Culture. Toxicol. Vitr. 1997, 11, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Maciaszek, K.; Gillies, S.; Kawichai, S.; Prapamontol, T.; Santijitpakdee, T.; Kliengchuay, W.; Sahanavin, N.; Mueller, W.; Vardoulakis, S.; Samutrtai, P.; et al. In Vitro Assessment of the Pulmonary Toxicity of Particulate Matter Emitted during Haze Events in Chiang Mai, Thailand via Investigation of Macrophage Responses. Environ. Res. Health 2022, 1, 025002. [Google Scholar] [CrossRef]
- Ferrero, H.; Soriano, S.; Quesada, I. Exploring the Effects of Metabolism-Disrupting Chemicals on Pancreatic α-Cell Viability, Gene Expression and Function: A Screening Testing Approach. Int. J. Mol. Sci. 2023, 24, 1044. [Google Scholar] [CrossRef]
- Soto-Bielicka, P.; Tejeda, I.; Peropadre, A.; Hazen, M.J.; Fernández Freire, P. Detrimental Effects of Individual versus Combined Exposure to Tetrabromobisphenol A and Polystyrene Nanoplastics in Fish Cell Lines. Environ. Toxicol. Pharmacol. 2023, 98, 104072. [Google Scholar] [CrossRef] [PubMed]
- Dove, A.S.; Dzurny, D.I.; Dees, W.R.; Qin, N.; Nunez Rodriguez, C.C.; Alt, L.A.; Ellward, G.L.; Best, J.A.; Rudawski, N.G.; Fujii, K.; et al. Silver Nanoparticles Enhance the Efficacy of Aminoglycosides against Antibiotic-Resistant Bacteria. Front. Microbiol. 2023, 13, 1064095. [Google Scholar] [CrossRef]
- Sun, W.; Sanderson, P.E.; Zheng, W. Drug Combination Therapy Increases Successful Drug Repositioning. Drug Discov. Today 2016, 21, 1189–1195. [Google Scholar] [CrossRef]
- Ghaffar, N.; Javad, S.; Farrukh, M.A.; Shah, A.A.; Gatasheh, M.K.; Al-Munqedhi, B.M.A.; Chaudhry, O. Metal Nanoparticles Assisted Revival of Streptomycin against MDRS Staphylococcus aureus. PLoS ONE 2022, 17, e0264588. [Google Scholar] [CrossRef]
- Ribeiro, A.I.; Dias, A.M.; Zille, A. Synergistic Effects Between Metal Nanoparticles and Commercial Antimicrobial Agents: A Review. ACS Appl. Nano Mater. 2022, 5, 3030–3064. [Google Scholar] [CrossRef]
- Markowska, K.; Grudniak, A.M.; Krawczyk, K.; Wróbel, I.; Wolska, K.I. Modulation of Antibiotic Resistance and Induction of a Stress Response in Pseudomonas aeruginosa by Silver Nanoparticles. J. Med. Microbiol. 2014, 63, 849–854. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.; Jyoti, K.; Patnaik, A.; Singh, A.; Chauhan, S.C. Spectroscopic, Microscopic Characterization of Cannabis sativa Leaf Extract Mediated Silver Nanoparticles and Their Synergistic Effect with Antibiotics against Human Pathogen. Alex. Eng. J. 2018, 57, 3043–3051. [Google Scholar] [CrossRef]
- Hussein, E.A.M.; Mohammad, A.A.; Harraz, F.A.; Ahsan, M.F. Biologically Synthesized Silver Nanoparticles for Enhancing Tetracycline Activity Against Staphylococcus aureus and Klebsiella pneumoniae. Braz. Arch. Biol. Technol. 2019, 62, e19180266. [Google Scholar] [CrossRef]
- Habash, M.B.; Goodyear, M.C.; Park, A.J.; Surette, M.D.; Vis, E.C.; Harris, R.J.; Khursigara, C.M. Potentiation of Tobramycin by Silver Nanoparticles against Pseudomonas aeruginosa Biofilms. Antimicrob. Agents Chemother. 2017, 61, e00415-17. [Google Scholar] [CrossRef] [PubMed]
- Bera, S.; Mondal, D. Antibacterial Efficacies of Nanostructured Aminoglycosides. ACS Omega 2022, 7, 4724–4734. [Google Scholar] [CrossRef] [PubMed]
- Trzcińska-Wencel, J.; Wypij, M.; Rai, M.; Golińska, P. Biogenic Nanosilver Bearing Antimicrobial and Antibiofilm Activities and Its Potential for Application in Agriculture and Industry. Front. Microbiol. 2023, 14, 1125685. [Google Scholar] [CrossRef] [PubMed]
- Wypij, M.; Czarnecka, J.; Świecimska, M.; Dahm, H.; Rai, M.; Golinska, P. Synthesis, Characterization and Evaluation of Antimicrobial and Cytotoxic Activities of Biogenic Silver Nanoparticles Synthesized from Streptomyces xinghaiensis OF1 Strain. World J. Microbiol. Biotechnol. 2018, 34, 23. [Google Scholar] [CrossRef]
- Li, P.; Li, J.; Wu, C.; Wu, Q.; Li, J. Synergistic Antibacterial Effects of β-lactam Antibiotic Combined with Silver Nanoparticles. Nanotechnology 2005, 16, 1912–1917. [Google Scholar] [CrossRef]
- Liu, X.; Ma, L.; Chen, F.; Liu, J.; Yang, H.; Lu, Z. Synergistic Antibacterial Mechanism of Bi2Te3 Nanoparticles Combined with the Ineffective β-lactam Antibiotic Cefotaxime against Methicillin-Resistant Staphylococcus aureus. J. Inorg. Biochem. 2019, 196, 110687. [Google Scholar] [CrossRef]
- Deng, H.; McShan, D.; Zhang, Y.; Sinha, S.S.; Arslan, Z.; Ray, P.C.; Yu, H. Mechanistic Study of the Synergistic Antibacterial Activity of Combined Silver Nanoparticles and Common Antibiotics. Environ. Sci. Technol. 2016, 50, 8840–8848. [Google Scholar] [CrossRef]
- Ahmed, B.; Hashmi, A.; Khan, M.S.; Musarrat, J. ROS Mediated Destruction of Cell Membrane, Growth and Biofilms of Human Bacterial Pathogens by Stable Metallic AgNPs Functionalized from Bell Pepper Extract and Quercetin. Adv. Powder Technol. 2018, 29, 1601–1616. [Google Scholar] [CrossRef]
- Mammari, N.; Lamouroux, E.; Boudier, A.; Duval, R.E. Current Knowledge on the Oxidative-Stress-Mediated Antimicrobial Properties of Metal-Based Nanoparticles. Microorganisms 2022, 10, 437. [Google Scholar] [CrossRef] [PubMed]
- Choi, O.; Hu, Z. Size Dependent and Reactive Oxygen Species Related Nanosilver Toxicity to Nitrifying Bacteria. Environ. Sci. Technol. 2008, 42, 4583–4588. [Google Scholar] [CrossRef] [PubMed]
- Martínez, S.R.; Aiassa, V.; Sola, C.; Becerra, M.C. Oxidative Stress Response in Reference and Clinical Staphylococcus aureus Strains under Linezolid Exposure. J. Glob. Antimicrob. Res. 2020, 22, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Bellanger, X.; Schneider, R.; Dezanet, C.; Arroua, B.; Balan, L.; Billard, P.; Merlin, C. Zn2+ Leakage and Photo-induced Reactive Oxidative Species Do Not Explain the Full Toxicity of ZnO Core Quantum Dots. J. Hazard. Mater. 2020, 39, 122616. [Google Scholar] [CrossRef] [PubMed]
- Quinteros, M.A.; Viviana, C.A.; Onnainty, R.; Mary, V.S.; Theumer, M.G.; Granero, G.E.; Paraje, M.G.; Paez, P.L. Biosynthesized Silver Nanoparticles: Decoding Their Mechanism of Action in Staphylococcus aureus and Escherichia coli. Int. J. Biochem. Cell Biol. 2018, 104, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kuk, E.; Yu, K.N.; Kim, J.H.; Park, S.J.; Lee, H.J.; Kim, S.H.; Park, Y.K.; Park, Y.H.; Hwang, C.Y.; et al. Antimicrobial Effects of Silver Nanoparticles. Nanomedicine 2007, 3, 95–101. [Google Scholar] [CrossRef]
- Stanier, R.Y.; Deruelles, J.; Rippka, R.; Herdman, M.; Waterbury, J.B. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. Microbiology 1979, 111, 1–61. [Google Scholar]
- ISO 10993-5:2009; Biological Evaluation of Medical Devices Part 5: Tests for In Vitro Cytotoxicity. International Organization for Standardization: Geneva, Switzerland, 2009.

| Nanoparticles (AgNPs) | Z-Potential (mV) | Diameter (DLS) (nm) | PDI (DLS) | Diameter (TEM) (nm) | PDI (TEM) |
|---|---|---|---|---|---|
| E4Cl-AgNPs | −36.88 ± 1.03 | 94.44 ± 2.74 | 0.29 | 14.88 ± 8.02 | 0.29 |
| L4Cl-AgNPs | −26.12 ± 0.81 | 75.58 ± 1.66 | 0.47 | 12.19 ± 5.93 | 0.24 |
| E4-AgNPs | −24.71 ± 2.19 | 98.53 ± 5.79 | 0.48 | 11.01 ± 6.45 | 0.34 |
| L4-AgNPs | −22.41 ± 2.66 | 83.20 ± 2.06 | 0.30 | 7.02 ± 3.19 | 0.21 |
| E7Cl-AgNPs | −35.37 ± 1.17 | 128.50 ± 3.98 | 0.42 | 20.41 ± 15.04 | 0.54 |
| L7Cl-AgNPs | −27.25 ± 1.28 | 63.88 ± 0.86 | 0.37 | 14.28 ± 10.87 | 0.58 |
| E7-AgNPs | −31.86 ± 0.16 | 79.20 ± 0.55 | 0.37 | 10.95 ± 7.03 | 0.41 |
| L7-AgNPs | −29.86 ± 0.11 | 89.16 ± 5.39 | 0.30 | 10.65 ± 6.07 | 0.32 |
| Bacteria | AgNPs | MIC (μg/mL) | MBC (μg/mL) | IC50 (μg/mL) | ICb50 (μg/mL) | MIC_AB (%) | MIC_CFDA-AM (%) | MIC_NRU (%) |
|---|---|---|---|---|---|---|---|---|
| E. coli ATCC 25922 | Sm | 16.00 | 16.00 | 5.40 ± 1.06 | 5.83 ± 1.52 | - | - | - |
| AgNO3 | 0.53 | 0.53 | 0.15 ± 0.01 | 0.33 ± 0.03 | 100.00 ± 0.00 | 93.39 ± 0.00 | 100.00 ± 0.00 | |
| E4Cl-AgNPs | 1.25 | 1.25 | 0.31 ± 0.07 */# | 0.56 ± 0.07 */#/• | 93.63 ± 2.58 | 89.32 ± 3.99 | 99.77 ± 0.69 | |
| L4Cl-AgNPs | 0.47 | 0.47 | 0.14 ± 0.05 */#/• | 0.29 ± 0.05 */#/• | 97.56 ± 2.13 | 94.90 ±2.79 | 99.94 ± 0.07 | |
| E4-AgNPs | 2.64 | 2.64 | 1.04 ± 0.17 # | 1.75 ± 0.10 #/• | 96.67 ± 3.32 | 87.69 ± 4.45 | 100.00 ± 0.00 | |
| L4-AgNPs | 1.84 | 1.84 | 0.99 ± 0.03 # | 1.47 ± 0.41 # | 95.56 ± 2.65 | 93.38 ± 4.17 | 100.00 ±0.00 | |
| E7Cl-AgNPs | 1.73 | 1.73 | 0.40 ± 0.09 # | 0.79 ± 0.11 */#/• | 76.39 ± 4.89 | 87.54 ± 6.00 | 89.52 ± 5.05 | |
| L7Cl-AgNPs | 0.84 | 0.84 | 0.43 ± 0.16 • | 0.43 ± 0.01 */#/• | 87.77 ± 4.45 | 89.92 ± 5.29 | 99.94 ± 0.20 | |
| E7-AgNPs | 5.78 | 5.78 | 1.44 ± 0.27 # | 3.83 ± 0.34 */#/• | 87.53 ± 4.12 | 68.76 ± 5.09 | 99.82 ± 1.04 | |
| L7-AgNPs | 2.63 | 2.63 | 0.87 ± 0.28 | 1.91 ± 0.45 */# | 99.99 ± 0.03 | 97.90 ± 1.90 | 100.00 ± 0.01 | |
| K. pneumoniae ATCC 29665 | Sm | 4.00 | 4.00 | 2.59 ± 0.18 | 2.29 ± 0.38 | - | - | - |
| AgNO3 | 0.53 | 0.53 | 0.27 ± 0.01 | 0.30 ± 0.07 | 100.00 ± 0.00 | 93.39 ± 0.00 | 100.00 ± 0.00 | |
| E4Cl-AgNPs | 1.25 | 1.25 | 0.51 ± 0.16 # | 0.76 ± 0.13 # | 93.63 ± 2.58 | 89.32 ± 3.99 | 99.77 ± 0.69 | |
| L4Cl-AgNPs | 0.93 | 0.93 | 0.46 ± 0.10 #/• | 0.66 ± 0.10 #/• | 89.03 ± 5.28 | 86.74 ± 4.65 | 98.88 ± 0.71 | |
| E4-AgNPs | 5.27 | 5.27 | 1.49 ± 0.12 */#/• | 2.24 ± 0.74 */#/• | 95.46 ± 3.07 | 84.83 ± 4.27 | 100.00 ± 0.00 | |
| L4-AgNPs | 3.69 | 3.69 | 0.93 ± 0.12 */#/• | 0.98 ± 0.25 */#/• | 94.22 ± 2.47 | 90.30 ± 4.09 | 100.00 ± 0.00 | |
| E7Cl-AgNPs | 1.73 | 1.73 | 0.92 ± 0.48 # | 0.90 ± 0.10 # | 76.39 ± 4.89 | 87.54 ± 6.00 | 89.52 ± 5.05 | |
| L7Cl-AgNPs | 1.67 | 1.67 | 1.00 ± 0.19 #/• | 0.99 ± 0.03 #/• | 77.39 ± 5.08 | 82.28 ± 6.17 | 99.49 ± 1.13 | |
| E7-AgNPs | 11.56 | 23.13 | 6.93 ± 0.80 */#/• | 8.41 ± 1.32 #/• | 76.06 ± 3.97 | 55.87 ± 4.32 | 96.78 ± 9.54 | |
| L7-AgNPs | 10.53 | 21.05 | 3.86 ± 0.42 */#/• | 4.73 ± 2.02 #/• | 96.97 ± 2.88 | 73.11 ± 6.85 | 96.28 ± 5.92 | |
| P. aeruginosa CECT 108 | Sm | 16.00 | 16.00 | 4.50 ± 0.27 | 6.43 ± 0.75 | - | - | - |
| AgNO3 | 0.27 | 0.53 | 0.03 ± 0.01 | 0.16 ± 0.02 | 100.00 ± 0.00 | 93.39 ± 0.00 | 100.00 ± 0.00 | |
| E4Cl-AgNPs | 0.63 | 0.63 | 0.15 ± 0.03 #/• | 0.36 ± 0.07 #/• | 96.05 ± 2.10 | 92.83 ± 3.58 | 99.93 ± 0.25 | |
| L4Cl-AgNPs | 0.47 | >0.93 | 0.18 ± 0.03 #/• | 0.36 ± 0.07 */• | 97.56 ± 2.13 | 94.90 ± 2.79 | 99.94 ± 0.07 | |
| E4-AgNPs | 1.32 | 2.64 | 0.08 ± 0.02 */#/• | 0.34 ± 0.02 */#/• | 97.57 ± 3.36 | 90.08 ± 4.78 | 100.00 ± 0.00 | |
| L4-AgNPs | 0.92 | 3.69 | 0.28 ± 0.04 */#/• | 0.70 ± 0.12 */#/• | 96.60 ± 2.67 | 95.52 ± 3.81 | 100.00 ± 0.00 | |
| E7Cl-AgNPs | 0.87 | 0.87 | 0.55 ± 0.23 */#/• | 0.56 ± 0.08 */#/• | 95.82 ± 2.16 | 96.48 ± 3.00 | 98.90 ± 1.09 | |
| L7Cl-AgNPs | 0.84 | 0.84 | 0.07 ± 0.02 */#/• | 0.73 ± 0.06 */#/• | 87.77 ± 4.45 | 89.92 ± 5.29 | 99.94 ± 0.20 | |
| E7-AgNPs | 11.56 | >23.13 | 1.62 ± 0.77 #/• | 6.38 ± 0.54 */#/• | 76.06 ± 3.97 | 55.87 ± 4.32 | 96.78 ± 9.54 | |
| L7-AgNPs | 2.63 | 2.63 | 0.77 ± 0.17 #/• | 1.45 ± 0.04 */#/• | 100.00 ± 0.03 | 97.90 ± 1.90 | 100.00 ± 0.01 | |
| S. aureus CECT 794 | Sm | 32.00 | 32.00 | 4.15 ± 0.11 | 5.45 ± 0.92 | - | - | - |
| AgNO3 | 2.12 | 4.24 | 0.56 ± 0.02 | 1.28 ± 0.15 | 100.00 ± 0.00 | 93.39 ± 0.00 | 100.00 ± 0.00 | |
| E4Cl-AgNPs | 5.00 | 5.00 | 2.68 ± 0.38 */#/• | 3.07 ± 0.42 */• | 84.13 ± 3.03 | 77.58 ± 4.16 | 97.36 ± 4.09 | |
| L4Cl-AgNPs | 1.87 | 3.73 | 0.72 ± 0.05 */#/• | 0.98 ± 0.11 */#/• | 61.33 ± 5.41 | 69.19 ± 5.23 | 81.07 ± 3.19 | |
| E4-AgNPs | 10.54 | 10.54 | 3.74 ± 0.40 */#/• | 4.39 ± 2.32 • | 93.83 ± 3.26 | 81.43 ± 4.95 | 100.00 ± 0.00 | |
| L4-AgNPs | 14.74 | 29.48 | 7.71 ± 0.71 */#/• | 7.61 ± 0.25 #/• | 90.38 ± 2.78 | 80.32 ± 3.67 | 100.00 ± 0.00 | |
| E7Cl-AgNPs | 3.47 | 3.47 | 1.02 ± 0.09 */#/• | 1.59 ± 0.12 #/• | 30.79 ± 5.92 | 63.91 ± 6.61 | 44.08 ± 7.00 | |
| L7Cl-AgNPs | 3.43 | 6.69 | 1.53 ± 0.10 */#/• | 2.13 ± 0.35 #/• | 61.18 ± 4.99 | 70.08 ± 6.14 | 95.35 ± 4.81 | |
| E7-AgNPs | 23.13 | 46.25 | 6.39 ± 0.64 #/• | 11.51 ± 1.58 #/• | 58.97 ± 4.75 | 42.13 ± 5.00 | 61.87 ± 8.28 | |
| L7-AgNPs | 21.05 | 21.05 | 6.03 ± 0.02 #/• | 10.15 ± 0.28 #/• | 66.41 ± 11.00 | 39.70 ± 7.81 | 33.27 ± 23.90 | |
| S. epidermidis ATCC 12228 | Sm | >256.0 | >256.0 | >256.0 | >256.0 | - | - | - |
| AgNO3 | 1.06 | 4.24 | 0.27 ± 0.01 | 0.90 ± 0.35 | 100.00 ± 0.00 | 93.39 ± 0.00 | 100.00 ± 0.00 | |
| E4Cl-AgNPs | 2.50 | 10.00 | 0.49 ± 0.11 # | 0.80 ± 0.35 | 89.82 ± 2.91 | 84.32 ± 4.12 | 99.21 ± 1.78 | |
| L4Cl-AgNPs | 0.93 | 3.73 | 0.31 ± 0.08 # | 0.55 ± 0.08 #/• | 89.03 ± 5.28 | 86.74 ± 4.65 | 98.88 ± 0.71 | |
| E4-AgNPs | 2.64 | 10.54 | 1.27 ± 0.37 #/• | 1.40 ± 0.20 • | 96.67 ± 3.32 | 87.69 ± 4.45 | 100.00 ± 0.00 | |
| L4-AgNPs | 1.84 | 14.74 | 1.25 ± 0.23 # | 1.61 ± 0.26 # | 95.56 ± 2.65 | 93.38 ± 4.17 | 100.00 ± 0.00 | |
| E7Cl-AgNPs | 1.73 | 3.47 | 0.42 ± 0.09 # | 0.81 ± 0.05 # | 76.39 ± 4.89 | 87.54 ± 6.00 | 89.52 ± 5.05 | |
| L7Cl-AgNPs | 1.67 | 3.34 | 0.47 ± 0.09 # | 0.82 ± 0.10 #/• | 77.39 ± 5.08 | 82.28 ± 6.17 | 99.49 ± 1.13 | |
| E7-AgNPs | 11.56 | 23.13 | 2.27 ± 0.28 */#/• | 5.28 ± 0.19 */#/• | 76.06 ± 3.97 | 55.87 ± 4.32 | 96.78 ± 9.54 | |
| L7-AgNPs | 5.26 | 42.10 | 1.30 ± 0.33 */# | 1.57 ± 0.01 */# | 99.81 ± 0.31 | 91.85 ± 4.55 | 99.93 ± 0.32 | |
| B. subtilis 168 | Sm | 64.00 | >256.00 | 7.58 ± 0.82 | 14.09 ± 5.80 | - | - | - |
| AgNO3 | 1.06 | 1.06 | 0.27 ± 0.08 | 0.40 ± 0.13 | 100.00 ± 0.00 | 93.39 ± 0.00 | 100.00 ± 0.00 | |
| E4Cl-AgNPs | 5.00 | 5.00 | 1.40 ± 0.31 */#/• | 2.01 ± 0.34 */• | 84.13 ± 3.03 | 77.58 ± 4.16 | 97.36 ± 4.09 | |
| L4Cl-AgNPs | 1.87 | 7.46 | 0.58 ± 0.06 */#/• | 0.84 ± 0.15 */#/• | 61.33 ± 5.41 | 69.19 ± 5.23 | 81.07 ± 3.19 | |
| E4-AgNPs | 2.64 | 10.54 | 0.76 ± 0.18 */#/• | 1.26 ± 0.45 */• | 96.67 ± 3.32 | 87.69 ± 4.45 | 100.00 ± 0.00 | |
| L4-AgNPs | 7.37 | 7.37 | 4.14 ± 1.29 */# | 6.37 ± 0.75 */#/• | 92.53 ± 2.29 | 86.05 ± 3.51 | 100.00 ± 0.00 | |
| E7Cl-AgNPs | 3.34 | 3.34 | 0.45 ± 0.03 */#/• | 0.86 ± 0.01 */#/• | 33.16 ± 5.91 | 65.63 ± 6.66 | 47.33 ± 6.99 | |
| L7Cl-AgNPs | 3.34 | 3.34 | 1.06 ± 0.10 */#/• | 1.28 ± 0.18 */#/• | 61.86 ± 4.99 | 70.60 ± 6.16 | 95.71 ± 4.63 | |
| E7-AgNPs | 23.13 | 23.13 | 4.57 ± 1.71 #/• | 6.20 ± 0.35 */#/• | 58.97 ± 4.75 | 42.13 ± 5.00 | 61.87 ± 8.28 | |
| L7-AgNPs | 10.52 | 21.05 | 3.14 ± 0.49 # | 3.35 ± 0.83 */#/• | 96.98 ± 2.87 | 73.15 ± 6.85 | 96.30 ± 5.91 |
| AgNPs | H. T. (h) | E. coli ATCC 25922 | S. aureus CECT 108 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC (µg/mL) | MBC (µg/mL) | IC50 (µg/mL) | ICb50 (µg/mL) | MIC (µg/mL) | MBC (µg/mL) | IC50 (µg/mL) | ICb50 (µg/mL) | ||
| L4Cl-AgNPs | 24 | 0.23 | 0.46 | 0.13 ± 0.01 * | 0.18 ± 0.04 | 0.93 | 2.78 | 0.65 ± 0.02 * | 0.89 ± 0.11 |
| 48 | 0.55 | 1.09 | 0.20 ± 0.01 | 0.36 ± 0.01 | 2.18 | >2.18 | 0.78 ± 0.01 | 0.92 ± 0.13 | |
| 72 | 0.44 | 0.87 | 0.26 ± 0.03 | 0.26 ± 0.02 | 1.74 | >1.74 | 0.79 ± 0.06 | 1.13 ± 0.04 | |
| 144 | 1.59 | >1.59 | 0.35 ± 0.07 | 0.70 ± 0.15 * | 6.12 | >6.12 | 1.98 ± 0.02 * | 2.88 ± 0.54 * | |
| L4-AgNPs | 48 | 1.49 | 1.49 | 0.42 ± 0.10 * | 0.44 ± 0.05 * | 5.95 | >5.95 | 3.19 ± 0.09 * | 3.41 ± 0.40 * |
| 100 | 1.58 | 1.58 | 1.14 ± 0.08 | 1.52 ± 0.38 | 12.65 | 25.30 | 4.64 ± 0.35 * | 4.66 ± 0.65 * | |
| 200 | 1.93 | 1.93 | 1.17 ± 0.34 | 1.74 ± 0.29 | 15.43 | 30.86 | 6.41 ± 0.28 | 7.44 ± 0.06 | |
| 400 | 2.24 | 2.24 | 1.44 ± 0.44 | 2.11 ± 0.65 | 17.90 | 35.80 | 9.52 ± 0.05 * | 9.61 ± 1.06 * | |
| AgNPs | Batch | E. coli ATCC 25922 | S. aureus CECT 108 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC (µg/mL) | MBC (µg/mL) | IC50 (µg/mL) | ICb50 (µg/mL) | MIC (µg/mL) | MBC (µg/mL) | IC50 (µg/mL) | ICb50 (µg/mL) | ||
| L4Cl-AgNPs | S1st | 0.47 | 0.47 | 0.14 ± 0.05 | 0.29 ± 0.05 | 1.87 | 3.73 | 0.72 ± 0.05 | 0.98 ± 0.11 |
| S2nd | 0.42 | 0.42 | 0.19 ± 0.02 | 0.30 ± 0.00 | 1.69 | 1.69 | 0.65 ± 0.01 | 1.18 ± 0.39 | |
| S3rd | 0.84 | 1.84 | 0.31 ± 0.03 | 0.42 ± 0.00 | 3.63 | 3.63 | 1.15 ± 0.06 * | 1.27 ± 0.19 | |
| S4th | 1.93 | 7.73 | 1.18 ± 0.21 * | 1.46 ± 0.14 * | 15.47 | >15.47 | 6.90 ± 0.89 * | 7.29 ± 0.89 * | |
| L7Cl-AgNPs | S1st | 0.84 | 0.84 | 0.43 ± 0.16 | 0.43 ± 0.01 | 3.43 | 6.69 | 1.53 ± 0.10 | 2.13 ± 0.35 |
| S2nd | 0.84 | 0.84 | 0.37 ± 0.07 | 0.36 ± 0.04 | 1.68 | 6.73 | 1.61 ± 0.07 | 1.56 ± 0.23 | |
| S3rd | 2.13 | 2.13 | 0.80 ± 0.13 | NC | 8.50 | >8.50 | 4.62 ± 0.29 * | 4.61 ± 0.34 * | |
| S4th | 4.26 | 8.16 | 3.16 ± 0.24 * | 4.57 ± 0.08 * | 17.24 | >17.24 | 7.51 ± 0.76 * | 8.20 ± 1.25 * | |
| AgNPs | IC30_AB (μg/mL) | IC30_CFDA-AM μg/mL) | IC30_NRU (μg/mL) |
|---|---|---|---|
| AgNO3 | 4.01 | 3.43 | 4.76 |
| E4Cl-AgNPs | 15.25 */#/• | 9.28 */#/• | 23.78 */• |
| L4Cl-AgNPs | 1.59 */# | 1.82 */# | 2.15 */#/• |
| E4-AgNPs | >25.00 #/• | >25.00 #/• | 26.18 |
| L4-AgNPs | >25.00 #/• | >25.00 #/• | 25.67/# |
| E7Cl-AgNPs | 1.94 #/• | 3.02/• | 2.53 */#/• |
| L7Cl-AgNPs | 2.38 # | 3.44/# | 6.85 */#/• |
| E7-AgNPs | 15.14 #/• | 5.37 */• | 21.22 # |
| L7-AgNPs | 20.20 #/• | 11.35 */#/• | 16.02 # |
| AgNPs | Bacteria | Ap | Cp | Ep | Km | Nx | Sm | Tc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FICI | MF | FICI | MF | FICI | MF | FICI | MF | FICI | MF | FICI | MF | FICI | MF | ||
| E4Cl-AgNPs | E. coli | 2.000 | 1 | 2.000 | 1 | 1.000 | 2 | 0.094 | 32 | 1.000 | 2 | 0.063 | 32 | 0.375 | 4 |
| S. aureus | 0.375 | 4 | 1.000 | 2 | 2.000 | 1 | 0.180 | 32 | 2.000 | 1 | 0.188 | 16 | 2.000 | 1 | |
| L4Cl-AgNPs | E. coli | 1.000 | 2 | 2.000 | 1 | 1.000 | 2 | 0.125 | 16 | 1.000 | 2 | 0.094 | 32 | 0.500 | 4 |
| S. aureus | 0.375 | 4 | 1.000 | 2 | 2.000 | 1 | 0.180 | 16 | 1.000 | 2 | 0.125 | 16 | 2.000 | 1 | |
| E4-AgNPs | E. coli | 2.000 | 1 | 2.000 | 1 | 1.000 | 2 | 0.094 | 32 | 2.000 | 1 | 0.125 | 16 | 0.375 | 4 |
| S. aureus | 0.375 | 4 | 0.750 | 2 | 2.000 | 1 | 0.180 | 16 | 2.000 | 1 | 0.156 | 8 | 1.000 | 2 | |
| L4-AgNPs | E. coli | 1.000 | 2 | 1.000 | 2 | 0.750 | 4 | 0.063 | 32 | 0.750 | 2 | 0.094 | 16 | 0.500 | 4 |
| S. aureus | 0.500 | 4 | 0.750 | 2 | 2.000 | 1 | 0.125 | 16 | 0.750 | 4 | 0.063 | 32 | 0.750 | 4 | |
| E7Cl-AgNPs | E. coli | 2.000 | 1 | 1.000 | 2 | 2.000 | 1 | 0.094 | 32 | 2.000 | 1 | 0.313 | 16 | 0.250 | 8 |
| S. aureus | 0.250 | 8 | 0.625 | 2 | 2.000 | 1 | 0.125 | 16 | 1.000 | 2 | 0.125 | 16 | 1.000 | 2 | |
| L7Cl-AgNPs | E. coli | 1.000 | 2 | 2.000 | 1 | 1.000 | 2 | 0.063 | 32 | 0.750 | 2 | 0.047 | 32 | 0.375 | 4 |
| S. aureus | 0.375 | 4 | 1.000 | 2 | 2.000 | 1 | 0.180 | 16 | 0.625 | 8 | 0.094 | 32 | 1.000 | 2 | |
| E7-AgNPs | E. coli | 0.625 | 8 | 1.000 | 2 | 0.750 | 4 | 0.156 | 32 | 1.000 | 2 | 0.125 | 16 | 0.500 | 4 |
| S. aureus | 0.625 | 8 | 2.000 | 1 | 2.000 | 1 | 0.375 | 4 | 2.000 | 1 | 0.125 | 16 | 2.000 | 1 | |
| L7-AgNPs | E. coli | 1.000 | 2 | 2.000 | 1 | 2.000 | 1 | 0.156 | 32 | 1.000 | 2 | 0.156 | 32 | 0.188 | 8 |
| S. aureus | 0.500 | 2 | 1.000 | 2 | 2.000 | 1 | 0.125 | 16 | 2.000 | 1 | 0.125 | 16 | 1.000 | 2 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pernas-Pleite, C.; Conejo-Martínez, A.M.; Fernández Freire, P.; Hazen, M.J.; Marín, I.; Abad, J.P. Microalga Broths Synthesize Antibacterial and Non-Cytotoxic Silver Nanoparticles Showing Synergy with Antibiotics and Bacterial ROS Induction and Can Be Reused for Successive AgNP Batches. Int. J. Mol. Sci. 2023, 24, 16183. https://doi.org/10.3390/ijms242216183
Pernas-Pleite C, Conejo-Martínez AM, Fernández Freire P, Hazen MJ, Marín I, Abad JP. Microalga Broths Synthesize Antibacterial and Non-Cytotoxic Silver Nanoparticles Showing Synergy with Antibiotics and Bacterial ROS Induction and Can Be Reused for Successive AgNP Batches. International Journal of Molecular Sciences. 2023; 24(22):16183. https://doi.org/10.3390/ijms242216183
Chicago/Turabian StylePernas-Pleite, Carlos, Amparo M. Conejo-Martínez, Paloma Fernández Freire, María José Hazen, Irma Marín, and José P. Abad. 2023. "Microalga Broths Synthesize Antibacterial and Non-Cytotoxic Silver Nanoparticles Showing Synergy with Antibiotics and Bacterial ROS Induction and Can Be Reused for Successive AgNP Batches" International Journal of Molecular Sciences 24, no. 22: 16183. https://doi.org/10.3390/ijms242216183
APA StylePernas-Pleite, C., Conejo-Martínez, A. M., Fernández Freire, P., Hazen, M. J., Marín, I., & Abad, J. P. (2023). Microalga Broths Synthesize Antibacterial and Non-Cytotoxic Silver Nanoparticles Showing Synergy with Antibiotics and Bacterial ROS Induction and Can Be Reused for Successive AgNP Batches. International Journal of Molecular Sciences, 24(22), 16183. https://doi.org/10.3390/ijms242216183






