The Identification of a Quantative Trait Loci-Allele System of Antixenosis against the Common Cutworm (Spodoptera litura Fabricius) at the Seedling Stage in the Chinese Soybean Landrace Population
Abstract
1. Introduction
2. Results
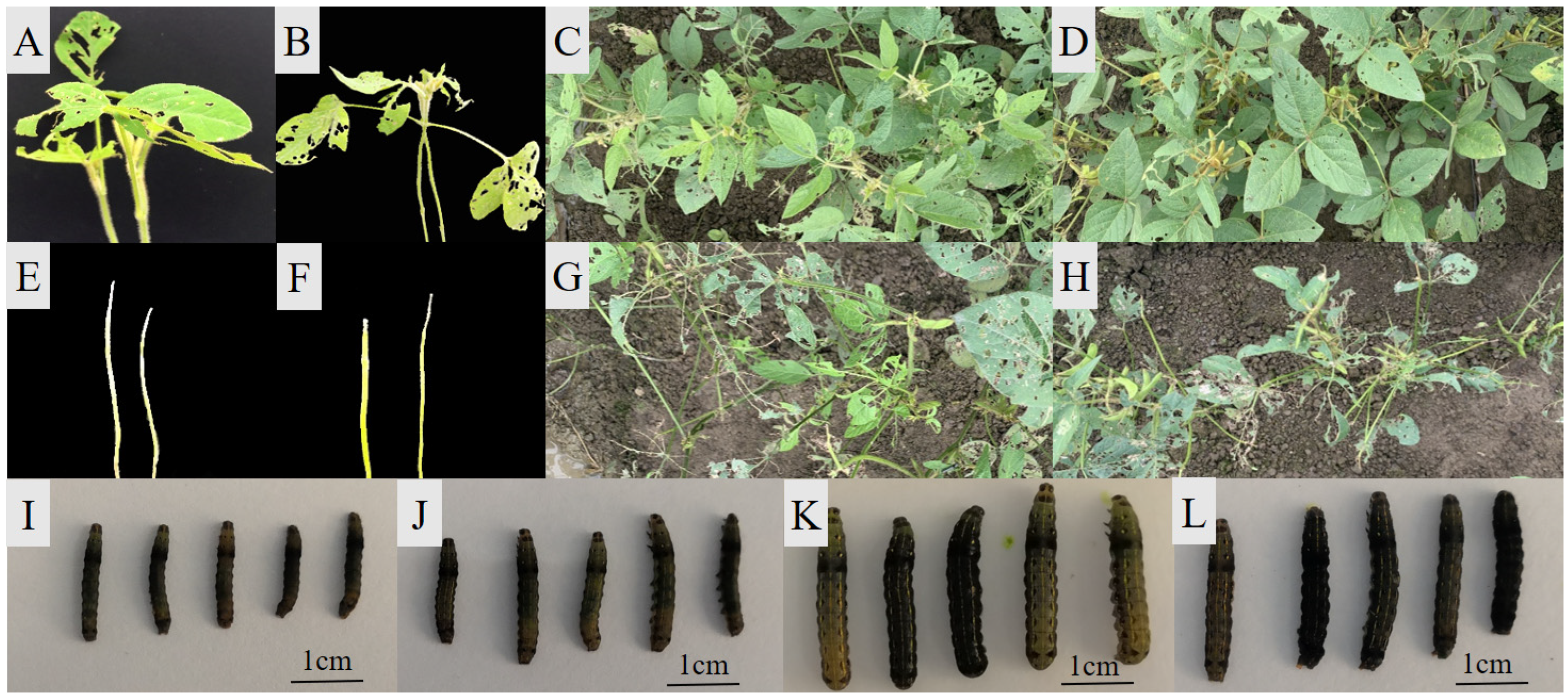
2.1. Antixenosis Variation in CSLRP
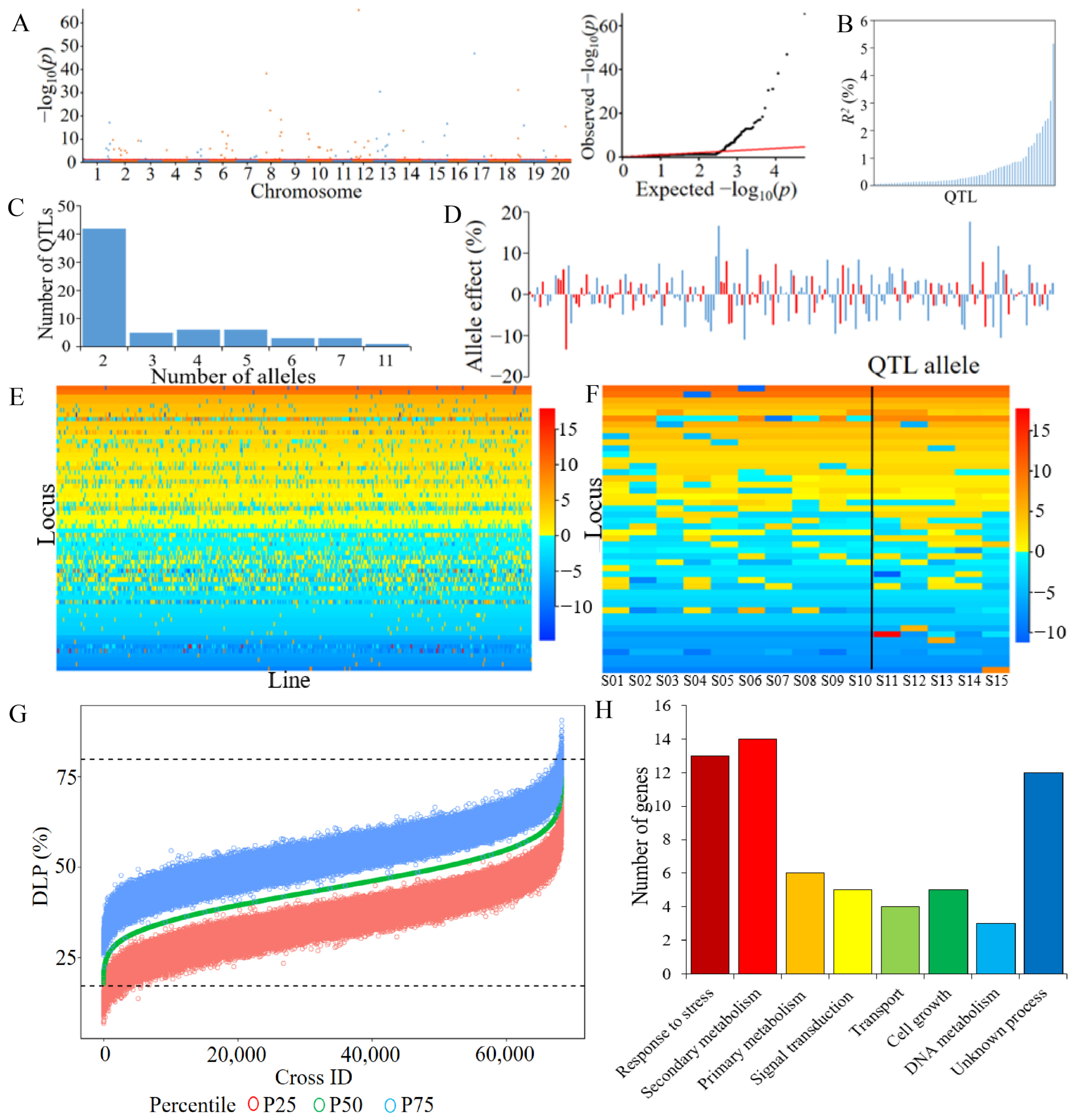
2.2. QTL-Allele System of Antixenosis in CSLRP
2.3. Prediction of Recombination Potential and Optimal Crosses for Antixenosis in CSLRP
2.4. Candidate Gene Annotation of Antixenosis in CSLRP
2.5. Genetic Structure Differentiation among Antixenosis Degree Groups in CSLRP
3. Discussion
3.1. Efficiency of the Seedling Stage Mini-Tray Identification System and Relative Consistency of Antixenosis between the Seedling and Adult Stage
3.2. Important QTLs-Alleles and Candidate Genes of Antixenosis against CCW in CSLRP
3.3. Promising Breeding Potential and the Novel Antixenosis Sources in CSLRP
4. Materials and Methods
4.1. Source of CCW Used
4.2. Plant Materials and Evaluation of Antixenosis to CCW at the Seedling Stage
4.3. Evaluation of Antixenosis and Antibiosis at the Adult Stage of Selected Materials
4.4. Statistical Analysis of Phenotypic Data
4.5. Genome-Wide Association Analysis Using RTM-GWAS
4.6. Optimal Cross Prediction
4.7. Candidate Gene Function Annotation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramakrishnan, N.; Saxena, V.S.; Dhingra, S. Insecticide resistance in the population of Spodoptera litura (Fabricius) in Andhra Pradesh. Pesticides 1984, 18, 23–27. [Google Scholar]
- Xing, G.; Liu, K.; Gai, J. A high-throughput phenotyping procedure for evaluation of antixenosis against common cutworm at early seedling stage in soybean. Plant Methods 2017, 13, 66. [Google Scholar] [CrossRef]
- Dhir, B.C.; Mohapatra, H.K.; Senapati, B. Assessment of crop loss in groundnut due to tobacco caterpillar, Spodoptera litura (F.). Indian J. Plant Prot. 1992, 2, 215–217. [Google Scholar]
- Tian, L.; Gao, X.; Zhang, S.; Zhang, Y.; Ma, D.; Cui, J. Dynamic changes of transcriptome of fifth-instar Spodoptera litura larvae in response to insecticide. 3 Biotech 2021, 11, 98. [Google Scholar] [CrossRef] [PubMed]
- Cardona, E.V.; Ligat, C.S.; Subang, M.P. Life history of common cutworm, Spodoptera litura Fabricius (Noctuidae: Lepidoptera) in Benguet. BSU Res. J. 2007, 56, 73–84. [Google Scholar]
- Kim, H.; Xing, G.; He, J.; Zhao, T.; Yang, S.; Li, Y.; Palmer, R.G.; Gai, J. An environmental differential association analysis of antibiosis to common cutworm in a Chinese soybean germplasm population and optimization of the cross design. Mol. Breed. 2015, 35, 76. [Google Scholar] [CrossRef]
- Chen, X.; Vosman, B.; Visser, R.G.F.; Der-Vlugt, R.V.; Broekgaarden, C. High throughput phenotyping for aphid resistance in large plant collections. Plant Methods 2012, 8, 33. [Google Scholar] [CrossRef]
- Komatsu, K.; Okuda, S.; Takahashi, M.; Matsunaga, R.; Nakazawa, Y. QTL mapping of antibiosis resistance to common cutworm (Spodoptera litura Fabricius) in soybean. Crop Sci. 2005, 45, 2044–2048. [Google Scholar] [CrossRef]
- Painter, R.H. Insect resistance in crop plants. Soil Sci. 1951, 72, 481. [Google Scholar] [CrossRef]
- Liu, H.; Che, Z.; Zeng, X.; Zhang, G.; Wang, H.; Yu, D. Identification of single nucleotide polymorphisms in soybean associated with resistance to common cutworm (Spodoptera litura Fabricius). Euphytica. 2016, 209, 49–62. [Google Scholar] [CrossRef]
- Oki, N.; Komatsu, K.; Takahashi, M.; Takahashi, M.; Kono, Y.; Ishimoto, M. Field assessment of resistance QTL to common cutworm in soybean. Crop Sci. 2015, 55, 624–630. [Google Scholar] [CrossRef]
- Kim, H.; Xing, G.; Wang, Y.; Zhao, T.; Yu, D.; Yang, S.; Li, Y.; Chen, S.; Palmer, R.G.; Gai, J. Constitution of resistance to common cutworm in terms of antibiosis and antixenosis in soybean RIL populations. Euphytica 2014, 196, 137–154. [Google Scholar] [CrossRef]
- Hatchett, J.H.; Beland, G.L.; Hartwig, E.E. Leaf-feeding resistance to bollworm and tobacco budworm in three soybean plant introductions. Crop Sci. 1976, 16, 277–280. [Google Scholar] [CrossRef]
- Reynolds, G.W.; Smith, C.M. Effects of leaf position, leaf wounding, and plant age of two soybean genotypes on soybean looper (Lepidoptera: Noctuidae) growth. Environ. Entomol. 1985, 14, 475–478. [Google Scholar] [CrossRef]
- Van, D.J.; Turnipseed, S.; Maxwell, J. Resistance in soybeans to the Mexican bean beetle: Sources of resistance. Crop Sci. 1971, 11, 572–573. [Google Scholar]
- Vishal, K.; Bantewad, S.; Kota, S.; Mishra, S.P.; Hingane, A.; Jaba, J. Antixenosis & antibiosis mechanisms of resistance to the pod borer, Helicoverpa armigera in pigeonpea cultigens and hybrids. Int. J. Trop. Insect Sci. 2023, 43, 665–675. [Google Scholar]
- Eduardo, W.I.; Junior, A.L.B.; Moraes, R.F.O.; Souza, B.H.S.; Louvandini, H.; Barbosa, J.C. Protocol for assessing soybean antixenosis to Heliothis virescens. Entomol. Exp. Appl. 2020, 168, 911–927. [Google Scholar] [CrossRef]
- Komatsu, K.; Okuda, S.; Takahashi, M.; Matsunaga, R. Antibiotic effect of insect-resistant soybean on common cutworm (Spodoptera litura) and its inheritance. Breed. Sci. 2004, 54, 27–32. [Google Scholar] [CrossRef][Green Version]
- Souza, B.H.S.; Silva, A.G.; Janini, J.C.; Boica-Júnior, A.L. Antibiosis in soybean genotypes and the resistance levels to Spodoptera eridania (Cramer) (Lepidoptera: Noctuidae). Neotrop. Entomol. 2014, 43, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008, 59, 41–66. [Google Scholar] [CrossRef] [PubMed]
- Hymowitz, T. On the domestication of the soybean. Econ. Bot. 1970, 24, 408–421. [Google Scholar] [CrossRef]
- Gai, J.; Chen, L.; Zhang, Y.; Zhao, T.; Xing, G.; Xing, H. Genome-wide genetic dissection of germplasm resources and implications for breeding by design in soybean. Breed. Sci. 2012, 61, 495–510. [Google Scholar] [CrossRef] [PubMed]
- Oki, N.; Komatsu, K.; Sayama, T.; Ishimoto, M.; Takahashi, M.; Takahashi, M. Genetic analysis of antixenosis resistance to the common cutworm (Spodoptera litura Fabricius) and its relationship with pubescence characteristics in soybean (Glycine max (L.) Merr.). Breed. Sci. 2012, 61, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Oki, N.; Kaga, A.; Shimizu, T.; Takahashi, M.; Kono, Y.; Takahashi, M. QTL mapping of antixenosis resistance to common cutworm (Spodoptera litura Fabricius) in wild soybean (Glycine soja). PLoS ONE 2017, 12, e0189440. [Google Scholar] [CrossRef] [PubMed]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler IV, E.S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Wang, Y.; Ren, H.; Du, W.; Wang, D.; Bao, R.; Yang, X.; Tian, Z.; Fu, L.; Cheng, Y.; et al. Genetic dynamics of earlier maturity group emergence in south-to-north extension of Northeast China soybeans. Theor. Appl. Genet. 2020, 133, 1839–1857. [Google Scholar] [CrossRef]
- Chang, H.; Hartman, G.L. Characterization of insect resistance loci in the USDA soybean germplasm collection using genome-wide association studies. Front. Plant Sci. 2017, 8, 670. [Google Scholar] [CrossRef]
- Wang, H.; Yan, H.; Du, H.; Chao, M.; Gao, Z.; Yu, D. Mapping quantitative trait loci associated with soybean resistance to common cutworm and soybean compensatory growth after defoliation using SNP marker-based genome-wide association analysis. Mol. Breed. 2015, 35, 168. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; Mcmullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- He, J.; Meng, S.; Zhao, T.; Xing, G.; Yang, S.; Li, Y.; Guan, R.; Lu, J.; Wang, Y.; Xia, Q.; et al. An innovative procedure of genome-wide association analysis fits studies on germplasm population and plant breeding. Theor. Appl. Genet. 2017, 130, 2327–2343. [Google Scholar] [CrossRef]
- Meng, S.; He, J.; Zhao, T.; Xing, G.; Li, Y.; Yang, S.; Liu, J.; Wang, Y.; Gai, J. Detecting the QTL-allele system of seed isoflavone content in Chinese soybean landrace population for optimal cross design and gene system exploration. Theor. Appl. Genet. 2016, 129, 1557–1576. [Google Scholar] [CrossRef]
- Zhang, Y.; He, J.; Xu, Y.; Wang, H.; Meng, S.; Xing, G.; Li, Y.; Yang, S.; Zhao, J.; Zhao, T.; et al. Detecting the QTL-allele system of seed oil traits using multi-locus genome-wide association analysis for population characterization and optimal cross prediction in soybean. Front. Plant Sci. 2018, 9, 1793. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W.; Zhang, L.; Dai, H.; Ci, D.; Xu, R. QTL mapping of soybean resistance to whitefly (Bemisia Tabaci Gennadius) under multi-environment conditions. Aust. J. Crop Sci. 2013, 7, 1212–1218. [Google Scholar]
- Narvel, J.M.; Walker, D.R.; Rector, B.G.; All, J.N.; Parrott, W.A.; Boerma, H.R. A retrospective DNA marker assessment of the development of insect resistant soybean. Crop Sci. 2001, 41, 1931–1939. [Google Scholar] [CrossRef]
- Xing, G.; Zhou, B.; Wang, Y.; Zhao, T.; Yu, D.; Chen, S.; Gai, J. Genetic components and major QTL confer resistance to bean pyralid (Lamprosema indicata Fabricius) under multiple environments in four RIL populations of soybean. Theor. Appl. Genet. 2012, 125, 859–875. [Google Scholar] [CrossRef] [PubMed]
- Jun, T.; Mian, M.A.R.; Michel, A.P. Genetic mapping of three quantitative trait loci for soybean aphid resistance in PI 567324. Heredity 2013, 111, 16–22. [Google Scholar] [CrossRef]
- Shan, A.Z.; Ma, C.; Zhang, Y.; Zhang, Q.; Xu, G.; Yang, G. Decoyinine induced resistance in rice against small brown planthopper Laodelphax striatellus. Insects 2022, 13, 104. [Google Scholar]
- Zhang, Y.; Guo, W.; Chen, L.; Shen, X.; Yang, H.; Fang, Y.; Ouyang, W.; Mai, S.; Chen, H.; Chen, S.; et al. CRISPR/Cas9-mediated targeted mutagenesis of GmUGT enhanced soybean resistance against leaf-chewing insects through flavonoids biosynthesis. Front. Plant Sci. 2022, 13, 802716. [Google Scholar] [CrossRef]
- Sun, Z.; Zang, Y.; Zhou, L.; Song, Y.; Chen, D.; Zhang, Q.; Liu, Y.; Yi, Y.; Zhu, B.; Fu, D.; et al. A tomato receptor-like cytoplasmic kinase, SIZRK1, acts as a negative regulator in wound-induced jasmonic acid accumulation and insect resistance. J. Exp. Bot. 2021, 72, 7285–7300. [Google Scholar] [CrossRef]
- Rector, B.G.; All, J.N.; Parrott, W.A.; Boerma, H.R. Identification of molecular markers linked to quantitative trait loci for soybean resistance to corn earworm. Theor. Appl. Genet. 1998, 96, 786–790. [Google Scholar] [CrossRef]
- Tabari, M.A.; Fathi, S.; Nouri-Ganbalani, G.; Moumeni, A.; Razmjou, J. Antixenosis and antibiosis resistance in rice cultivars against Chilo suppressalis (walker) (Lepidoptera: Crambidae). Neotrop. Entomol. 2017, 46, 452–460. [Google Scholar] [CrossRef]
- Kuswantoro, H.; Bayu, M.S.Y.I.; Baliadi, Y.; Tengkano, W. Resistance of advanced soybean lines to pod borrer (Etiella zinckenella). Biosaintifika 2017, 9, 317–324. [Google Scholar] [CrossRef]
- Hu, X.; Liu, Y.; Wang, Y.; Wang, Z.; Yu, X.; Wang, B.; Zhang, G.; Liu, X.; Hu, Z.; Zhao, H.; et al. Resistance of wheat accessions to the english grain aphid Sitobion avenae. PLoS ONE 2016, 11, e0156158. [Google Scholar] [CrossRef] [PubMed]
- Panda, N.; Heinrichs, E.A. Levels of tolerance and antibiosis in rice varieties having moderate resistance to the brown planthopper, Nilaparvata lugens (Stal; Hemiptera: Delphacidae). Environ. Entomol. 1983, 12, 1204–1214. [Google Scholar] [CrossRef]
- Prahalada, G.D.; Shivakumar, N.; Lohithaswa, H.C.; Siddle-Gowda, D.K.; Ramkumar, G.; Kim, S.R.; Ramachandra, C.; Hittalmani, S.; Mohapatra, T.; Jena, K.K. Identification and fine mapping of a new gene, BPH31 conferring resistance to brown planthopper biotype 4 of India to improve rice, Oryza sativa L. Rice 2017, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Phi, N.P.; Fujita, D.; Yamagata, Y.; Yoshimura, A.; Yasui, H. High-resolution mapping of GRH6, a gene from Oryza nivara (Sharma et Shastry) conferring resistance to green rice leafhopper (Nephotettix cincticeps Uhler). Breed. Sci. 2019, 69, 439–446. [Google Scholar] [CrossRef]
- Xu, X.; Li, G.; Carver, B.F.; Armstrong, J.S. Gb8, a new gene conferring resistance to economically important greenbug biotypes in wheat. Theor. Appl. Genet. 2020, 133, 615–622. [Google Scholar] [CrossRef]
- Zhao, L.; Abdelsalam, N.R.; Xu, Y.; Chen, M.; Feng, Y.; Kong, L.; Bai, G. Identification of two novel hessian fly resistance genes H35 and H36 in a hard winter wheat line SD06165. Theor. Appl. Genet. 2020, 133, 2343–2353. [Google Scholar] [CrossRef]
- Huang, X.; Jing, D.; Zhang, T.; Wang, Z. Analysis of midgut transcriptome and ABC gene expression in Spodoptera frugiperda (Lepidoptera: Noctuidae) after feeding Cry1 Ab and Cry Fa proteins. Acta Entomol. Sin. 2022, 65, 1127–1135. [Google Scholar]
- Bittner, S. When quinones meet amino acids: Chemical, physical and biological consequences. Amino Acids 2006, 30, 205–224. [Google Scholar] [CrossRef]
- Viswanath, K.K.; Varakumar, P.; Pamuru, R.R.; Basha, S.J.; Mehta, S.; Rao, A.D. Plant lipoxygenases and their role in plant physiology. J. Plant Biol. 2020, 63, 83–95. [Google Scholar] [CrossRef]
- Oki, N.; Takagi, K.; Ishimoto, M.; Takahashi, M.; Takahashi, M. Evaluation of the resistance effect of QTLs derived from wild soybean (Glycine soja) to common cutworm (Spodoptera litura Fabricius). Breed. Sci. 2019, 69, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, E.E.; Lambert, L.; Kilen, T.C. Registration of ‘Lamar’ soybean. Crop Sci. 1990, 30, 231. [Google Scholar] [CrossRef]
- Wang, H.; Gao, Z.; Zhang, D.; Cheng, H.; Yu, D. Identification of genes with soybean resistance to common cutworm by association analysis. Chin. Bull. Bot. 2011, 46, 514–524. [Google Scholar] [CrossRef]
- Hu, Z.; Xu, X.; Pan, L.; Li, M.; Zeng, J.; Muhammad, K.R.; Xing, G.; Gai, J. Resistance analyses of soybean organs to common cutworm (Spodoptera litura) at different reproductive stages. Soybean Sci. 2020, 39, 932–939. [Google Scholar]
- Fehr, W.R.; Caviness, C.E.; Burmood, D.T.; Pennington, J.S. Stage of development descriptions for soybeans, Glycine Max (L.) Merrill1. Crop Sci. 1971, 11, 929–931. [Google Scholar] [CrossRef]
- Hanson, C.; Robinson, H.; Comstock, R. Biometrical studies of yield in segregating populations of korean lespedeza. Agron. J. 1956, 48, 268–272. [Google Scholar] [CrossRef]
- Yi, X.; Liang, Y.; Huerta-Sanchez, E.; Jin, X.; Cuo, Z.X.P.; Pool, J.E.; Xu, X.; Jiang, H.; Vinckenbosch, N.; Korneliussen, T.S.; et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 2010, 329, 75–78. [Google Scholar] [CrossRef]
- Scheet, P.; Stephens, M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006, 78, 629–644. [Google Scholar] [CrossRef]
| Eco-Region | Class Midpoint of DLP | N | Mean | Min. | Max. | F | h2 | GCV | CV | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 20 | 25 | 30 | 35 | 40 | 45 | 50 | 55 | 60 | 65 | 70 | 75 | 80 | |||||||||
| All | 1 | 6 | 28 | 30 | 45 | 62 | 47 | 50 | 44 | 27 | 18 | 7 | 4 | 1 | 370 | 44.5 | 17.2 | 79.9 | 6.2 ** | 77.3 | 24.1 | 19.1 |
| ER-I | 1 | 2 | 5 | 4 | 7 | 6 | 6 | 7 | 3 | 2 | 43 | 44.6 ab | 18.9 | 74.9 | 3.7 ** | 63.3 | 21.7 | 24.7 | ||||
| ER-II | 1 | 1 | 11 | 11 | 14 | 18 | 12 | 12 | 14 | 7 | 2 | 103 | 42.0 b | 17.2 | 69.5 | 5.3 ** | 72.1 | 22.9 | 20.3 | |||
| ER-III | 3 | 8 | 16 | 8 | 8 | 10 | 6 | 10 | 2 | 71 | 49.3 a | 30.2 | 70.3 | 5.9 ** | 75.9 | 19.7 | 16.2 | |||||
| ER-IV | 2 | 6 | 4 | 9 | 9 | 10 | 13 | 10 | 5 | 7 | 2 | 2 | 1 | 80 | 46.9 a | 19.8 | 79.9 | 9.3 ** | 83.3 | 13.6 | 16.3 | |
| ER-V | 3 | 2 | 6 | 8 | 4 | 8 | 3 | 34 | 41.7 b | 25 | 62.1 | 5.7 ** | 77.5 | 20.1 | 16.6 | |||||||
| ER-VI | 2 | 6 | 5 | 4 | 4 | 7 | 3 | 3 | 3 | 1 | 1 | 39 | 40.2 b | 20.3 | 67.5 | 6.6 ** | 80.1 | 28.4 | 21.4 | |||
| QTL | AN | QTL Main | QTL × Env | Reported QTL/Marker | ||
|---|---|---|---|---|---|---|
| −log10p | R2 (%) | −log10p | R2 (%) | |||
| q-DLP-01-1 | 2 | 1.66 | 0.08 | |||
| q-DLP-01-2 | 2 | 1.55 | 0.07 | 4.24 | 0.28 | Gm01sv037 [6] |
| q-DLP-01-3 | 2 | 2.84 | 0.15 | Satt436 [6] | ||
| q-DLP-01-4 | 5 | 4.71 | 0.39 | |||
| q-DLP-01-5 | 2 | 1.42 | 0.09 | |||
| q-DLP-01-6 | 4 | 8.56 | 0.62 | 4.84 | 0.46 | |
| q-DLP-01-7 | 2 | 3.54 | 0.19 | Satt147 [6]/qRWF-1 [33] | ||
| q-DLP-02-1 | 6 | 1.41 | 0.17 | 4.83 | 0.58 | |
| q-DLP-02-2 | 4 | 1.67 | 0.14 | 2.77 | 0.30 | |
| q-DLP-02-3 | 2 | 9.45 | 0.57 | |||
| q-DLP-02-4 | 2 | 2.78 | 0.14 | 2.17 | 0.14 | |
| q-DLP-02-5 | 5 | 9.32 | 0.71 | 1.85 | 0.27 | |
| q-DLP-02-6 | 2 | 3.57 | 0.24 | Satt005 [6]/SIR-D1b [34] | ||
| q-DLP-02-7 | 2 | 2.92 | 0.15 | 4.10 | 0.27 | Sat_289 [6] |
| q-DLP-03-1 | 3 | 4.10 | 0.27 | 1.79 | 0.17 | |
| q-DLP-03-2 | 2 | 2.87 | 0.15 | Gm03sv093 [6] | ||
| q-DLP-04-1 | 2 | 1.97 | 0.09 | 1.81 | 0.12 | BP4-1 [35] |
| q-DLP-04-2 | 2 | 2.44 | 0.16 | |||
| q-DLP-05-1 | 2 | 2.29 | 0.15 | |||
| q-DLP-05-2 | 2 | 3.73 | 0.25 | |||
| q-DLP-06-1 | 3 | 7.14 | 0.47 | Satt681 [6] | ||
| q-DLP-06-2 | 2 | 1.59 | 0.07 | 2.03 | 0.13 | Satt489 [6] |
| q-DLP-06-3 | 2 | 4.66 | 0.26 | 3.82 | 0.25 | Satt489 [6] |
| q-DLP-06-4 | 2 | 1.87 | 0.12 | |||
| q-DLP-06-5 | 2 | 5.01 | 0.28 | 4.74 | 0.31 | |
| q-DLP-06-6 | 7 | 8.74 | 0.75 | 2.67 | 0.44 | Sat_312 [6] |
| q-DLP-06-7 | 2 | 9.00 | 0.54 | 3.28 | 0.22 | BP6-2 [35] |
| q-DLP-06-8 | 2 | 2.27 | 0.15 | |||
| q-DLP-07-1 | 2 | 2.92 | 0.15 | |||
| q-DLP-07-2 | 2 | 2.67 | 0.18 | |||
| q-DLP-07-3 | 2 | 3.78 | 0.25 | |||
| q-DLP-08-1 | 3 | 2.87 | 0.19 | 2.59 | 0.23 | |
| q-DLP-08-2 | 6 | 29.24 | 2.16 | 3.60 | 0.48 | |
| q-DLP-08-3 | 5 | 11.46 | 0.86 | 2.68 | 0.35 | |
| q-DLP-08-4 | 2 | 1.33 | 0.06 | |||
| q-DLP-08-5 | 2 | 1.86 | 0.12 | |||
| q-DLP-08-6 | 2 | 11.21 | 0.68 | |||
| q-DLP-08-7 | 2 | 24.2 | 1.55 | |||
| q-DLP-08-8 | 5 | 8.59 | 0.66 | 4.50 | 0.50 | |
| q-DLP-10-1 | 4 | 10.64 | 0.76 | 2.8 | 0.31 | |
| q-DLP-10-2 | 2 | 5.01 | 0.33 | |||
| q-DLP-10-3 | 2 | 14.02 | 0.87 | |||
| q-DLP-10-4 | 6 | 19.29 | 1.46 | 2.90 | 0.42 | |
| q-DLP-10-5 | 2 | 3.60 | 0.19 | BP10-2 [35] | ||
| q-DLP-10-6 | 2 | 2.03 | 0.13 | |||
| q-DLP-10-7 | 2 | 6.21 | 0.41 | |||
| q-DLP-11-1 | 2 | 3.01 | 0.20 | |||
| q-DLP-11-2 | 5 | 2.56 | 0.23 | |||
| q-DLP-11-3 | 2 | 2.04 | 0.13 | |||
| q-DLP-12-1 | 2 | 5.98 | 0.34 | |||
| q-DLP-12-2 | 2 | 2.71 | 0.14 | |||
| q-DLP-12-3 | 2 | 1.87 | 0.09 | |||
| q-DLP-12-4 | 5 | 71.48 | 5.16 | 2.43 | 0.33 | |
| q-DLP-12-5 | 2 | 2.38 | 0.16 | |||
| q-DLP-12-6 | 2 | 6.53 | 0.38 | |||
| q-DLP-12-7 | 2 | 5.37 | 0.30 | 2.71 | 0.18 | |
| q-DLP-12-8 | 2 | 2.48 | 0.12 | 3.44 | 0.23 | |
| q-DLP-13-1 | 2 | 13.42 | 0.83 | |||
| q-DLP-13-2 | 2 | 2.69 | 0.14 | |||
| q-DLP-13-3 | 11 | 22.1 | 1.88 | 7.25 | 1.06 | |
| q-DLP-13-4 | 2 | 6.05 | 0.35 | 2.84 | 0.19 | |
| q-DLP-13-5 | 2 | 3.10 | 0.20 | |||
| q-DLP-13-6 | 2 | 5.67 | 0.32 | |||
| q-DLP-13-7 | 2 | 7.12 | 0.47 | |||
| q-DLP-13-8 | 3 | 14.96 | 1.00 | 2.52 | 0.23 | QTL_13_1 [36] |
| q-DLP-14-1 | 7 | 17.82 | 1.40 | 2.95 | 0.47 | |
| q-DLP-14-2 | 2 | 1.98 | 0.09 | |||
| q-DLP-14-3 | 2 | 3.25 | 0.17 | Satt020 [6] | ||
| q-DLP-15-1 | 2 | 35.9 | 2.35 | 1.86 | 0.12 | |
| q-DLP-15-2 | 4 | 43.98 | 3.08 | 8.68 | 0.75 | Gm15sv557 [6] |
| q-DLP-15-3 | 2 | 3.87 | 0.21 | 5.13 | 0.34 | |
| q-DLP-15-4 | 2 | 1.98 | 0.13 | |||
| q-DLP-17-1 | 4 | 27.4 | 1.91 | 4.60 | 0.45 | |
| q-DLP-17-2 | 2 | 1.33 | 0.06 | 2.27 | 0.15 | |
| q-DLP-18-1 | 2 | 2.44 | 0.16 | |||
| q-DLP-18-2 | 2 | 2.06 | 0.10 | 3.79 | 0.25 | Gm18sv664 [6] |
| q-DLP-18-3 | 2 | 6.62 | 0.38 | Gm18sv704 [6] | ||
| q-DLP-18-4 | 7 | 32.42 | 2.44 | Gm18sv704 [6] | ||
| q-DLP-18-5 | 2 | 3.75 | 0.20 | 1.31 | 0.09 | Gm18sv704 [6] |
| q-DLP-18-6 | 2 | 2.81 | 0.14 | |||
| q-DLP-18-7 | 2 | 1.67 | 0.08 | 3.14 | 0.21 | |
| q-DLP-19-1 | 4 | 15.13 | 1.07 | 3.76 | 0.38 | |
| q-DLP-19-2 | 2 | 2.46 | 0.12 | 2.35 | 0.16 | |
| q-DLP-19-3 | 2 | 2.33 | 0.11 | |||
| q-DLP-20-1 | 2 | 2.02 | 0.10 | |||
| q-DLP-20-2 | 3 | 13.17 | 0.88 | GMES0205 [6] | ||
| LC-QTL | 12 (61) a | 25.45 | 1 (6) | 1.06 | ||
| SC-QTL | 54 (142) | 16.55 | 56 (166) | 14.79 | ||
| Total 86 | 243 | 66 (203) | 42.00 | 57 (172) | 15.85 | |
| ER | Minimum | Total No. of Crosses | Predicted DLP (%) | No. of Optimal Crosses | ||
|---|---|---|---|---|---|---|
| DLP (%) | Mean | Min. | Max. | |||
| ER-I | 18.9 | 903 | 36.6 (36.3) | 12.6 (10.4) | 62.3 (60.6) | 9 (16) |
| ER-II | 17.2 | 5253 | 34.3 (33.9) | 9.4 (10.3) | 62.4 (60.3) | 79 (85) |
| ER-III | 30.2 | 2485 | 42.1 (41.4) | 20.2 (16.8) | 65.5 (65.5) | 140 (191) |
| ER-IV | 19.8 | 3160 | 39.1 (39.1) | 5.1 (8.9) | 71.1 (71.3) | 106 (83) |
| ER-V | 25.0 | 561 | 35.3 (35.1) | 20.4 (20.5) | 55.8 (56.5) | 23 (30) |
| ER-VI | 20.3 | 741 | 33.0 (32.4) | 5.7 (9.5) | 59.1 (59.9) | 70 (73) |
| Within | 17.2 | 13,103 | 37.1 (36.7) | 5.1 (8.9) | 71.1 (71.3) | 180 (171) |
| Between | 17.2 | 55,162 | 37.0 (36.6) | 3.0 (6.7) | 71.7 (70.6) | 695 (659) |
| All | 17.2 | 68,265 | 37.0 (36.6) | 3.0 (6.7) | 71.7 (71.3) | 875 (830) |
| Code | Parent 1 | Parent 2 | DLP of Parent 1 | DLP of Parent 2 | Predicted DLP | ||
|---|---|---|---|---|---|---|---|
| Mean | 25th Percentile | 20th Percentile | |||||
| 1 | S04 | 4L367 | 17.2 | 21.1 | 18.5 (19.3) | 3.0 (6.7) | 0.1 (3.9) |
| 2 | S03 | 4L367 | 19.8 | 21.1 | 20.4 (20.0) | 3.7 (7.5) | 0.2 (4.3) |
| 3 | S03 | S06 | 19.8 | 23.1 | 20.5 (20.6) | 5.1 (8.9) | 1.3 (6.1) |
| 4 | S04 | S06 | 17.2 | 23.1 | 19.8 (19.6) | 5.6 (7.0) | 2.4 (4.2) |
| 5 | 4L262 | 4L367 | 20.3 | 21.1 | 21.1 (20.4) | 5.7 (9.5) | 3.3 (7.2) |
| 6 | 4L250 | 4L367 | 24.1 | 21.1 | 23.0 (22.1) | 5.9 (10.3) | 3.0 (7.5) |
| 7 | 4L187 | 4L367 | 25.6 | 21.1 | 22.9 (24.1) | 6.2 (11.5) | 3.1 (8.8) |
| 8 | 4L088 | 4L367 | 25.7 | 21.1 | 23.5 (23.2) | 6.3 (9.7) | 2.8 (6.9) |
| 9 | S03 | 4L242 | 19.8 | 29.4 | 24.0 (24.8) | 6.6 (12.1) | 3.4 (8.9) |
| 10 | S04 | 4L242 | 17.2 | 29.4 | 23.5 (23.5) | 7.1 (10.9) | 3.7 (7.9) |
| 11 | 4L074 | 4L367 | 23.3 | 21.1 | 21.8 (22.4) | 7.3 (11.2) | 5.6 (8.3) |
| 12 | 4L263 | 4L367 | 26.1 | 21.1 | 23.3 (23.3) | 7.5 (11.8) | 4.7 (9.4) |
| 13 | 4L261 | 4L367 | 23.6 | 21.1 | 22.0 (22.5) | 7.6 (10.2) | 3.8 (6.9) |
| 14 | S06 | 4L250 | 23.1 | 24.1 | 23.4 (23.2) | 7.6 (11.0) | 5.6 (8.7) |
| 15 | 4L090 | 4L367 | 28.2 | 21.1 | 24.8 (24.3) | 7.8 (12.3) | 4.8 (8.9) |
| 16 | 4L041 | S04 | 27.0 | 17.2 | 21.5 (22.8) | 7.9 (8.7) | 4.7 (5.8) |
| 17 | S06 | 4L262 | 23.1 | 20.3 | 21.8 (22.6) | 7.9 (11.6) | 5.1 (9.0) |
| 18 | S05 | 4L367 | 27.2 | 21.1 | 24.7 (24.4) | 7.9 (13.5) | 5.9 (11.0) |
| 19 | S10 | 4L367 | 24.2 | 21.1 | 22.6 (22.9) | 8.0 (11.2) | 5.5 (8.2) |
| 20 | S02 | 4L367 | 18.9 | 21.1 | 19.8 (19.9) | 8.0 (8.6) | 5.1 (5.8) |
| QTL | Position (bp) | Candidate Gene | Gene Ontology Description (Category) |
|---|---|---|---|
| q-DLP-01-1 | 2,018,307 | Glyma01g02525 | Xylan biosynthetic process (II) |
| q-DLP-01-2 | 45,358,479 | ||
| q-DLP-01-3 | 48,727,687 | Glyma01g36200 | Response to salicylic acid stimulus (I) |
| q-DLP-01-4 | 49,414,457_49,512,678 | Glyma01g37020 | Regulation of meristem growth (VI) |
| q-DLP-01-5 | 50,563,918_50,575,958 | Glyma01g38470 | Response to auxin stimulus (I) |
| q-DLP-01-6 | 51,562,541_51,627,699 | Glyma01g39590 | Xylan biosynthetic process (II) |
| q-DLP-01-7 | 52,647,305 | Glyma01g41110 | Hormone-mediated signaling pathway (IV) |
| q-DLP-02-1 | 1,973,000_2,157,204 | Glyma02g02860 | Toxin catabolic process (II) |
| q-DLP-02-2 | 4,018,617_4,025,103 | Glyma02g04965 | Response to chitin (I) |
| q-DLP-02-3 | 14,648,440_14,667,968 | Glyma02g16260 | Response to abscisic acid stimulus (I) |
| q-DLP-02-4 | 17,944,684 | ||
| q-DLP-02-5 | 25,406,263_25,479,114 | ||
| q-DLP-02-6 | 29,142,262_29,263,830 | Glyma02g27950 | Unknown process (VIII) |
| q-DLP-02-7 | 51,455,660 | Glyma02g48060 | Transmembrane transport (V) |
| q-DLP-03-1 | 5,122,536_5,122,820 | Glyma03g04920 | Lipid transport (V) |
| q-DLP-03-2 | 14,208,130 | ||
| q-DLP-04-1 | 7,402,649 | Glyma04g09220 | Unknown process (VIII) |
| q-DLP-04-2 | 46,268,205_46,268,222 | Glyma04g40121 | Regulation of transcription, DNA-dependent (VII) |
| q-DLP-05-1 | 18,014,789 | ||
| q-DLP-05-2 | 24,588,612 | ||
| q-DLP-06-1 | 420,122_487,621 | Glyma06g00730 | Response to insect (I) |
| q-DLP-06-2 | 22,275,863 | Glyma06g24701 | Unknown process (VIII) |
| q-DLP-06-3 | 24,692,151 | ||
| q-DLP-06-4 | 29,026,459 | ||
| q-DLP-06-5 | 32,468,737 | ||
| q-DLP-06-6 | 35,040,678_35,236,874 | Glyma06g33880 | Protein glycosylation (III) |
| q-DLP-06-7 | 44,178,948 | ||
| q-DLP-06-8 | 50,304,823_50,305,172 | Glyma06g47920 | Unknown process (VIII) |
| q-DLP-07-1 | 28,717,244 | ||
| q-DLP-07-2 | 36,293,327_36,297,011 | Glyma07g31320 | Protein complex assembly (III) |
| q-DLP-07-3 | 40,096,883 | Glyma07g35023 | Unknown process (VIII) |
| q-DLP-08-1 | 14,753,730_14,760,906 | Glyma08g19560 | Response to wounding (I) |
| q-DLP-08-2 | 14,777,419_14,822,821 | Glyma08g19650 | Sugar-mediated signaling pathway (IV) |
| q-DLP-08-3 | 22,259,705_22,457,223 | Glyma08g28111 | Transcription, DNA-dependent (VII) |
| q-DLP-08-4 | 31,137,298 | ||
| q-DLP-08-5 | 40,375,262 | Glyma08g40646 | Signal transduction (IV) |
| q-DLP-08-6 | 41,582,869 | Glyma08g41670 | Hydrogen peroxide biosynthetic process (II) |
| q-DLP-08-7 | 43,612,649 | Glyma08g43830 | Transmembrane transport (V) |
| q-DLP-08-8 | 45,217,877_45,266,178 | Glyma08g45950 | Response to wounding (I) |
| q-DLP-10-1 | 2,137,913_2,266,216 | Glyma10g03140 | Regulation of flavonoid biosynthetic process (II) |
| q-DLP-10-2 | 2,777,471 | Glyma10g03720 | Response to jasmonic acid stimulus (I) |
| q-DLP-10-3 | 4,019,592 | Glyma10g05160 | Xylan biosynthetic process (II) |
| q-DLP-10-4 | 24,526,775_24,706,774 | Glyma10g19710 | Unknown process (VIII) |
| q-DLP-10-5 | 39,555,777 | Glyma10g30930 | Unknown process (VIII) |
| q-DLP-10-6 | 45,227,388 | ||
| q-DLP-10-7 | 46,996,455 | Glyma10g39250 | Unknown process (VIII) |
| q-DLP-11-1 | 4,931,380 | Glyma11g07080 | Indoleacetic acid biosynthetic process (II) |
| q-DLP-11-2 | 28,787,840_28,938,957 | Glyma11g28580 | Unknown process (VIII) |
| q-DLP-11-3 | 35,388,102 | ||
| q-DLP-12-1 | 4,069,210_4,160,826 | Glyma12g05960 | Sugar-mediated signaling pathway (IV) |
| q-DLP-12-2 | 5,802,081 | Glyma12g08080 | Regulation of meristem growth (VI) |
| q-DLP-12-3 | 6,664,148_6,664,175 | Glyma12g08900 | Response to oxidative stress (I) |
| q-DLP-12-4 | 10,441,681_10,513,287 | ||
| q-DLP-12-5 | 14,076,555 | ||
| q-DLP-12-6 | 21,170,928 | ||
| q-DLP-12-7 | 28,906,122 | ||
| q-DLP-12-8 | 31,830,778 | Glyma12g28490 | Selenium compound metabolic process (II) |
| q-DLP-13-1 | 5,519,025 | ||
| q-DLP-13-2 | 7,661,807_7,661,825 | Glyma13g07530 | Unknown process (VIII) |
| q-DLP-13-3 | 11,608,283_11,644,846 | Glyma13g10070 | Positive regulation of flavonoid biosynthetic process (II) |
| q-DLP-13-4 | 16,616,018_16,616,049 | ||
| q-DLP-13-5 | 22,285,871 | ||
| q-DLP-13-6 | 22,414,892 | Glyma13g18720 | Plant-type cell wall modification (VI) |
| q-DLP-13-7 | 25,945,441 | Glyma13g22450 | Sugar-mediated signaling pathway (IV) |
| q-DLP-13-8 | 29,302,932_29,302,990 | Glyma13g26030 | Unknown process (VIII) |
| q-DLP-14-1 | 13,354,054_13,553,649 | Glyma14g13792 | Histone H3-K4 methylation (III) |
| q-DLP-14-2 | 15,737,492 | Glyma14g14990 | Jasmonic acid biosynthetic process (II) |
| q-DLP-14-3 | 43,771,273 | ||
| q-DLP-15-1 | 1,215,528 | Glyma15g01790 | Protein ubiquitination (III) |
| q-DLP-15-2 | 42,911,867_42,923,525 | Glyma15g37276 | Defense response to fungus (I) |
| q-DLP-15-3 | 47,989,791_47,999,025 | Glyma15g41020 | Regulation of meristem growth (VI) |
| q-DLP-15-4 | 48,785,322_48,785,485 | Glyma15g41640 | Oxidation-reduction process (II) |
| q-DLP-17-1 | 13,426,556_13,441,932 | Glyma17g16641 | Regulation of transcription, DNA-dependent (VII) |
| q-DLP-17-2 | 32,299,189 | Glyma17g29620 | Protein N-linked glycosylation (III) |
| q-DLP-18-1 | 3,801,888 | Glyma18g05090 | Positive regulation of cell proliferation (VI) |
| q-DLP-18-2 | 12,477,590 | ||
| q-DLP-18-3 | 55,745,667 | Glyma18g46050 | Defense response (I) |
| q-DLP-18-4 | 55,988,107_55,988,755 | Glyma18g46220 | Response to cold (I) |
| q-DLP-18-5 | 56,055,671 | Glyma18g46286 | Transmembrane transport (V) |
| q-DLP-18-6 | 58,793,224 | Glyma18g49390 | Unknown process (VIII) |
| q-DLP-18-7 | 60,245,212 | Glyma18g51380 | Polyamine biosynthetic process (II) |
| q-DLP-19-1 | 5,454,340_5,454,561 | ||
| q-DLP-19-2 | 43,592,061 | Glyma19g36280 | Xylan biosynthetic process (II) |
| q-DLP-19-3 | 47,642,370 | Glyma19g41400 | Protein glycosylation (III) |
| q-DLP-20-1 | 16,840,184 | Glyma20g11950 | Oxidation-reduction process (II) |
| q-DLP-20-2 | 35,128,616_35,247,501 | Glyma20g25418 | Systemic acquired resistance (I) |
| Antixenosis | Total Alleles | Inherent Alleles | Changed Alleles | Allele Increased | Allele Decreased | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Degree | Allele No. | QTL No. | Allele No. | QTL No. | Allele No. | QTL No. | Allele No. | QTL No. | Allele No. | QTL No. |
| SV | 186 (93, 93) | 66 | ||||||||
| MRV vs. SV | 202 (104, 98) | 66 | 185 (93, 92) | 66 | 18 (11, 7) | 15 | 17 (11, 6) | 14 | 1 (0, 1) | 1 |
| RV vs. MRV | 187 (102, 85) | 66 | 186 (102, 84) | 66 | 17 (2, 15) | 16 | 1 (0, 1) | 1 | 16 (2, 14) | 15 |
| RV vs. SV | 187 (102, 85) | 66 | 172 (92, 80) | 66 | 29 (11, 18) | 23 | 15 (10, 5) | 12 | 14 (1, 13) | 13 |
| QTL | R2 (%) | Allele | Effect | Frequency (%) | Annotated Gene (Category) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Entire | SV | MRV | RV | MRV + RV | |||||
| q-DLP-01-7 | 0.19 | a1 | −7.01 | 1.08 | 0 | 0.90 | 2.70 | 1.35 | Glyma01g41110 (IV) |
| q-DLP-02-1 | 0.17 | a4 | −0.26 | 2.97 | 0 | 3.15 | 5.41 | 3.72 | Glyma02g02860 (II) |
| q-DLP-02-4 | 0.14 | a1 | −2.35 | 1.35 | 0 | 2.25 | 0 | 1.69 | |
| q-DLP-02-5 | 0.71 | a3 | 0.36 | 6.22 | 0 | 4.05 | 18.92 | 7.77 | |
| q-DLP-02-7 | 0.15 | a2 | 4.92 | 1.62 | 2.70 | 1.80 | 0 | 1.35 | Glyma02g48060 (V) |
| q-DLP-04-1 | 0.09 | a2 | 2.65 | 1.35 | 2.70 | 1.35 | 0 | 1.01 | Glyma04g09220 (VIII) |
| q-DLP-06-3 | 0.26 | a2 | 7.47 | 1.35 | 4.05 | 0.90 | 0 | 0.68 | |
| q-DLP-06-6 | 0.75 | a1 | −7.95 | 1.35 | 0 | 1.80 | 1.35 | 1.69 | Glyma06g33880 (III) |
| a2 | −1.42 | 1.89 | 1.35 | 2.70 | 0 | 2.03 | |||
| a7 | 5.88 | 1.89 | 2.70 | 2.25 | 0 | 1.69 | |||
| q-DLP-08-4 | 0.06 | a1 | −2.76 | 1.08 | 0 | 0.90 | 2.70 | 1.35 | |
| q-DLP-08-7 | 1.55 | a1 | −11.02 | 2.16 | 0 | 0.90 | 8.11 | 2.70 | Glyma08g43830 (V) |
| q-DLP-08-8 | 0.66 | a5 | 4.76 | 1.35 | 4.05 | 0.90 | 0 | 0.68 | Glyma08g45950 (I) |
| q-DLP-10-1 | 0.76 | a1 | −5.06 | 5.68 | 0 | 6.76 | 8.11 | 7.09 | Glyma10g03140 (II) |
| a2 | −1.41 | 2.70 | 0 | 1.80 | 8.11 | 3.38 | |||
| q-DLP-10-3 | 0.87 | a2 | 7.38 | 1.35 | 4.05 | 0.90 | 0 | 0.68 | Glyma10g05160 (II) |
| q-DLP-10-4 | 1.46 | a1 | −6.51 | 1.89 | 0 | 1.35 | 5.41 | 2.36 | Glyma10g19710 (VIII) |
| a6 | 5.92 | 1.08 | 0 | 1.35 | 1.35 | 1.35 | |||
| q-DLP-11-2 | 0.23 | a5 | 4.46 | 2.43 | 4.05 | 2.70 | 0 | 2.03 | Glyma11g28580 (VIII) |
| q-DLP-12-6 | 0.38 | a2 | 7.09 | 1.08 | 2.70 | 0.90 | 0 | 0.68 | |
| q-DLP-12-7 | 0.30 | a2 | 6.38 | 1.08 | 4.05 | 0 | 1.35 | 0.34 | |
| q-DLP-12-8 | 0.12 | a2 | 2.53 | 1.89 | 2.70 | 2.25 | 0 | 1.69 | Glyma12g28490 (II) |
| q-DLP-13-1 | 0.83 | a1 | −8.46 | 1.08 | 0 | 0.45 | 4.05 | 1.35 | |
| q-DLP-13-3 | 1.88 | a1 | −6.53 | 1.62 | 0 | 1.35 | 4.05 | 2.03 | Glyma13g10070 (II) |
| a10 | 5.06 | 2.70 | 2.70 | 3.60 | 0 | 2.70 | |||
| a11 | 6.60 | 2.70 | 0 | 2.70 | 5.41 | 3.38 | |||
| q-DLP-14-1 | 1.40 | a7 | 3.65 | 1.35 | 0 | 1.80 | 1.35 | 1.69 | Glyma14g13792 (III) |
| q-DLP-17-2 | 0.06 | a2 | 2.42 | 1.62 | 2.70 | 1.80 | 0 | 1.35 | Glyma17g29620 (III) |
| q-DLP-18-3 | 0.38 | a2 | 7.88 | 1.08 | 1.35 | 1.35 | 0 | 1.01 | Glyma18g46050 (I) |
| q-DLP-18-4 | 2.44 | a6 | 5.93 | 1.35 | 0 | 1.35 | 2.70 | 1.69 | Glyma18g46220 (I) |
| q-DLP-19-2 | 0.12 | a2 | 2.75 | 1.35 | 0 | 2.25 | 0 | 1.69 | Glyma19g36280 (II) |
| q-DLP-19-3 | 0.11 | a2 | 2.92 | 1.62 | 2.70 | 1.80 | 0 | 1.35 | Glyma19g41400 (III) |
| 26 | 32 | 19 genes (3 I, 6 II, 4 III, 1 IV, 2 V, 3 VIII) 25 QTL-alleles (3 I, 9 II, 6 III, 1 IV, 2 V, 4 VIII) | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, L.; Gai, J.; Xing, G. The Identification of a Quantative Trait Loci-Allele System of Antixenosis against the Common Cutworm (Spodoptera litura Fabricius) at the Seedling Stage in the Chinese Soybean Landrace Population. Int. J. Mol. Sci. 2023, 24, 16089. https://doi.org/10.3390/ijms242216089
Pan L, Gai J, Xing G. The Identification of a Quantative Trait Loci-Allele System of Antixenosis against the Common Cutworm (Spodoptera litura Fabricius) at the Seedling Stage in the Chinese Soybean Landrace Population. International Journal of Molecular Sciences. 2023; 24(22):16089. https://doi.org/10.3390/ijms242216089
Chicago/Turabian StylePan, Lin, Junyi Gai, and Guangnan Xing. 2023. "The Identification of a Quantative Trait Loci-Allele System of Antixenosis against the Common Cutworm (Spodoptera litura Fabricius) at the Seedling Stage in the Chinese Soybean Landrace Population" International Journal of Molecular Sciences 24, no. 22: 16089. https://doi.org/10.3390/ijms242216089
APA StylePan, L., Gai, J., & Xing, G. (2023). The Identification of a Quantative Trait Loci-Allele System of Antixenosis against the Common Cutworm (Spodoptera litura Fabricius) at the Seedling Stage in the Chinese Soybean Landrace Population. International Journal of Molecular Sciences, 24(22), 16089. https://doi.org/10.3390/ijms242216089








