Template-Assisted Assembly of Hybrid DNA/RNA Nanostructures Using Branched Oligodeoxy- and Oligoribonucleotides
Abstract
:1. Introduction
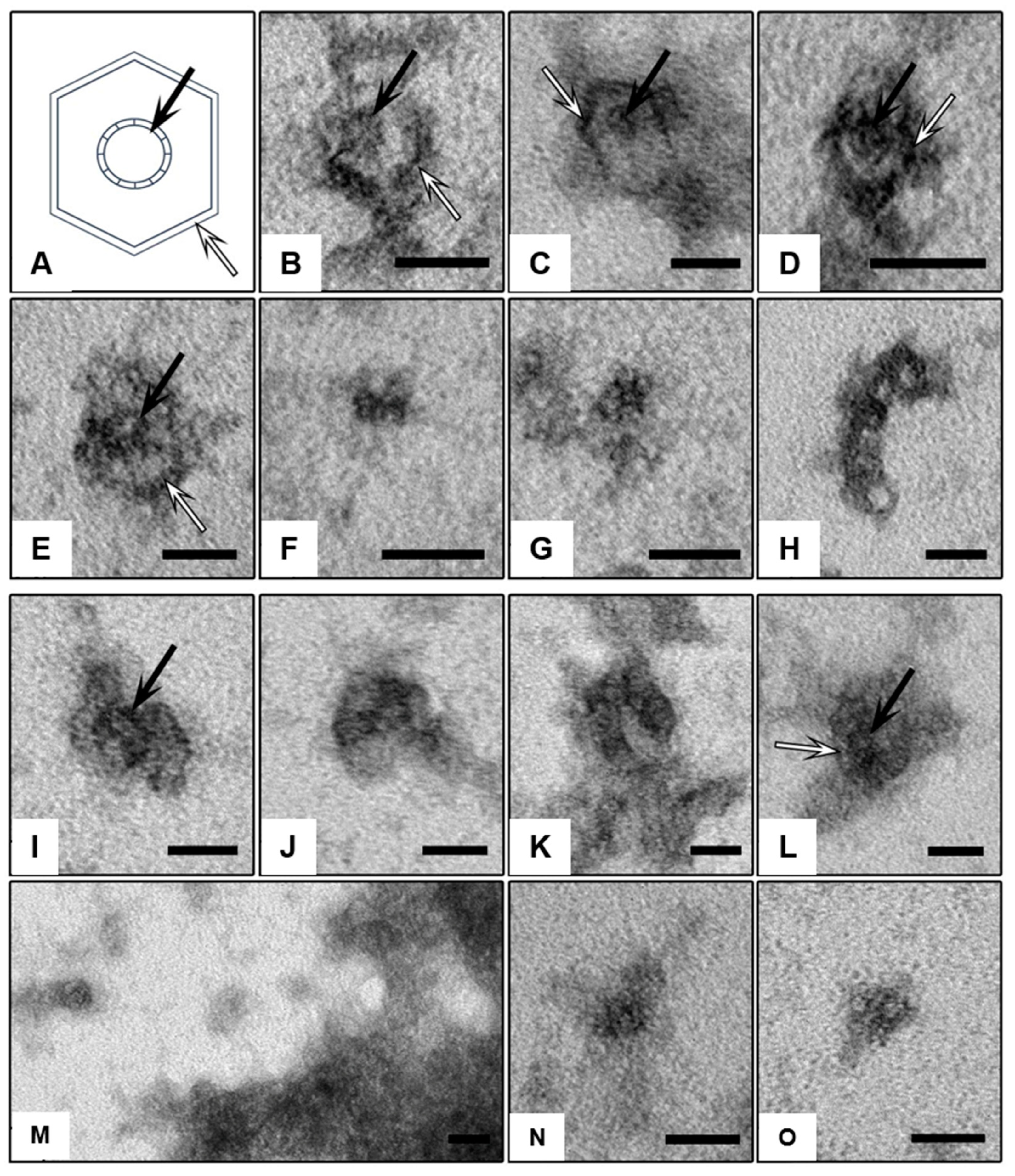
2. Results
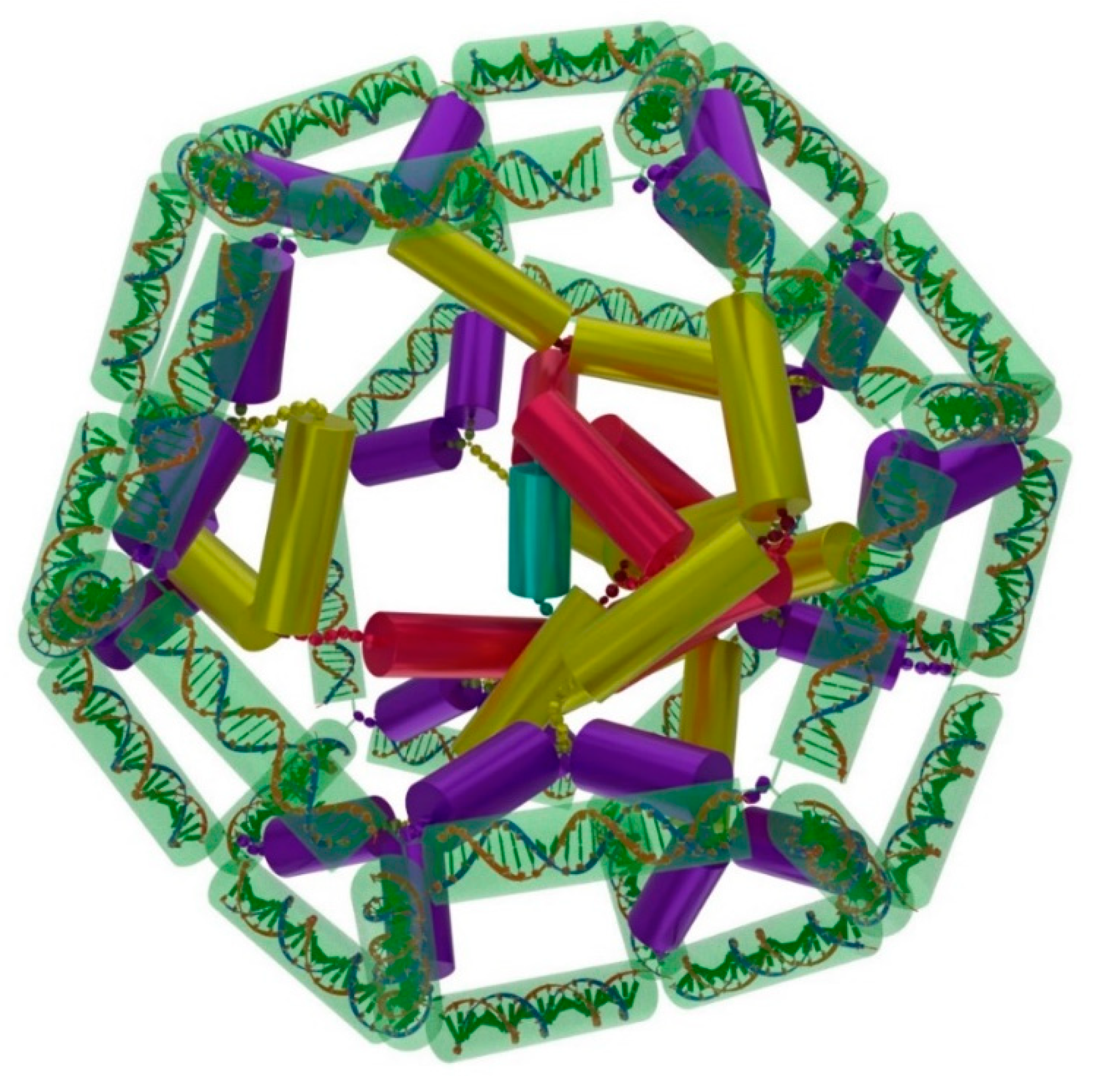
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seeman, N.C.; Sleiman, H.F. DNA Nanotechnology. Nat. Rev. Mater. 2018, 3, 17068. [Google Scholar] [CrossRef]
- Seeman, N.C. DNA Nanotechnology at 40. Nano Lett. 2020, 20, 1477–1478. [Google Scholar] [CrossRef]
- Govindaraju, T. (Ed.) Templated DNA Nanotechnology: Functional DNA Nanoarchitectonics; Pan Stanford Publishing Pte. Ltd.: Singapore, 2019; ISBN 978-0-429-42866-1. [Google Scholar]
- Ghosh, D.; Datta, L.P.; Govindaraju, T. Molecular architectonics of DNA for functional nanoarchitectures. Beilstein J. Nanotechnol. 2020, 11, 124–140. [Google Scholar] [CrossRef]
- Rajwar, A.; Kharbanda, S.; Chandrasekaran, A.R.; Gupta, S.; Bhatia, D. Designer, Programmable 3D DNA Nanodevices to Probe Biological Systems. ACS Appl. Bio Mater. 2020, 3, 7265–7277. [Google Scholar] [CrossRef]
- Bhatia, D.; Wunder, C.; Johannes, L. Self-assembled, Programmable DNA Nanodevices for Biological and Biomedical Applications. ChemBioChem 2021, 22, 763–778. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z.; Wu, X.; Liu, S.; Liu, F.; Jiang, Q.; Ding, B. DNA Nanodevices: From Mechanical Motions to Biomedical Applications. Curr. Top. Med. Chem. 2022, 22, 640–651. [Google Scholar] [CrossRef]
- Huang, J.; Gambietz, S.; Saccà, B. Self-Assembled Artificial DNA Nanocompartments and Their Bioapplications. Small 2022, 19, e2202253. [Google Scholar] [CrossRef]
- Angell, C.; Kai, M.; Xie, S.; Dong, X.; Chen, Y. Bioderived DNA Nanomachines for Potential Uses in Biosensing, Diagnostics, and Therapeutic Applications. Adv. Healthc. Mater. 2018, 7, 1701189. [Google Scholar] [CrossRef]
- Li, J.; Huang, J. Fuel-Powered DNA Nanomachines for Biosensing and Cancer Therapy. ChemPlusChem 2022, 87, e202200098. [Google Scholar] [CrossRef]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Anderson, G.J.; Han, J.-Y.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef]
- Mirzaiebadizi, A.; Ravan, H.; Dabiri, S.; Mohammadi, P.; Shahba, A.; Ziasistani, M.; Khatami, M. An intelligent DNA nanorobot for detection of MiRNAs cancer biomarkers using molecular programming to fabricate a logic-responsive hybrid nanostructure. Bioprocess Biosyst. Eng. 2022, 45, 1781–1797. [Google Scholar] [CrossRef]
- Wang, M.; Li, X.; He, F.; Li, J.; Wang, H.H.; Nie, Z. Advances in Designer DNA Nanorobots Enabling Programmable Functions. ChemBioChem 2022, 23, e202200119. [Google Scholar] [CrossRef]
- Hu, Y.; Cecconello, A.; Idili, A.; Ricci, F.; Willner, I. Triplex DNA Nanostructures: From Basic Properties to Applications. Angew. Chem. Int. Ed. Engl. 2017, 56, 15210–15233. [Google Scholar] [CrossRef]
- Lat, P.K.; Schultz, C.W.; Yu, H.Z.; Sen, D. A Long and Reversibly Self-Assembling 1D DNA Nanostructure Built from Triplex and Quadruplex Hybrid Tiles. Angew. Chem. Int. Ed. Engl. 2021, 60, 8722–8727. [Google Scholar] [CrossRef]
- Lin, P.Y.; Chi, R.; Wu, Y.L.; Ho, J.A. Applications of triplex DNA nanostructures in sensor development. Anal. Bioanal. Chem. 2022, 414, 5217–5237. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef]
- Wang, P.; Meyer, T.A.; Pan, V.; Dutta, P.K.; Yonggang Ke, Y. The Beauty and Utility of DNA Origami. Chem 2017, 2, 359–382. [Google Scholar] [CrossRef]
- Dey, S.; Fan, C.; Gothelf, K.V.; Li, J.; Lin, C.; Liu, L.; Liu, N.; Nijenhuis, M.A.D.; Saccà, B.; Simmel, F.C.; et al. DNA origami. Nat. Rev. Methods Primers. 2021, 1, 13. [Google Scholar] [CrossRef]
- Liu, W.; Duan, H.; Zhang, D.; Zhang, X.; Luo, Q.; Xie, T.; Yan, H.; Peng, L.; Hu, Y.; Liang, L.; et al. Concepts and Application of DNA Origami and DNA Self-Assembly: A Systematic Review. Appl. Bionics Biomech. 2021, 2021, 9112407. [Google Scholar] [CrossRef]
- Arrabito, G.; Wang, L. (Eds.) DNA Nanotechnology for Bioanalysis: From Hybrid DNA Nanostructures to Functional Devices; World Scientific Publishing Europe Ltd.: Singapore, 2018. [Google Scholar] [CrossRef]
- Yu, S.; Chen, T.; Zhang, Q.; Zhoua, M.; Zhu, X. Application of DNA nanodevices for biosensing. Analyst 2020, 145, 3481–3489. [Google Scholar] [CrossRef]
- Loretan, M.; Domljanovic, I.; Lakatos, M.; Rüegg, C.; Acuna, G.P. DNA Origami as Emerging Technology for the Engineering of Fluorescent and Plasmonic-Based Biosensors. Materials 2020, 13, 2185. [Google Scholar] [CrossRef]
- Muhammad, M.; Baig, F.A.; Lai, W.-F.; Akhtar, M.F.; Saleem, A.; Ahmed, S.A.; Xia, X.-H. DNA nanotechnology as a tool to develop molecular tension probes for bio-sensing and bio-imaging applications: An up-to-date review. Nano-Struct. Nano-Objects 2020, 23, 100523. [Google Scholar] [CrossRef]
- Li, F.; Li, J.; Dong, B.; Wang, F.; Fan, C.; Zuo, X. DNA nanotechnology-empowered nanoscopic imaging of biomolecules. Chem. Soc. Rev. 2021, 50, 5650–5667. [Google Scholar] [CrossRef]
- Yang, Q.; Chang, X.; Lee, J.Y.; Olivera, T.R.; Saji, M.; Wisniewski, H.; Kim, S.; Zhang, F. Recent Advances in Self-Assembled DNA Nanostructures for Bioimaging. ACS Appl. Bio Mater. 2022, 5, 4652–4667. [Google Scholar] [CrossRef]
- Kuzyk, A.; Jungmann, R.; Acuna, G.P.; Liu, N. DNA Origami Route for Nanophotonics. ACS Photonics 2018, 5, 1151–1163. [Google Scholar] [CrossRef]
- Dai, L.; Liu, P.; Hu, X.; Zhao, X.; Shao, G.; Tian, Y. DNA origami: An outstanding platform for functions in nanophotonics and cancer therapy. Analyst 2021, 146, 1807–1819. [Google Scholar] [CrossRef]
- Chi, Q.; Yang, Z.; Xu, K.; Wang, C.; Liang, H. DNA Nanostructure as an Efficient Drug Delivery Platform for Immunotherapy. Front. Pharmacol. 2019, 10, 1585. [Google Scholar] [CrossRef]
- Wang, Y.; Chen-Mayfield, T.J.; Li, Z.; Younis, M.H.; Cai, W.; Hu, Q. Harnessing DNA for immunotherapy: Cancer, infectious diseases, and beyond. Adv. Funct. Mater. 2022, 32, 2112273. [Google Scholar] [CrossRef]
- Wu, X.; Wu, T.; Liu, J.; Ding, B. Gene Therapy Based on Nucleic Acid Nanostructure. Adv. Healthc. Mater. 2020, 9, e2001046. [Google Scholar] [CrossRef]
- Gangrade, A.; Stephanopoulos, N.; Bhatia, D. Programmable, self-assembled DNA nanodevices for cellular programming and tissue engineering. Nanoscale 2021, 13, 16834–16846. [Google Scholar] [CrossRef]
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev. 2019, 119, 6459–6506. [Google Scholar] [CrossRef]
- Xu, F.; Xia, Q.; Wang, P. Rationally Designed DNA Nanostructures for Drug Delivery. Front. Chem. 2020, 8, 751. [Google Scholar] [CrossRef] [PubMed]
- Readman, J.B.; Dickson, G.; Coldham, N.G. Tetrahedral DNA Nanoparticle Vector for Intracellular Delivery of Targeted Peptide Nucleic Acid Antisense Agents to Restore Antibiotic Sensitivity in Cefotaxime-Resistant Escherichia coli. Nucleic Acid Ther. 2017, 27, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.M.; Keller, A. DNA Nanostructures in the Fight Against Infectious Diseases. Adv. Nanobiomed. Res. 2021, 1, 2000049. [Google Scholar] [CrossRef]
- Lv, Z.; Zhu, Y.; Li, F. DNA Functional Nanomaterials for Controlled Delivery of Nucleic Acid-Based Drugs. Front. Bioeng. Biotechnol. 2021, 9, 720291. [Google Scholar] [CrossRef]
- Bush, J.; Singh, S.; Vargas, M.; Oktay, E.; Hu, C.-H.; Veneziano, R. Synthesis of DNA Origami Scaffolds: Current and Emerging Strategies. Molecules 2020, 25, 3386. [Google Scholar] [CrossRef]
- Madsen, M.; Gothelf, K.V. Chemistries for DNA Nanotechnology. Chem. Rev. 2019, 119, 6384–6458. [Google Scholar] [CrossRef]
- Henry, S.J.W.; Stephanopoulos, N. Functionalizing DNA nanostructures for therapeutic applications. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2021, 13, e1729. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, X.; Wu, X.; Li, Y.; Tang, W.; Yang, C.; Liu, J.; Ding, B. Chemically modified DNA nanostructures for drug delivery. Innovation 2022, 3, 100217. [Google Scholar] [CrossRef]
- Frtús, A.; Smolková, B.; Uzhytchak, M.; Lunova, M.; Jirsa, M.; Henry, S.J.W.; Dejneka, A.; Stephanopoulos, N.; Lunov, O. The interactions between DNA nanostructures and cells: A critical overview from a cell biology perspective. Acta Biomater. 2022, 146, 10–22. [Google Scholar] [CrossRef]
- Balakrishnan, D.; Wilkens, G.D.; Heddle, J.D. Delivering DNA origami to cells. Nanomedicine 2019, 14, 911–925. [Google Scholar] [CrossRef]
- Rajwar, A.; Shetty, S.R.; Vaswani, P.; Morya, V.; Barai, A.; Sen, S.; Sonawane, M.; Bhatia, D. Geometry of a DNA Nanostructure Influences Its Endocytosis: Cellular Study on 2D, 3D, and in Vivo Systems. ACS Nano 2022, 16, 10496–10508. [Google Scholar] [CrossRef]
- Chandrasekaran, A.R. Nuclease resistance of DNA nanostructures. Nat. Rev. Chem. 2021, 5, 225–239. [Google Scholar] [CrossRef]
- Bakulina, A.Y.; Rad’kova, Z.V.; Burakova, E.A.; Benassi, E.; Zatsepin, T.S.; Fokina, A.A.; Stetsenko, D.A. Design and Visualization of DNA/RNA Nanostructures from Branched Oligonucleotides Using Blender Software. Russ. J. Bioorg. Chem. 2019, 45, 608–618. [Google Scholar] [CrossRef]
- Fokina, A.A.; Poletaeva, Y.E.; Burakova, E.A.; Bakulina, A.Y.; Zatsepin, T.S.; Ryabchikova, E.I.; Stetsenko, D.A. Template-Assisted Assembly of DNA Nanostructures from Branched Oligonucleotides. Russ. J. Bioorg. Chem. 2021, 47, 700–712. [Google Scholar] [CrossRef]
- Silant’ev, A.V. Energy Spectrum and Optical Properties of C24 Fullerene within the Hubbard Model. Phys. Metals Metallogr. 2020, 121, 195–201. [Google Scholar] [CrossRef]
- Mahani, N.M.; Sabermahani, F.; Shamsolmaali, A. C24 fullerene as drug delivery for anticancer activity of pyridine derivatives: A density functional theory approach. Pak. J. Pharm. Sci. 2019, 2, 2741–2744. [Google Scholar]
- Tomić, B.T.; Abraham, C.S.; Pelemiš, S.; Armaković, S.J.; Armaković, S. Fullerene C24 as a potential carrier of ephedrine drug—A computational study of interactions and influence of temperature. Phys. Chem. Chem. Phys. 2019, 21, 23329–23337. [Google Scholar] [CrossRef]
- Blender 3.6 LTS. Available online: http://www.blender.org (accessed on 17 July 2023).
- Chan, K.F.; Poh, J.J.; Wu, W.L.; Gan, S.K.E. Augmented reality in scientific visualization and communications: A new dawn of looking at antibody interactions. Antib. Ther. 2020, 3, 221–226. [Google Scholar] [CrossRef]
- Yeh, C.T.; Obendorf, L.; Parmeggiani, F. Elfin UI: A Graphical Interface for Protein Design with Modular Building Blocks. Front. Bioeng. Biotechnol. 2020, 23, 568318. [Google Scholar] [CrossRef]
- Brown, J.W.; Alford, R.G.; Walsh, J.C.; Spinney, R.E.; Xu, S.Y.; Hertel, S.; Berengut, J.F.; Spenkelink, L.M.; Van Oijen, A.M.; Bocking, T.; et al. Rapid exchange of stably bound protein and DNA cargo on a DNA origami receptor. ACS Nano 2022, 16, 6455–6467. [Google Scholar] [CrossRef] [PubMed]
- Azizyan, R.A.; Garro, A.; Radkova, Z.; Anikeenko, A.; Bakulina, A.; Dumas, C.; Kajava, A.V. Establishment of Constraints on Amyloid Formation Imposed by Steric Exclusion of Globular Domains. J. Mol. Biol. 2018, 430, 3835–3846. [Google Scholar] [CrossRef] [PubMed]
- Aviñó, A.; Ocampo, S.M.; Perales, J.C.; Eritja, R. Branched RNA: A New Architecture for RNA Interference. J. Nucleic Acids. 2011, 2021, 586935. [Google Scholar] [CrossRef]
- Prikazchikova, T.A.; Abakumova, T.O.; Sergeeva, O.V.; Zatsepin, T.S. Design and Validation of siRNA Targeting Gankyrin in the Murine Liver. Russ. J. Bioorg. Chem. 2021, 47, 441–446. [Google Scholar] [CrossRef]
- Lai, Y.; Kong, Z.; Zeng, T.; Xu, S.; Duan, X.; Li, S.; Cai, C.; Zhao, Z.; Wu, W. PARP1-siRNA suppresses human prostate cancer cell growth and progression. Oncol. Rep. 2018, 39, 1901–1909. [Google Scholar] [CrossRef]
- Nakano, S.; Kirihata, T.; Fujii, S.; Sakai, H.; Kuwahara, M.; Sawai, H.; Sugimoto, N. Influence of cationic molecules on the hairpin to duplex equilibria of self-complementary DNA and RNA oligonucleotides. Nucleic Acids Res. 2007, 35, 486–494. [Google Scholar] [CrossRef]
- Young, D.D.; Deiters, A. Photochemical control of biological processes. Org. Biomol. Chem. 2007, 5, 999–1005. [Google Scholar] [CrossRef]
- Lusic, H.; Young, D.D.; Lively, M.O.; Deiters, A. Photochemical DNA activation. Org Lett. 2007, 9, 1903–1906. [Google Scholar] [CrossRef]
- Deiters, A. Light activation as a method of regulating and studying gene expression. Curr. Opin. Chem. Biol. 2009, 13, 678–686. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
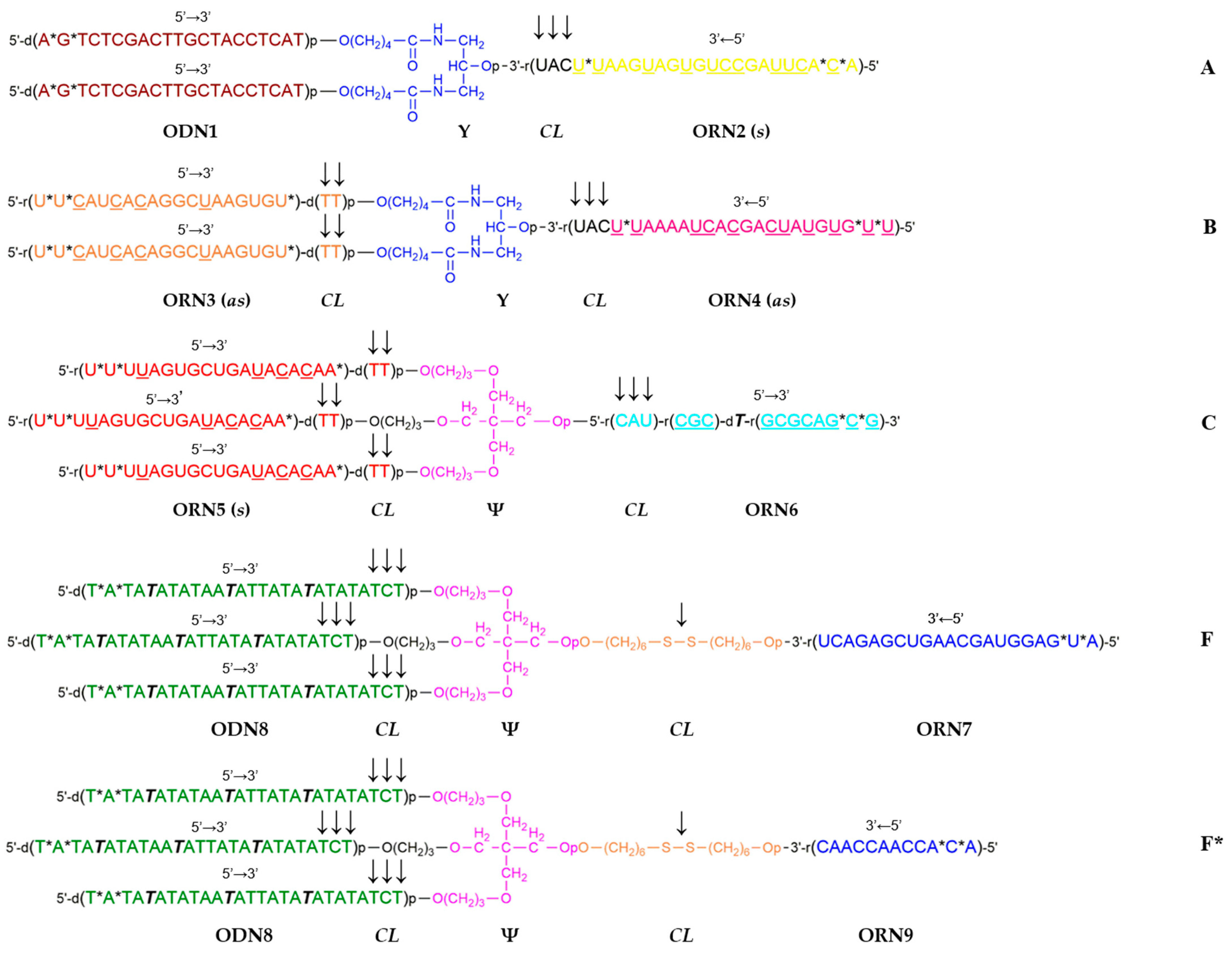

| Description | Sequence | Code 1 |
|---|---|---|
| msGankyrin-4 [57] | Sense (s) 3′-U*UAAGUAGUGUCCGAUUCA*C*A-5′ Antisense (as) 5′-U*U*CAUCACAGGCUAAGUGU*tt-3′ | ORN2 ORN3 |
| msPARP-1-4 [58] | Sense (s) 3′-U*UAAAAUCACGACUAUGUG*U*U-5′ Antisense (as) 5′-U*U*UUAGUGCUGAUACACAA*tt-3′ | ORN4 ORN5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fokina, A.; Poletaeva, Y.; Dukova, S.; Klabenkova, K.; Rad’kova, Z.; Bakulina, A.; Zatsepin, T.; Ryabchikova, E.; Stetsenko, D. Template-Assisted Assembly of Hybrid DNA/RNA Nanostructures Using Branched Oligodeoxy- and Oligoribonucleotides. Int. J. Mol. Sci. 2023, 24, 15978. https://doi.org/10.3390/ijms242115978
Fokina A, Poletaeva Y, Dukova S, Klabenkova K, Rad’kova Z, Bakulina A, Zatsepin T, Ryabchikova E, Stetsenko D. Template-Assisted Assembly of Hybrid DNA/RNA Nanostructures Using Branched Oligodeoxy- and Oligoribonucleotides. International Journal of Molecular Sciences. 2023; 24(21):15978. https://doi.org/10.3390/ijms242115978
Chicago/Turabian StyleFokina, Alesya, Yulia Poletaeva, Svetlana Dukova, Kristina Klabenkova, Zinaida Rad’kova, Anastasia Bakulina, Timofei Zatsepin, Elena Ryabchikova, and Dmitry Stetsenko. 2023. "Template-Assisted Assembly of Hybrid DNA/RNA Nanostructures Using Branched Oligodeoxy- and Oligoribonucleotides" International Journal of Molecular Sciences 24, no. 21: 15978. https://doi.org/10.3390/ijms242115978
APA StyleFokina, A., Poletaeva, Y., Dukova, S., Klabenkova, K., Rad’kova, Z., Bakulina, A., Zatsepin, T., Ryabchikova, E., & Stetsenko, D. (2023). Template-Assisted Assembly of Hybrid DNA/RNA Nanostructures Using Branched Oligodeoxy- and Oligoribonucleotides. International Journal of Molecular Sciences, 24(21), 15978. https://doi.org/10.3390/ijms242115978







