Abstract
The overuse and misuse of antibiotics have led to the emergence and spread of multidrug-resistant (MDR), extensively drug-resistant (XDR), and pan-drug-resistant (PDR) bacteria strains, usually associated with poorer patient outcomes and higher costs. In order to preserve the usefulness of these life-saving drugs, it is crucial to use them appropriately, as also recommended by the WHO. Moreover, innovative, safe, and more effective approaches are being investigated, aiming to revise drug treatments to improve their pharmacokinetics and distribution and to reduce the onset of drug resistance. Globally, to reduce the burden of antimicrobial resistance (AMR), guidelines and indications have been developed over time, aimed at narrowing the use and diminishing the environmental spread of these life-saving molecules by optimizing prescriptions, dosage, and times of use, as well as investing resources into obtaining innovative formulations with better pharmacokinetics, pharmacodynamics, and therapeutic results. This has led to the development of new nano-formulations as drug delivery vehicles, characterized by unique structural properties, biocompatible natures, and targeted activities such as state-of-the-art phospholipid particles generally grouped as liposomes, virosomes, and functionalized exosomes, which represent an attractive and innovative delivery approach. Liposomes and virosomes are chemically synthesized carriers that utilize phospholipids whose nature is predetermined based on their use, with a long track record as drug delivery systems. Exosomes are vesicles naturally released by cells, which utilize the lipids present in their cellular membranes only, and therefore, are highly biocompatible, with investigations as a delivery system having a more recent origin. This review will summarize the state of the art on microvesicle research, liposomes, virosomes, and exosomes, as useful and effective tools to tackle the threat of antibiotic resistance.
1. Introduction
Antibiotics are the staple of treatments against bacterial infections: since their discovery in the early 20th century, the death rates from threatening microbes lowered drastically. Antibiotics’ therapeutic effects involve either inhibiting proliferation (bacteriostatic) or killing pathogens (bactericidal) with the lowest affinity for the host’s cells; the mechanisms thanks to which antibiotics achieve their effect are diverse, extending from a disruption in the bacterial wall to an impairment in vital internal processes such as DNA decoding, RNA translation, and protein synthesis. Bacteria, however, can develop antimicrobial resistance (AMR) to antibiotics via simple natural selection: when they stay in enough contact with a chemotherapeutic drug, selected strains can produce genes that grant themselves endurance against it; this event occurs in a relatively quick timespan, and sadly, the survival of multi-resistant strains has been fostered in recent years by the massive and more than often erroneous employment of antibiotics and by the worsening of the hygienic criteria in hospitals due to monetary cuts to healthcare budgets [1,2,3].
Nowadays, multidrug-resistant (MDR) bacteria represent one of the biggest threats to human health because of their lethality and innate capacity to pass over resistance-inducing genes [4]; even during the COVID-19 pandemic, MDR bacteria were detected as a co-infection factor in a large portion of hospitalized COVID patients and resulted in being one of the major contributing causes of worse outcomes and, eventually, death [4,5,6]. To prevent the chemo-resistance phenomenon and, consequentially, the rise of MDR bacteria, innovative phospholipid carriers (IPCs) were demonstrated to be promising in the improvement in the efficacy profile of antibiotics.
IPCs are third-generation, phospholipid-based pharmaceutical formulations that act as carriers for the delivery of a pharmacologically active substance. Their development aims at achieving both spatial and temporal modulation of the release of the active compound, control of the quantity of the substance to be encapsulated, and protection of the drug from metabolization and degradation reactions [7]. Among them, liposomes, functionalized or not; exosomes; and virosomes found broad application in the fight against chemo-resistance in both experimental and clinical settings: their principal advantage against traditional methods to overcome bacterial resistance resides in their capacity to make old, ineffective molecules efficacious again versus MDR bacteria, thus reducing the need for the synthesis of new antibiotics.
This review will evaluate the most recent advances against chemo-resistance by incorporating antibiotics or other molecules in IPCs.
1.1. Antibiotics and Chemo-Resistance: A Worrying Phenomenon
Antibiotics were formerly discovered by Sir Alexander Fleming in 1928, who noticed the antibacterial activity of a certain mold, Penicillium notatum, on different pathogenic microbes; his studies on molds were inspired by his precedent research on lysozyme and by the findings of medical captain Vincenzo Tiberio on the prominent efficacy of Penicillium glaucum against a wide range of bacteria [8].
In 1943, Fleming won the Nobel Prize for medicine. Only two years later, he started giving the world an important message that remained unheard: large-scale use, especially when erroneous, of penicillin and antibiotics, in general, could lead to the end of the miracle since it would accentuate the selection of resistant bacteria [9].
In the recent past, the situation escalated to unthinkable levels: chemo-resistance became one of the greatest concerns of the 21st century, to the point that some microbial species are completely immune to last-line antibiotics like vancomycin, thus re-acquiring an unprecedented lethality [10].
Antibiotics suppress microbial infection through a wide variety of bacteriostatic and bactericidal mechanisms (Figure 1 and Table 1): e.g., beta-lactams, like penicillins and cephalosporins, bind irreversibly to D-alanyl-D-alanine-transpeptidase (DD-TPs) and other carboxypeptidases (commonly known as penicillin-binding proteins, PBPs), inhibiting the formation of cross-links and so causing the disruption the bacterial wall, which leaves the target vulnerable to osmotic and molecular pressures [11].
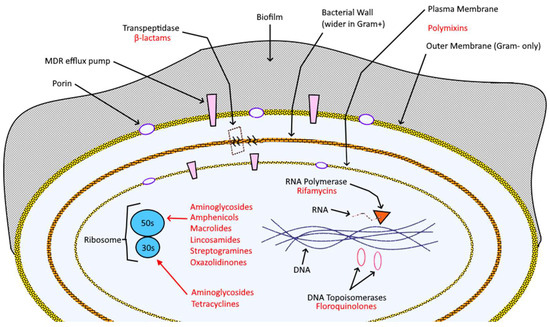
Figure 1.
Bacterial cell composition, key structures (in black), and targeting antibiotics (red).

Table 1.
Specific mechanism of action (M.O.A.) and chemo-resistance mechanisms (C.M.) per class of antibiotic, with references.
AC stems from microbial selection because of the large-scale use of antibiotics but, most importantly, for their clinical misusage (which permits the bacteria to stay in contact with a non-lethal dose of antibiotic for enough time to develop resistance) [44] and environmental misusage (farms that feed the animals huge amounts of antibiotics make the selection progress faster both in animals and in consumers of the final product) [45]. Moreover, most antibiotics are excreted unmodified from animals or from industrial waste, and both byproducts are sadly poured into rivers, lakes, and terrains, which become gyms for bacteria to train in. MDR bacteria can be consistently found in nature [46,47]. Add the hygiene- and protocol-related problems in hospitals to make up the perfect storm, with the rise of multi-resistant nosocomial infections that nowadays spread even out of hospitals to become serious community infections [48,49].
Even if the awareness of the chemo-resistance problem is low, scientists looked for solutions against it. The first logical approach was to search for more natural and synthesized small molecules that retained their activity against resistant microbes. This strategy, although functional, proved in recent years to be destined for failure since even the newest drugs will eventually encounter resistance due to the bacterial high capacity in resistance gene development and transmission. The risk of untreatable microbial infections is so impervious that it requires a multidisciplinary, coordinated action of the scientific community and the final users, doctors, patients, farmers, or industries.
Antimicrobial peptide-based therapy, suggested by and developed from the multitude of insects, plants, and other organisms that make daily use of them for survival [50,51,52,53], showed a minor insurgence of antibiotic resistance due to the intrinsic multi-modal, multi-target action derived from the more complex structure of these compounds, and a plethora of co-adjuvant properties such as antioxidant, anti-inflammatory, and pro-healing effects [54,55]. However, their application in therapy is restricted because of their side effects, caused by off-target reactions with human epitopes and by the high concentration (derived from their conspicuous molecular weight) at which they need to be administered to exert their properties.
Another molecule-based approach, often involving peptides, is to recognize and thus inhibit the principal factors that trigger antibiotic resistance: notable examples are the elimination of biofilms, blockage of multidrug resistance pumps (which could be over-expressed both for genetic reasons or due to heavy metal contamination), amelioration of bacterial membrane permeability, and disruption of vital bacterial processes like dihydrofolate reductase (DHFR) and translational elongation of protein inhibition [56,57,58]. While useful, this solution alone in the long term is insufficient since it faces the same obstacles as the principal resistance mechanism, namely target mutation and escaping, thanks to the innate adaptability of the bacteria.
Recent renewed interest arose in the employment of metals, despite non-specificity, against resistant bacteria: not only have they been demonstrated to be exceptional carriers for traditional and innovative antibiotics since they possess intrinsic antibacterial properties, targeting capacity, and low immune response, but even metals alone as such, in particular metal ions, could be of synergistic usage to combat microbes [59,60,61].
From the engineering and agro-industrial point of view, removing pharmaceutics and antibiotic resistance genes (ARGs) from water, soil, animal feed, and manure would greatly help reduce antibiotic resistance. This feature can be achieved by combining metal-based filtering systems [62,63], biological and biotechnological agents [64,65], and better maintenance and management [66,67]; a remarkable, somewhat different example in the field is brought up by the use of cultivated meat, which not only drastically reduces CO2, biological, and chemical waste with respect to farms but also almost deletes the need for antibiotics [68,69].
Last but not least, awareness campaigns [70,71], surveys [72], and technological integration in work [73,74] and learning [75,76] environments are crucial to stop an otherwise announced disastrous, unstoppable plague: communication is the key to unified, efficacious actions, especially when the enemy to defeat is so protean, diffused, and challenging.
1.2. Innovative Phospholipid Carriers (IPCs)
In recent years, nanomedicine significantly ameliorated therapy against a wide variety of illnesses, thanks to the capacity of these innovative drug delivery systems to obtain a time-controlled, precise, and quantitatively major transport of actives to the pharmacological targets: such a heavy impact on pharmacokinetics permitted the resurrection of old molecules that became inactive for years or that were not even considered for clinical usage because of their high toxicity, and even alone, nanoparticles contribute to fostering innovation in the fields they are applied into [77,78,79].
Lipid nanoparticles are one of the most promising vehicles of the family thanks to their elevated degree of biocompatibility, encapsulation efficiency, and customization [80]. Among them, the two types frequently used for the treatment of microbial infections are solid lipid nanoparticles and liposomes [81,82]; this review will discuss the main features and some of their technological evolutions, namely functionalized liposomes and exosomes, under the label of innovative phospholipid carriers (IPCs).
1.2.1. Liposomes
Liposomes are vesicles composed of phospholipids, which are naturally formed when the latter are inserted in aqueous solutions. They can be used both as biomembrane models to preliminarily assess the interaction of compounds in terms of quantity and quality and as drug carriers [83,84].
Phospholipids are amphiphilic compounds that sport a glyceryl backbone linked by ester bonds with a polar phosphate head and two chains of fatty acids. The characteristics of these two moieties, along with the cholesterol percentage content, contribute to the properties of the final liposome preparation, such as permeability, phase transitions, hydrophilic–lipophilic balance (HLB), dimensions, and affinity to a variety of substrates.
Liposomes are classified by size and number of bilayers (Figure 2); the most used drug delivery systems are unilamellar nanoliposomes in the 50–200 nm range, depending on the targeted tissue [85].
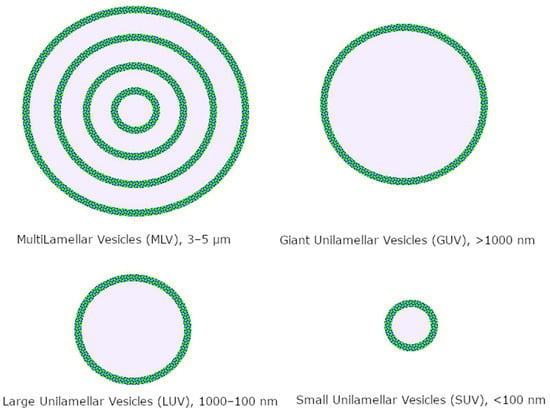
Figure 2.
Classification of liposomes.
Several preparation methods exist for these vehicles: the thin layer hydration—extrusion method and the Mozafari method [86,87]. Liposomes can encapsulate various active molecules since they can be highly optimized in size. They can naturally contain lipophilic and hydrophilic compounds as well, and the great range of preparation methods can account for the weaknesses of the drugs, e.g., thermolability, sterical instability, and susceptibility to solvents and pH.
Even if simple liposomes had great success as phospholipid carriers, the need for more precise targeting, escape mechanisms from macrophages, longer half-life, and other crucial factors made scientists develop functionalized drug delivery systems that could satisfy these necessities.
1.2.2. Functionalized Liposomes
PEGylation, the coating of liposomes with a thin layer of polyethylene glycol or the insertion of long PEG chains in the nano lipid structure, is a wide used technique to make “stealth” nanoparticles that avoid human immune surveillance systems retaining the original biocompatibility: bringing in the macrophage example, the thin layer of PEG negates protein adsorption, thus inhibiting opsonization, whereas long PEG chains heavily affect phagocytosis thanks to sterical and structural incompatibilities with the pseudopods [88]. Both PEGylation methods, mostly when used in synergy, considerably improve liposome half-life, but they are not exempt from downsides: drug unloading can be more challenging when PEG is included in phospholipids, rendering targeting less efficient and side effects more frequent, and the human body can also produce anti-PEG IgM when blood comes in contact with the polymer for too long; most recent studies found that these inconveniences can be mitigated by the regulating PEG shedding rate [89].
Fusogenic liposomes, as their name suggests, boast a peculiar fusion mechanism with cellular membranes triggered by microenvironmental pH [90]. The specific phospholipid composition (that comprises a mixture of charged lipids necessary for the additional phase transition, the inverted hexagonal phase, responsible for the fusion) ensures that they display optimal stability in physiological conditions, but when they encounter an element whose extracellular pH is acidic—e.g., cells suffering from tumoral diseases or bacterial infections and the outer membrane of Gram-negative bacteria [91,92]—a conformational change in lipids disposition make them able to merge with biomembranes and to release their content selectively; however, this type of targeting is not absolute and the surface charge renders them more susceptible to faster excretion.
Liposomes can be modified with different types of molecules to achieve better targeting properties or to monitor their bio-distribution. Passive targeting can be obtained by conjugating molecules that naturally accumulate in specific sites, like biotin for breast cancer cells and folate for FR-rich tumoral regions [93,94]. Active targeting is mostly reached by decorating phospholipids with monoclonal antibodies and viral glycoproteins, which are engineered to reach very specific targets [95,96]. Conjugation and decoration come with the major downside of an improved immunogenic response from the host system and thus faster degradation/resistance reactions, which can be partially surpassed by combination with PEGylation (Figure 3).
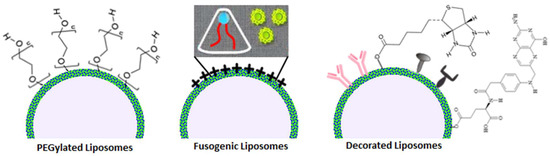
Figure 3.
Visualization of different types of functionalized liposomes.
1.2.3. Exosomes
Extracellular vesicles (EVs) are subcellular structures delimited by a lipid bilayer and shed by cells into their surrounding environment. These vesicles are involved in cell-to-cell communication and play important roles in various physiological and pathological processes. They are further categorized in the following three main types of extracellular vesicles:
- Exosomes are the smallest type of EVs, typically ranging in size from 30 to 200 nanometers. They originate from the endocytic pathway within the cell. Exosomes contain a diverse array of biomolecules, including proteins, nucleic acids (such as RNA and DNA), and lipids.
- Microvesicles are larger than exosomes, typically ranging from 100 to 1000 nanometers in size. They are formed by the outward budding of the cell’s plasma membrane and contain a similar assortment of biomolecules, serving as carriers for intercellular communication.
- Apoptotic bodies are the largest of the extracellular vesicles, typically exceeding 1000 nanometers in size. They are produced during the process of cell apoptosis (programmed cell death) and contain cellular debris and organelles from the dying cell.
These three subtypes vary in size, function, biological origin, and other attributes.
Exosomes, which are the smaller extracellular vesicles (EVs), are accumulated into multi-vesicular bodies (MVBs) before secretion. Exosome are shed by a wide array of cell types, including dendritic cells (DCs), lymphocytes, epithelial cells, endothelial cells, neurons, etc. They are detectable in an extensive range of bodily fluids, including blood, urine, saliva, amniotic fluid, breast milk, hydrothoracic fluid, ascitic fluid, and the culture medium of most cell types [97]. Various factors can induce the release of exosomes: microbial agents, external stimuli, and various stressors can elicit these responses [98]. Exosomes encompass sugars, lipids, proteins, nucleic acids, and bioactive substances within the extracellular matrix. Initially, their role was believed to be the removal of metabolic waste [99].
The composition of exosomes differs depending on their source cell (Figure 1). So far, research has identified nearly 350,000 proteins, 40,000 nucleic acids, and 600 lipids in various exosomes [100]. This extensive flexibility offers numerous possibilities for practical clinical diagnosis and treatment scenarios involving exosomes.
Exosomes contain a wide range of proteins, including transmembrane proteins, lipid-anchored membrane proteins, peripherally adsorbed membrane proteins, and soluble proteins within the exosome lumen [101]. The most commonly found proteins in exosomes include membrane transporters and fusion proteins (e.g., GTPases, annexins, and flotillin), heat shock proteins (e.g., HSC70), tetraspanins (e.g., CD9, CD63, and CD81, which are typically considered exosome markers), proteins involved in multi-vesicular body (MVB) biogenesis (e.g., Alix and TSG101), and lipid-related proteins and phospholipases. Some proteins are recognized as specific markers of exosomes, with CD63 and CD81 tetraspanins being the most commonly used ones. Noteworthy, exosomes are also rich in lipids, primarily cholesterol, sphingolipids, phospholipids, and bisphosphates [102].
Exosomes are flexible enough to carry a variety of nucleic acids such as RNAs (mRNA, microRNA, and other non-coding RNAs) and DNAs (mitochondrial DNA, double-stranded DNA, single-stranded DNA, and viral DNA), suggesting that exosomes could act as carriers of genetic information [103]. Despite many of the RNAs in exosomes being degraded fragments with lengths of less than 200 nucleotides, some full-length RNAs may be present and delivered to recipient cells through endocytosis, potentially influencing protein production in those cells. In this context, exosomal miRNAs are also associated with specific diseases [104].
Beyond their cargo, exosomes exhibit an intricate array of biological agents on their membranes. Adhesion molecules, signaling molecules, immunomodulatory factors, receptors, antibodies, lipids, proteins, transporters, and channels collectively contribute to the complexity of these vesicles, facilitating their interaction with the target cells.
Recent attention has focused on exosomes as an auspicious drug delivery system. Their inherent biocompatibility, efficient delivery mechanisms, and minimal immunogenicity have elevated their standing. Extensive research has revealed the role of exosomes in mediating intercellular communication and participating in various physiological and pathological processes in the body. Their functions span a wide spectrum, encompassing immune responses, antigen presentation, cell migration, differentiation, angiogenesis, inflammation induction, apoptosis, atherosclerosis, tumor development, invasion, metastasis, and drug resistance. The capacity of exosomes to transport bioactive substances holds great potential for deciphering an enigma of diseases such as cancer, neurological disorders, cardiovascular conditions, and metabolic disorders, as well as for disease diagnosis using biomarkers [98,105].
Therefore, exosomes as a drug delivery system stand out for their several advantages over existing synthetic delivery systems like liposomes. Their reduced likelihood of provoking immune reactions and toxicity, coupled with their inherent precision in targeting specific cells, renders them an attractive choice. In particular, small RNA therapeutics, anti-inflammatory agents, and anticancer drugs are among the drugs that could particularly benefit from delivery via exosomes. Researchers have explored two approaches to loading exosomes with therapeutic small RNA molecules: post-loading after EV isolation (known as the exogenous method) and pre-loading during EV formation (known as the endogenous method). However, the effectiveness of these methods has yet to be fully demonstrated [106].
2. Innovative Phospholipid Carriers versus Antimicrobial Resistance
IPCs have found profuse employment in treating microbial infections as adjuvants and carriers, especially in therapeutic and theragnostic applications against resistant and multi-resistant bacteria. Starting from “simple” liposomes, some formulations were so successful and incisive that they even hit the market, revolutionizing the battle against certain illnesses.
Liposomal antibiotic preparations can be traced back to the 1990s when workgroups like Lagacé et al. tried to deal with complicate bacterial infections like the one sustained by Pseudomonas Aeruginosa. This notoriously difficult-to-deal-with microbe also tends to produce a potent biofilm and to internalize into the host cells: they found that the employment of a fairly plain distearoylphosphatidylcholine/1,2-dimyristoylphosphatidylglycerol 10:1 liposome dramatically enhanced the sensitivity of resistant P. Aeruginosa to ticarcillin and tobramycin [107].
In line with these results, other groups have investigated the potential amelioration of treatments against potent bacteria with the encapsulation of traditional, not-more-efficacious antibiotics in liposomal formulations, obtaining in this way promising results [108,109,110]: among them, a case study was represented by two liposomal ciprofloxacin products, Lipoquin and Pulmaquin, for inhalation use, which permits ciprofloxacin to be utilized again as a first-line choice when the patient is facing complicated lung infections, even in cystic fibrosis-based (CF) scenarios [111,112]; sadly, both formulations were discontinued in 2022 since decisive data on their functionality were not achieved after an ORBIT phase 3 study. A different, brilliant destiny was instead achieved by liposomal formulations of amikacin (Arikayce), which have been tested with remarkable outcomes in CF, particularly in non-CF severe lung illnesses alone, and in combination with colistin [81,113].
There are even particular cases in which non-antibiotic molecules, both alone or co-encapsulated with antibiotics, proved to be useful to treat collateral symptoms, to adjuvant in the circumvention of common resistance mechanisms such as MDR pumps, to obtain clear imaging and theragnostic effects, to control ocular microflora after cataract surgery, and to preemptively stimulate innate immunity for major protection from bacterial infections, hinting at the diverse out-of-the-box possibilities offered by liposomal preparations [114,115,116,117,118].
Since most bacteria present a negatively charged outer layer, cationic and fusogenic liposomes proved to possess a certain selectivity and thus better delivery thanks to their net positive charge, favoring electrostatic interactions but also, especially in the case of fusogenic liposomes, triggering the fusion mechanism only in that determined microenvironment [119,120]. Similarly to their basic counterpart, but with the added benefits described above, these liposomal preparations enhanced and broadened the spectrum of action of classic antibiotics [121,122] and have found interesting applications in photodynamic therapy disinfection and photo-inactivation with aluminum chloride phthalocyanine and a porphyrinic compound, respectively [123,124]. An interesting work also showed how the fusogenic abilities of these carriers can be amped up by further decoration with cell-penetrating peptides like HIV-derived Tat surface protein, hinting at the strong potential of liposomal functionalization in therapy [125].
The most remarkable goals in antimicrobial (and not) applications of nanotherapy were indeed reached by functionalized liposomes, especially antibody-decorated ones, thanks to their precise targeting and the possibility of combining this technology with other techniques such as PEGylation, which proved to be extremely useful on its own due to the masking and stabilizing properties exerted on liposomes [126]. Surface-engineered liposomes can use a variety of ligands to properly direct therapeutics to sites of interest, whether the target is bacteria themselves or tissues affected by them while adopting synergistic mechanisms like cell penetration, mucoadhesion, tetraether-lipid-based stabilization, hetero-multivalent targeting, and the previously cited PEGylation [127,128,129,130].
Antibody-conjugated liposomes (ACL) have been thoroughly tested in a variety of applications, ranging from imaging-guided theragnostic activity [131] and maintenance of bacterial homeostasis [132] to the targeted release of antibiotics and, more generally, antimicrobial molecules. Natural extracts with known antimicrobial properties, like clove essential oil, have been successfully enclosed in ACL and showed a more precise release but even prolonged activity and bacteria-concentration-dependent action [133]. A detailed study by Krivic et al. demonstrated how antibody-conjugated hybrid erythrocyte liposomes encapsulating polymyxin B were capable of maintaining the unaltered activity of this antibiotic; drastically reducing common side effects (in vitro model) like hemolysis and nephrotoxicity; considerably improving drug retention and half-life; and, finally, obtaining selectivity against certain bacterial strains, namely E. coli and P. aeruginosa [134]. ACL can even deliver novel antisense oligonucleotides (composed of nucleic acid mimics with antimicrobial properties, paving the way to additional therapeutic options [135].
Virosomes, while promising and innovative, are a rather new technology that has not been extensively tested in the antimicrobial field yet [136]. On the other hand, exosomes, also recently employed as a drug delivery system, have already obtained landmark results against resistant bacteria.
The higher biocompatibility and membrane complexity of exosomes and exosome-like vesicles make sure that these carriers achieve a better intracellular uptake, evading lysosomal degradation in the cytoplasm and, in some cases, inducing macropinocytosis, thanks also to avant-garde synergism with other techniques such as decoration with cell-penetrating peptides [137]. Loadings of exosomes with natural toxins, like bee venom and mycobacterial antigens, have proven efficacious in eradicating mortal E. coli K99 infections in calves and provoking immunization with antigen-specific IFN-gamma and lymphocytic response against Mycobacterium tuberculosis, respectively [138,139]. While certain exosomes alone proved to exert an immunomodulatory and anti-inflammatory effect in cells during bacterial infections, being then a great aid in the co-administration with regular antibiotics to reduce side effects and pathological inflammatory [140], the most interesting applications are indeed in the ameliorated suppression of MRSA-sustained infections by loading antibiotics, namely linezolid and vancomycin in conjunction with lysostaphin [141,142].
3. Discussion
Antimicrobial resistance is undoubtedly one of the major health threats that should be overcome in the field of infectious diseases. Since its etiologies are so broad and diverse in terms of implicated molecular mechanisms and since bacteria can rapidly exchange this kind of information with each other, even among different species, AMR represents a worldwide problem that should be eradicated to avoid the insurgence of MDR-bacteria-driven illnesses, the latter of which proved to be highly deadly and resilient to complete eradication [143].
Apart from the natural occurrence of the spontaneous selection of resistant bacterial strains, the augmented appearance of MDR bacteria in recent decades has been amped up principally by a steady misusage and over-usage of antibiotics, both in community and nosocomial settings: the longer, unnecessary exposure to the drug and/or the employment of inefficacious dosages of pharmaceutics considerably speed up the bacterial selection process, favoring the flourishment of multi-resistant species; furthermore, the constant presence of antibiotics in food and in the environment due to livestock-related malpractice and mishandling of industrial waste, respectively, led to additional reinforcement of dangerous selection and exchange processes that can easily make a great portion of antibiotics virtually useless [144].
Even if nowadays the cohorts principally subjected to strong and often deadly MDR-bacteria-led infections are somewhat small, mostly relegated to hospitals and represented by elder and immunodeficient people, an increasing number of victims between healthy, immunological-sturdy patients is showing up in recent times—an alarming signal of the marked virulence that these pathogens possess and could increasingly exert in the near future if not contained appropriately [145].
The principal exponents of MDR bacteria are indeed methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Staphylococcus aureus (VRSA), Vancomycin-resistant Enterococcus (VRE), Mycobacterium tuberculosis MDR, extended-spectrum β-lactamase-producing Enterobacterales (ESBL), Klebsiella pneumoniae carbapenemase (KPC), and New Delhi Metallo-beta-lactamase-producing Pseudomonas aeruginosa (NDM) [146]. Through the development of resistance mechanisms that ensure bacterial survival against various concentrations of antimicrobial agents and other bacteria, genetic changes such as horizontal gene transfer by mobile genetic elements are to blame for the higher incidence of susceptibility loss, favoring the growth, colonization, and progression of the infectious process [145].
Strategies for overcoming these persistent infections include the following: the employment of multidrug protocols and pharmaceutics that do not trigger resistance mechanisms, increased social awareness, and correct use of conventional antibiotics, combined with vehicles or co-drugs that surpass specific resistance mechanisms [147]. Among these, IPCs, other than improving the overall pharmacokinetics of most antibiotics, both in terms of bettered metabolic profile and target selectivity, help in negating resistance mechanisms related to biofilm formation, outer membrane/plasmatic membrane modifications, and bacterial inclusion into host cells (i.e., P. Aeruginosa and Mycobacterium tuberculosis). This trend is proven beyond the existence of commercial or under clinical evaluation products (e.g., Arikayce, Lipoquin) and establishes the role of IPCs as a valid instrument against AMR [148].
4. Conclusions and Future Perspectives
The objective of this review is to underline the important synergy between non-/conventional antibiotics and IPCs in order to fight chemo-resistance phenomena properly, to achieve better targeting, and to reduce a plethora of annoying side effects; even if this research field is thriving, with some notable examples of pharmaceutical formulations which successfully hit the market, the necessity of carrying on more studies and experimentations on IPCs specifically tailored to carry antibiotics and other essential antimicrobial molecules is clearly inferable from the scientific evidence collected in this work. However, this alone obviously cannot be the solution to a complex problem such as AMR, which indeed needs to be approached in a multi-strategical and multidisciplinary way: starting from the conventional therapies, multi-therapy protocols should be properly adopted in conjunction with precise antibiograms, pondered dosages of antibiotics, and molecular diversification to reduce, at minimum, the risk of resistance derived by nosocomial and community misusage. The employment of antimicrobial peptides and other molecules (i.e., oligonucleotides, metals, etc.) that naturally do not trigger chemo-resistance mechanisms can definitely help reduce the burden on conventional antibiotics; other carriers like solid lipid nanoparticles (SLN) and nanostructured lipid carriers (NLC), and innovative biocarriers such as bacteriophages can offer additional solutions to formulation, chemical stability, and delivery problems; last but not least, the strong need for more and more social awareness in communities, hospitals, and industries regarding correct handling of antibiotics and the strong impact that multidrug-resistant superbugs (MSRBs) are already exerting on our health and environment should be communicated at local, national, and international levels.
To sum up, this review wants to be an additional voice in the already loud, worried chorus about the rampage of microbial resistance and its dangerous outcomes: if common action is not taken properly starting from the present, the world will experience pandemics much worse than the recent COVID-19 outbreak, as claimed by the World Health Organization (WHO), which predicts 5.2 million due to AMR deaths in the Western Pacific alone by 2030 [149]. In this scenario, IPCs are a well-known, precious weapon against AMR that should be implemented more, given the promising results it produced over time.
Author Contributions
Conceptualization, D.N.; software, S.R.; validation, D.N., G.P.P. and R.D.M.; investigation, S.R. and M.D.N.; data curation, S.R., G.P.P. and M.A.C.; writing—original draft preparation, D.N., S.R. and M.D.N.; writing—review and editing, G.P.P., D.N., R.D.M., S.R. and C.R.; visualization, D.N.; supervision, D.N., G.P.P. and R.D.M.; project administration, R.D.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial Resistance: A Global Emerging Threat to Public Health Systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6, 521–547. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, G.; Girish, M. Antibiotic Resistance—A Cause for Reemergence of Infections. Indian J. Pediatr. 2020, 87, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Antimicrobial Resistance Collaborators; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Ukuhor, H.O. The Interrelationships between Antimicrobial Resistance, COVID-19, Past, and Future Pandemics. J. Infect. Public Health 2021, 14, 53–60. [Google Scholar] [CrossRef]
- Rizvi, S.G.; Ahammad, S.Z. COVID-19 and Antimicrobial Resistance: A Cross-Study. Sci. Total Environ. 2022, 807, 150873. [Google Scholar] [CrossRef]
- Singh, R.P.; Gangadharappa, H.V.; Mruthunjaya, K. Phospholipids: Unique Carriers for Drug Delivery Systems. J. Drug Deliv. Sci. Technol. 2017, 39, 166–179. [Google Scholar] [CrossRef]
- Neu, H.C.; Gootz, T.D. Antimicrobial Chemotherapy. In Medical Microbiology; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; ISBN 978-0-9631172-1-2. [Google Scholar]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular Mechanisms of Antibiotic Resistance Revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef]
- Stogios, P.J.; Savchenko, A. Molecular Mechanisms of Vancomycin Resistance. Protein Sci. Publ. Protein Soc. 2020, 29, 654–669. [Google Scholar] [CrossRef]
- Lima, L.M.; da Silva, B.N.M.; Barbosa, G.; Barreiro, E.J. β-Lactam Antibiotics: An Overview from a Medicinal Chemistry Perspective. Eur. J. Med. Chem. 2020, 208, 112829. [Google Scholar] [CrossRef]
- Turk, S.; Verlaine, O.; Gerards, T.; Živec, M.; Humljan, J.; Sosič, I.; Amoroso, A.; Zervosen, A.; Luxen, A.; Joris, B. New Noncovalent Inhibitors of Penicillin-Binding Proteins from Penicillin-Resistant Bacteria. PLoS ONE 2011, 6, e19418. [Google Scholar] [CrossRef] [PubMed]
- Craft, K.M.; Nguyen, J.M.; Berg, L.J.; Townsend, S.D. Methicillin-Resistant Staphylococcus Aureus (MRSA): Antibiotic-Resistance and the Biofilm Phenotype. MedChemComm 2019, 10, 1231–1241. [Google Scholar] [CrossRef]
- Elshamy, A.A.; Aboshanab, K.M. A Review on Bacterial Resistance to Carbapenems: Epidemiology, Detection and Treatment Options. Future Sci. OA 2020, 6, FSO438. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Junghare, V.; Dutta, S.; Hazra, S.; Basu, S. Differential Binding of Carbapenems with the AdeABC Efflux Pump and Modulation of the Expression of AdeB Linked to Novel Mutations within Two-Component System AdeRS in Carbapenem-Resistant Acinetobacter Baumannii. mSystems 2022, 7, e00217-22. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ezpeleta-Lobato, G.; Han, X.; Carmona-Cartaya, Y.; Quiñones-Pérez, D. Carbapenamase-Producing Acinetobacter Baumannii in China, Latin America and the Caribbean: A Systematic Review and Meta-Analysis. MEDICC Rev. 2022, 24, 59–69. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline Antibiotics and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef]
- Milanović, V.; Maoloni, A.; Belleggia, L.; Cardinali, F.; Garofalo, C.; Cesaro, C.; Aquilanti, L.; Osimani, A. Tetracycline Resistance Genes in the Traditional Swedish Sour Herring Surströmming as Revealed Using qPCR. Genes 2022, 14, 56. [Google Scholar] [CrossRef]
- Doi, Y.; Wachino, J.; Arakawa, Y. Aminoglycoside Resistance. Infect. Dis. Clin. N. Am. 2016, 30, 523–537. [Google Scholar] [CrossRef]
- Jaimee, G.; Halami, P.M. Emerging Resistance to Aminoglycosides in Lactic Acid Bacteria of Food Origin—An Impending Menace. Appl. Microbiol. Biotechnol. 2016, 100, 1137–1151. [Google Scholar] [CrossRef]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter Baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef]
- Li, P.; Zhu, T.; Zhou, D.; Lu, W.; Liu, H.; Sun, Z.; Ying, J.; Lu, J.; Lin, X.; Li, K.; et al. Analysis of Resistance to Florfenicol and the Related Mechanism of Dissemination in Different Animal-Derived Bacteria. Front. Cell. Infect. Microbiol. 2020, 10, 369. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, C. Positive Association between the Use of Macrolides in Food-Producing Animals and Pneumococcal Macrolide Resistance: A Global Ecological Analysis. Int. J. Infect. Dis. IJID 2022, 116, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wu, P.; Tang, R.; Zhang, W. Global Prevalence of Resistance to Macrolides in Mycoplasma Pneumoniae: A Systematic Review and Meta-Analysis. J. Antimicrob. Chemother. 2022, 77, 2353–2363. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.S.; Cusini, M.; Gomberg, M.; Moi, H.; Wilson, J.; Unemo, M. 2021 European Guideline on the Management of Mycoplasma Genitalium Infections. J. Eur. Acad. Dermatol. Venereol. JEADV 2022, 36, 641–650. [Google Scholar] [CrossRef]
- Feßler, A.T.; Wang, Y.; Wu, C.; Schwarz, S. Mobile Lincosamide Resistance Genes in Staphylococci. Plasmid 2018, 99, 22–31. [Google Scholar] [CrossRef]
- Hurst-Hess, K.R.; Rudra, P.; Ghosh, P. Ribosome Protection as a Mechanism of Lincosamide Resistance in Mycobacterium Abscessus. Antimicrob. Agents Chemother. 2021, 65, e0118421. [Google Scholar] [CrossRef]
- Yu, R.; Xu, Y.; Schwarz, S.; Shang, Y.; Yuan, X.; Zhang, Y.; Li, D.; Du, X.-D. Erm(T)-Mediated Macrolide-Lincosamide Resistance in Streptococcus suis. Microbiol. Spectr. 2022, 10, e0165721. [Google Scholar] [CrossRef]
- Krüger, H.; Ji, X.; Hanke, D.; Schink, A.K.; Fiedler, S.; Kaspar, H.; Wang, Y.; Schwarz, S.; Wu, C.; Feßler, A.T. Novel Macrolide-Lincosamide-Streptogramin B Resistance Gene erm(54) in MRSA ST398 from Germany. J. Antimicrob. Chemother. 2022, 77, 2296–2298. [Google Scholar] [CrossRef]
- Marosevic, D.; Kaevska, M.; Jaglic, Z. Resistance to the Tetracyclines and Macrolide-Lincosamide-Streptogramin Group of Antibiotics and Its Genetic Linkage—A Review. Ann. Agric. Environ. Med. AAEM 2017, 24, 338–344. [Google Scholar] [CrossRef]
- Mišić, M.; Kocić, B.; Arsović, A.; Čukić, J.; Vidanović, D.; Šekler, M.; Baskić, D. Human Enterococcal Isolates as Reservoirs for Macrolide-Lincosamide-Streptogramin and Other Resistance Genes. J. Antibiot. 2022, 75, 396–402. [Google Scholar] [CrossRef]
- Crowe-McAuliffe, C.; Murina, V.; Turnbull, K.J.; Huch, S.; Kasari, M.; Takada, H.; Nersisyan, L.; Sundsfjord, A.; Hegstad, K.; Atkinson, G.C.; et al. Structural Basis for PoxtA-Mediated Resistance to Phenicol and Oxazolidinone Antibiotics. Nat. Commun. 2022, 13, 1860. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.-H.; Cha, M.-H.; Woo, G.-J.; Chi, Y.-M. Characterization of Oxazolidinone and Phenicol Resistance Genes in Non-Clinical Enterococcal Isolates from Korea. J. Glob. Antimicrob. Resist. 2021, 24, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Kerschner, H.; Rosel, A.C.; Hartl, R.; Hyden, P.; Stoeger, A.; Ruppitsch, W.; Allerberger, F.; Apfalter, P. Oxazolidinone Resistance Mediated by optrA in Clinical Enterococcus faecalis Isolates in Upper Austria: First Report and Characterization by Whole Genome Sequencing. Microb. Drug Resist. 2021, 27, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Ganapathy, U.S.; Sharma, S.; Ahn, Y.-M.; Zimmerman, M.; Molodtsov, V.; Hegde, P.; Gengenbacher, M.; Ebright, R.H.; Dartois, V.; et al. Redesign of Rifamycin Antibiotics to Overcome ADP-Ribosylation-Mediated Resistance. Angew. Chem. Int. Ed. 2022, 61, e202211498. [Google Scholar] [CrossRef] [PubMed]
- Surette, M.D.; Spanogiannopoulos, P.; Wright, G.D. The Enzymes of the Rifamycin Antibiotic Resistome. Acc. Chem. Res. 2021, 54, 2065–2075. [Google Scholar] [CrossRef]
- Cuypers, W.L.; Jacobs, J.; Wong, V.; Klemm, E.J.; Deborggraeve, S.; Van Puyvelde, S. Fluoroquinolone Resistance in Salmonella: Insights by Whole-Genome Sequencing. Microb. Genom. 2018, 4, e000195. [Google Scholar] [CrossRef]
- Ezelarab, H.A.A.; Abbas, S.H.; Hassan, H.A.; Abuo-Rahma, G.E.-D.A. Recent Updates of Fluoroquinolones as Antibacterial Agents. Arch. Pharm. 2018, 351, 1800141. [Google Scholar] [CrossRef]
- Sayadi, M.; Zare, H.; Jamedar, S.A.; Hashemy, S.I.; Meshkat, Z.; Soleimanpour, S.; Hoffner, S.; Ghazvini, K. Genotypic and Phenotypic Characterization of Mycobacterium tuberculosis Resistance against Fluoroquinolones in the Northeast of Iran. BMC Infect. Dis. 2020, 20, 390. [Google Scholar] [CrossRef]
- Mohapatra, S.S.; Dwibedy, S.K.; Padhy, I. Polymyxins, the Last-Resort Antibiotics: Mode of Action, Resistance Emergence, and Potential Solutions. J. Biosci. 2021, 46, 85. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Philippon, A.; Arlet, G.; Jacoby, G.A. Plasmid-Determined AmpC-Type β-Lactamases. Antimicrob. Agents Chemother. 2002, 46, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bennett, P.M. Plasmid Encoded Antibiotic Resistance: Acquisition and Transfer of Antibiotic Resistance Genes in Bacteria. Br. J. Pharmacol. 2008, 153, S347–S357. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic Resistance—The Need for Global Solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [PubMed]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Mol. J. Synth. Chem. Nat. Prod. Chem. 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.Q.; Vu, H.P.; Nguyen, L.N.; Wang, Q.; Djordjevic, S.P.; Donner, E.; Yin, H.; Nghiem, L.D. Monitoring Antibiotic Resistance Genes in Wastewater Treatment: Current Strategies and Future Challenges. Sci. Total Environ. 2021, 783, 146964. [Google Scholar] [CrossRef] [PubMed]
- Junaid, M.; Liu, S.; Liao, H.; Liu, X.; Wu, Y.; Wang, J. Wastewater Plastisphere Enhances Antibiotic Resistant Elements, Bacterial Pathogens, and Toxicological Impacts in the Environment. Sci. Total Environ. 2022, 841, 156805. [Google Scholar] [CrossRef]
- Farr, B.M.; Salgado, C.D.; Karchmer, T.B.; Sherertz, R.J. Can Antibiotic-Resistant Nosocomial Infections Be Controlled? Lancet Infect. Dis. 2001, 1, 38–45. [Google Scholar] [CrossRef]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32, e00001-19. [Google Scholar] [CrossRef]
- Yang, L.; Luo, X.; Sun, J.; Ma, X.; Ren, Q.; Wang, Y.; Wang, W.; He, Y.; Li, Q.; Han, B.; et al. The Antimicrobial Potential and Aquaculture Wastewater Treatment Ability of Penaeidins 3a Transgenic Duckweed. Plants 2023, 12, 1715. [Google Scholar] [CrossRef]
- Zhang, L.-M.; Yang, M.; Zhou, S.-W.; Zhang, H.; Feng, Y.; Shi, L.; Li, D.-S.; Lu, Q.-M.; Zhang, Z.-H.; Zhao, M. Blapstin, a Diapause-Specific Peptide-Like Peptide from the Chinese Medicinal Beetle Blaps rhynchopetera, Has Antifungal Function. Microbiol. Spectr. 2023, 11, e0308922. [Google Scholar] [CrossRef]
- Shang, C.; Ye, T.; Zhou, Q.; Chen, P.; Li, X.; Li, W.; Chen, S.; Hu, Z.; Zhang, W. Genome-Wide Identification and Bioinformatics Analyses of Host Defense Peptides Snakin/GASA in Mangrove Plants. Genes 2023, 14, 923. [Google Scholar] [CrossRef] [PubMed]
- Lima, R.M.; Rathod, B.B.; Tiricz, H.; Howan, D.H.O.; Al Bouni, M.A.; Jenei, S.; Tímár, E.; Endre, G.; Tóth, G.K.; Kondorosi, É. Legume Plant Peptides as Sources of Novel Antimicrobial Molecules Against Human Pathogens. Front. Mol. Biosci. 2022, 9, 870460. [Google Scholar] [CrossRef]
- Lima, W.G.; de Lima, M.E. Therapeutic Prospection of Animal Venoms-Derived Antimicrobial Peptides against Infections by Multidrug-Resistant Acinetobacter Baumannii: A Systematic Review of Pre-Clinical Studies. Toxins 2023, 15, 268. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.T.; Hansford, K.A.; Gong, Y.; Butler, M.S.; Muldoon, C.; Huang, J.X.; Ramu, S.; Silva, A.B.; Cheng, M.; Kavanagh, A.M.; et al. Protein-Inspired Antibiotics Active against Vancomycin- and Daptomycin-Resistant Bacteria. Nat. Commun. 2018, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.T.; Nguyen, H.D.; Le, M.H.; Nguyen, T.T.H.; Nguyen, T.D.; Nguyen, D.L.; Nguyen, Q.H.; Nguyen, T.K.O.; Michalet, S.; Dijoux-Franca, M.-G.; et al. Efflux Pump Inhibitors in Controlling Antibiotic Resistance: Outlook under a Heavy Metal Contamination Context. Molecules 2023, 28, 2912. [Google Scholar] [CrossRef] [PubMed]
- Masoudi-Sobhanzadeh, Y.; Pourseif, M.M.; Khalili-Sani, A.; Jafari, B.; Salemi, A.; Omidi, Y. Deciphering Anti-Biofilm Property of Arthrospira Platensis-Origin Peptides against Staphylococcusaureus aureus. Comput. Biol. Med. 2023, 160, 106975. [Google Scholar] [CrossRef]
- Haque, M.A.; Marathakam, A.; Rana, R.; Almehmadi, S.J.; Tambe, V.B.; Charde, M.S.; Islam, F.; Siddiqui, F.A.; Culletta, G.; Almerico, A.M.; et al. Fighting Antibiotic Resistance: New Pyrimidine-Clubbed Benzimidazole Derivatives as Potential DHFR Inhibitors. Molecules 2023, 28, 501. [Google Scholar] [CrossRef]
- Frei, A.; Verderosa, A.D.; Elliott, A.G.; Zuegg, J.; Blaskovich, M.A.T. Metals to Combat Antimicrobial Resistance. Nat. Rev. Chem. 2023, 7, 202–224. [Google Scholar] [CrossRef]
- Raj, R.; Bhattu, M.; Verma, M.; Acevedo, R.; Duc, N.D.; Singh, J. Biogenic Silver Based Nanostructures: Synthesis, Mechanistic Approach and Biological Applications. Environ. Res. 2023, 231, 116045. [Google Scholar] [CrossRef]
- Zafar, N.; Uzair, B.; Menaa, F.; Khan, B.A.; Niazi, M.B.K.; Alaryani, F.S.; Majrashi, K.A.; Sajjad, S. Moringa Concanensis-Mediated Synthesis and Characterizations of Ciprofloxacin Encapsulated into Ag/TiO2/Fe2O3/CS Nanocomposite: A Therapeutic Solution against Multidrug Resistant E. Coli Strains of Livestock Infectious Diseases. Pharmaceutics 2022, 14, 1719. [Google Scholar] [CrossRef]
- Li, X.; Zheng, S.; Li, Y.; Ding, J.; Qin, W. Effectively Facilitating the Degradation of Chloramphenicol by the Synergism of Shewanella oneidensis MR-1 and the Metal-Organic Framework. J. Hazard. Mater. 2023, 454, 131545. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhang, L.; Wang, X.; Gu, J.; Song, Z.; Wei, S.; Guo, H.; Xu, L.; Qian, X. Reductions in Abundances of Intracellular and Extracellular Antibiotic Resistance Genes by SiO2 Nanoparticles during Composting Driven by Mobile Genetic Elements. J. Environ. Manag. 2023, 341, 118071. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Chang, Y.; Liu, J.; Sangeetha, T.; Feng, Y.; Liu, D.; Xu, C. Removal of Antibiotic Resistance Genes and Mobile Genetic Elements in a Three-Stage Pig Manure Management System: The Implications of Microbial Community Structure. J. Environ. Manag. 2022, 323, 116185. [Google Scholar] [CrossRef]
- Yang, S.; Lu, C.; Qin, C.; Lu, C.; Pan, Z.; Zhao, L.; Bai, M.; Li, X.; Sun, Y.; Weng, L.; et al. Mitigation Effects and Microbial Mechanism of Two Ecological Earthworms on the Uptake of Chlortetracycline and Antibiotic Resistance Genes in Lettuce. Sci. Total Environ. 2023, 885, 163907. [Google Scholar] [CrossRef]
- Mthiyane, Z.L.; Makhubela, N.; Nyoni, H.; Madikizela, L.M.; Maseko, B.R.; Ncube, S. Determination of Antibiotics during Treatment of Hospital Wastewater Using Automated Solid-Phase Extraction Followed by UHPLC-MS: Occurrence, Removal and Environmental Risks. Environ. Technol. 2023, 2023, 2209741. [Google Scholar] [CrossRef] [PubMed]
- Gajdoš, S.; Zuzáková, J.; Pacholská, T.; Kužel, V.; Karpíšek, I.; Karmann, C.; Šturmová, R.; Bindzar, J.; Smrčková, Š.; Sýkorová, Z.; et al. Synergistic Removal of Pharmaceuticals and Antibiotic Resistance from Ultrafiltered WWTP Effluent: Free-Floating ARGs Exceptionally Susceptible to Degradation. J. Environ. Manag. 2023, 340, 117861. [Google Scholar] [CrossRef]
- McNamara, E.; Bomkamp, C. Cultivated Meat as a Tool for Fighting Antimicrobial Resistance. Nat. Food 2022, 3, 791–794. [Google Scholar] [CrossRef]
- Saied, A.A.; Chandran, D.; Chopra, H.; Dey, A.; Emran, T.B.; Dhama, K. Cultivated Meat Could Aid in Reducing Global Antimicrobial Resistance Burden—Producing Meat without Antibiotics as a Safer Food System for the Future. Int. J. Surg. 2023, 109, 189–190. [Google Scholar] [CrossRef]
- Ghiga, I.; Sidorchuk, A.; Pitchforth, E.; Stålsby Lundborg, C.; Machowska, A. ‘If You Want to Go Far, Go Together’—Community-Based Behaviour Change Interventions to Improve Antibiotic Use: A Systematic Review of Quantitative and Qualitative Evidence. J. Antimicrob. Chemother. 2023, 78, 1344–1353. [Google Scholar] [CrossRef]
- Allerton, F.; Jamieson, C.; Aggarwal, R.; Barker, A.; Work, M.; Cooper, D.; Ramsey, I. An Antibiotic Amnesty Can Be a One Health Tool to Tackle Antimicrobial Resistance. Nat. Med. 2023, 29, 1046–1047. [Google Scholar] [CrossRef]
- McCracken, C.M.; Tucker, K.J.; Tallman, G.B.; Holmer, H.K.; Noble, B.N.; McGregor, J.C. General Perceptions and Knowledge of Antibiotic Resistance and Antibiotic Use Behavior: A Cross-Sectional Survey of US Adults. Antibiotics 2023, 12, 672. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-González, A.; Zanin, M.; Menasalvas-Ruiz, E. Public Health and Epidemiology Informatics: Can Artificial Intelligence Help Future Global Challenges? An Overview of Antimicrobial Resistance and Impact of Climate Change in Disease Epidemiology. Yearb. Med. Inform. 2019, 28, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Kula, C.; Arga, K.Y. Systems Biomarkers, Artificial Intelligence, and One Health Vision Can Help Fight Antimicrobial Resistance. Omics J. Integr. Biol. 2023, 27, 191–192. [Google Scholar] [CrossRef] [PubMed]
- Lau, H.J.; Lim, C.H.; Foo, S.C.; Tan, H.S. The Role of Artificial Intelligence in the Battle against Antimicrobial-Resistant Bacteria. Curr. Genet. 2021, 67, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Pineros, N.; Tenaillon, K.; Marin, J.; Berry, V.; Jaureguy, F.; Ghelfenstein-Ferreira, T.; Carbonnelle, E.; Lescat, M. Using Gamification to Improve Engagement and Learning Outcomes in Medical Microbiology: The Case Study of “BacteriaGame”. FEMS Microbiol. Lett. 2023, 370, fnad034. [Google Scholar] [CrossRef]
- Mehrabi, M.R.; Soltani, M.; Chiani, M.; Raahemifar, K.; Farhangi, A. Nanomedicine: New Frontiers in Fighting Microbial Infections. Nanomaterials 2023, 13, 483. [Google Scholar] [CrossRef]
- Yi, W.; Yan, D.; Wang, D.; Li, Y. Smart Drug Delivery Systems to Overcome Drug Resistance in Cancer Immunotherapy. Cancer Biol. Med. 2023, 20, 248–267. [Google Scholar] [CrossRef]
- Yang, P.; Ren, J.; Yang, L. Nanoparticles in the New Era of Cardiovascular Therapeutics: Challenges and Opportunities. Int. J. Mol. Sci. 2023, 24, 5205. [Google Scholar] [CrossRef]
- Maddiboyina, B.; Ramaiah, R.; Nakkala, R.K.; Roy, H. Perspectives on Cutting-Edge Nanoparticulate Drug Delivery Technologies Based on Lipids and Their Applications. Chem. Biol. Drug Des. 2023, 102, 377–394. [Google Scholar] [CrossRef]
- Vairo, C.; Villar Vidal, M.; Maria Hernandez, R.; Igartua, M.; Villullas, S. Colistin- and Amikacin-Loaded Lipid-Based Drug Delivery Systems for Resistant Gram-Negative Lung and Wound Bacterial Infections. Int. J. Pharm. 2023, 635, 122739. [Google Scholar] [CrossRef]
- Kiymaci, M.E.; Topal, G.R.; Esim, O.; Bacanli, M.; Ozkan, C.K.; Erdem, O.; Savaser, A.; Ozkan, Y. Evaluation of Bacterial Uptake, Antibacterial Efficacy against Escherichia coli, and Cytotoxic Effects of Moxifloxacin-Loaded Solid Lipid Nanoparticles. Arh. Hig. Rada Toksikol. 2022, 73, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Russo, S.; Torrisi, C.; Cardullo, N.; Muccilli, V.; La Mantia, A.; Castelli, F.; Acquaviva, R.; Sarpietro, M.G. Ethyl Protocatechuate Encapsulation in Solid Lipid Nanoparticles: Assessment of Pharmacotechnical Parameters and Preliminary In Vitro Evaluation for Colorectal Cancer Treatment. Pharmaceutics 2023, 15, 394. [Google Scholar] [CrossRef] [PubMed]
- Magnifico, I.; Petronio Petronio, G.; Venditti, N.; Cutuli, M.A.; Pietrangelo, L.; Vergalito, F.; Mangano, K.; Zella, D.; Di Marco, R. Atopic Dermatitis as a Multifactorial Skin Disorder. Can the Analysis of Pathophysiological Targets Represent the Winning Therapeutic Strategy? Pharmaceuticals 2020, 13, 411. [Google Scholar] [CrossRef] [PubMed]
- Maja, L.; Željko, K.; Mateja, P. Sustainable Technologies for Liposome Preparation. J. Supercrit. Fluids 2020, 165, 104984. [Google Scholar] [CrossRef]
- Rasti, B.; Jinap, S.; Mozafari, M.R.; Abd-Manap, M.Y. Optimization on Preparation Condition of Polyunsaturated Fatty Acids Nanoliposome Prepared by Mozafari Method. J. Liposome Res. 2014, 24, 99–105. [Google Scholar] [CrossRef]
- Zhang, H. Thin-Film Hydration Followed by Extrusion Method for Liposome Preparation. Methods Mol. Biol. 2017, 1522, 17–22. [Google Scholar] [CrossRef]
- Nosova, A.S.; Koloskova, O.O.; Nikonova, A.A.; Simonova, V.A.; Smirnov, V.V.; Kudlay, D.; Khaitov, M.R. Diversity of PEGylation Methods of Liposomes and Their Influence on RNA Delivery. MedChemComm 2019, 10, 369–377. [Google Scholar] [CrossRef]
- Suzuki, T.; Suzuki, Y.; Hihara, T.; Kubara, K.; Kondo, K.; Hyodo, K.; Yamazaki, K.; Ishida, T.; Ishihara, H. PEG Shedding-Rate-Dependent Blood Clearance of PEGylated Lipid Nanoparticles in Mice: Faster PEG Shedding Attenuates Anti-PEG IgM Production. Int. J. Pharm. 2020, 588, 119792. [Google Scholar] [CrossRef]
- Kolašinac, R.; Kleusch, C.; Braun, T.; Merkel, R.; Csiszár, A. Deciphering the Functional Composition of Fusogenic Liposomes. Int. J. Mol. Sci. 2018, 19, 346. [Google Scholar] [CrossRef]
- Kato, Y.; Ozawa, S.; Miyamoto, C.; Maehata, Y.; Suzuki, A.; Maeda, T.; Baba, Y. Acidic Extracellular Microenvironment and Cancer. Cancer Cell Int. 2013, 13, 89. [Google Scholar] [CrossRef]
- Kromer, C.; Schwibbert, K.; Gadicherla, A.K.; Thiele, D.; Nirmalananthan-Budau, N.; Laux, P.; Resch-Genger, U.; Luch, A.; Tschiche, H.R. Monitoring and Imaging pH in Biofilms Utilizing a Fluorescent Polymeric Nanosensor. Sci. Rep. 2022, 12, 9823. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.J.; Low, P.S. Folate-Mediated Tumor Cell Targeting of Liposome-Entrapped Doxorubicin in Vitro. Biochim. Biophys. Acta BBA Biomembr. 1995, 1233, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Peng, Y.; Yue, Q.; Pu, Y.; Li, R.; Zhao, Y.; Hai, L.; Guo, L.; Wu, Y. Design, Preparation and Evaluation of Different Branched Biotin Modified Liposomes for Targeting Breast Cancer. Eur. J. Med. Chem. 2020, 193, 112204. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Gastfriend, B.D.; Umlauf, B.J.; Lynn, D.M.; Shusta, E.V. Antibody-Targeted Liposomes for Enhanced Targeting of the Blood-Brain Barrier. Pharm. Res. 2022, 39, 1523–1534. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, R.; Jain, V.K.; Nagpal, S. Preparation and Characterization of Nanocurcumin Based Hybrid Virosomes as a Drug Delivery Vehicle with Enhanced Anticancerous Activity and Reduced Toxicity. Sci. Rep. 2021, 11, 368. [Google Scholar] [CrossRef]
- Batrakova, E.V.; Kim, M.S. Using Exosomes, Naturally-Equipped Nanocarriers, for Drug Delivery. J. Control. Release 2015, 219, 396–405. [Google Scholar] [CrossRef]
- Si, Q.; Wu, L.; Pang, D.; Jiang, P. Exosomes in Brain Diseases: Pathogenesis and Therapeutic Targets. MedComm 2023, 4, e287. [Google Scholar] [CrossRef]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on Strategies and Technologies for Exosome Isolation and Purification. Front. Bioeng. Biotechnol. 2021, 9, 811971. [Google Scholar] [CrossRef]
- Vesiclepedia: Home: Extracellular Vesicles Database. Available online: http://microvesicles.org/ (accessed on 26 August 2023).
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Fagone, P.; Mangano, K.; Di Marco, R.; Reyes-Castillo, Z.; Muñoz-Valle, J.F.; Nicoletti, F. Altered Expression of TSPAN32 during B Cell Activation and Systemic Lupus Erythematosus. Genes 2021, 12, 931. [Google Scholar] [CrossRef]
- Avgoulas, D.I.; Tasioulis, K.S.; Papi, R.M.; Pantazaki, A.A. Therapeutic and Diagnostic Potential of Exosomes as Drug Delivery Systems in Brain Cancer. Pharmaceutics 2023, 15, 1439. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.-M.; Xu, Y.-M.; Huang, L.-F.; Wang, X.-Z. Exosomes: Novel Biomarkers for Clinical Diagnosis. Sci. World J. 2015, 2015, 657086. [Google Scholar] [CrossRef] [PubMed]
- Tenchov, R.; Sasso, J.M.; Wang, X.; Liaw, W.-S.; Chen, C.-A.; Zhou, Q.A. Exosomes—Nature’s Lipid Nanoparticles, a Rising Star in Drug Delivery and Diagnostics. ACS Nano 2022, 16, 17802–17846. [Google Scholar] [CrossRef]
- Lener, T.; Gimona, M.; Aigner, L.; Börger, V.; Buzas, E.; Camussi, G.; Chaput, N.; Chatterjee, D.; Court, F.A.; Del Portillo, H.A.; et al. Applying Extracellular Vesicles Based Therapeutics in Clinical Trials—An ISEV Position Paper. J. Extracell. Vesicles 2015, 4, 30087. [Google Scholar] [CrossRef] [PubMed]
- Lagacé, J.; Dubreuil, M.; Montplaisir, S. Liposome-Encapsulated Antibiotics: Preparation, Drug Release and Antimicrobial Activity against Pseudomonas aeruginosa. J. Microencapsul. 2008, 8, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-Y.; Yang, S.-C.; Sung, C.T.; Weng, Y.-H.; Fang, J.-Y. Anti-MRSA Malleable Liposomes Carrying Chloramphenicol for Ameliorating Hair Follicle Targeting. Int. J. Nanomed. 2017, 12, 8227–8238. [Google Scholar] [CrossRef] [PubMed]
- Bapolisi, A.M.; Nkanga, C.I.; Walker, R.B.; Krause, R.W.M. Simultaneous Liposomal Encapsulation of Antibiotics and Proteins: Co-Loading and Characterization of Rifampicin and Human Serum Albumin in Soy-Liposomes. J. Drug Deliv. Sci. Technol. 2020, 58, 101751. [Google Scholar] [CrossRef]
- Halwani, M.; Mugabe, C.; Azghani, A.O.; Lafrenie, R.M.; Kumar, A.; Omri, A. Bactericidal Efficacy of Liposomal Aminoglycosides against Burkholderia cenocepacia. J. Antimicrob. Chemother. 2007, 60, 760–769. [Google Scholar] [CrossRef][Green Version]
- Hamblin, K.A.; Armstrong, S.J.; Barnes, K.B.; Davies, C.; Laws, T.; Blanchard, J.D.; Harding, S.V.; Atkins, H.S. Inhaled Liposomal Ciprofloxacin Protects against a Lethal Infection in a Murine Model of Pneumonic Plague. Front. Microbiol. 2017, 8, 91. [Google Scholar] [CrossRef]
- d’Angelo, I.; Conte, C.; La Rotonda, M.I.; Miro, A.; Quaglia, F.; Ungaro, F. Improving the Efficacy of Inhaled Drugs in Cystic Fibrosis: Challenges and Emerging Drug Delivery Strategies. Adv. Drug Deliv. Rev. 2014, 75, 92–111. [Google Scholar] [CrossRef]
- Bilton, D.; Pressler, T.; Fajac, I.; Clancy, J.P.; Sands, D.; Minic, P.; Cipolli, M.; Galeva, I.; Solé, A.; Quittner, A.L.; et al. Amikacin Liposome Inhalation Suspension for Chronic Pseudomonas aeruginosa Infection in Cystic Fibrosis. J. Cyst. Fibros. 2020, 19, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.; Mandal, M.; Pinho, J.; Catalão, M.J.; Almeida, A.J.; Azevedo-Pereira, J.M.; Gaspar, M.M.; Anes, E. Liposomal Delivery of Saquinavir to Macrophages Overcomes Cathepsin Blockade by Mycobacterium tuberculosis and Helps Control the Phagosomal Replicative Niches. Int. J. Mol. Sci. 2023, 24, 1142. [Google Scholar] [CrossRef] [PubMed]
- Khameneh, B.; Iranshahy, M.; Ghandadi, M.; Ghoochi Atashbeyk, D.; Fazly Bazzaz, B.S.; Iranshahi, M. Investigation of the Antibacterial Activity and Efflux Pump Inhibitory Effect of Co-Loaded Piperine and Gentamicin Nanoliposomes in Methicillin-Resistant Staphylococcus aureus. Drug Dev. Ind. Pharm. 2015, 41, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Hedges, J.F.; Snyder, D.T.; Robison, A.; Thompson, M.A.; Aspelin, K.; Plewa, J.; Baldridge, J.; Jutila, M.A. A TLR4 Agonist Liposome Formulation Effectively Stimulates Innate Immunity and Enhances Protection from Bacterial Infection. Innate Immun. 2023, 29, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Giannaccare, G.; Comis, S.; Jannuzzi, V.; Camposampiero, D.; Ponzin, D.; Cambria, S.; Santocono, M.; Pallozzi Lavorante, N.; Del Noce, C.; Scorcia, V.; et al. Effect of Liposomal-Lactoferrin-Based Eye Drops on the Conjunctival Microflora of Patients Undergoing Cataract Surgery. Ophthalmol. Ther. 2023, 12, 1315–1326. [Google Scholar] [CrossRef]
- Karpuz, M.; Temel, A.; Ozgenc, E.; Tekintas, Y.; Erel-Akbaba, G.; Senyigit, Z.; Atlihan-Gundogdu, E. 99mTc-Labeled, Colistin Encapsulated, Theranostic Liposomes for Pseudomonas aeruginosa Infection. AAPS PharmSciTech 2023, 24, 77. [Google Scholar] [CrossRef]
- Guo, R.; Liu, Y.; Li, K.; Tian, B.; Li, W.; Niu, S.; Hong, W. Direct Interactions between Cationic Liposomes and Bacterial Cells Ameliorate the Systemic Treatment of Invasive Multidrug-Resistant Staphylococcus aureus Infections. Nanomed. Nanotechnol. Biol. Med. 2021, 34, 102382. [Google Scholar] [CrossRef]
- Patil, R.; Torris, A.; Bhat, S.; Patil, S. Mapping Fusogenicity of Ciprofloxacin-Loaded Liposomes with Bacterial Cells. AAPS PharmSciTech 2019, 20, 180. [Google Scholar] [CrossRef]
- Nicolosi, D.; Scalia, M.; Nicolosi, V.M.; Pignatello, R. Encapsulation in Fusogenic Liposomes Broadens the Spectrum of Action of Vancomycin against Gram-Negative Bacteria. Int. J. Antimicrob. Agents 2010, 35, 553–558. [Google Scholar] [CrossRef]
- Darvishi, M.; Farahani, S.; Haeri, A. Moxifloxacin-Loaded Lipidic Nanoparticles for Antimicrobial Efficacy. Curr. Pharm. Des. 2021, 27, 135–140. [Google Scholar] [CrossRef]
- Longo, J.P.F.; Leal, S.C.; Simioni, A.R.; de Fátima Menezes Almeida-Santos, M.; Tedesco, A.C.; Azevedo, R.B. Photodynamic Therapy Disinfection of Carious Tissue Mediated by Aluminum-Chloride-Phthalocyanine Entrapped in Cationic Liposomes: An in Vitro and Clinical Study. Lasers Med. Sci. 2012, 27, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Ferro, S.; Ricchelli, F.; Monti, D.; Mancini, G.; Jori, G. Efficient Photoinactivation of Methicillin-Resistant Staphylococcus aureus by a Novel Porphyrin Incorporated into a Poly-Cationic Liposome. Int. J. Biochem. Cell Biol. 2007, 39, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Bartomeu Garcia, C.; Shi, D.; Webster, T.J. Tat-Functionalized Liposomes for the Treatment of Meningitis: An in Vitro Study. Int. J. Nanomed. 2017, 12, 3009–3021. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Tang, Y.; Jiang, B.; Xu, Y.; Liu, S.; Huang, C. Functional Liposome Loaded Curcumin for the Treatment of Streptococcus mutans Biofilm. Front. Chem. 2023, 11, 1160521. [Google Scholar] [CrossRef]
- Vanamala, K.; Bhise, K.; Sanchez, H.; Kebriaei, R.; Luong, D.; Sau, S.; Abdelhady, H.; Rybak, M.J.; Andes, D.; Iyer, A.K. Folate Functionalized Lipid Nanoparticles for Targeted Therapy of Methicillin-Resistant Staphylococcus aureus. Pharmaceutics 2021, 13, 1791. [Google Scholar] [CrossRef]
- Rani, N.N.; Chen, X.Y.; Al-Zubaidi, Z.M.; Azhari, H.; Khaitir, T.M.N.; Ng, P.Y.; Buang, F.; Tan, G.C.; Wong, Y.P.; Said, M.M.; et al. Surface-Engineered Liposomes for Dual-Drug Delivery Targeting Strategy against Methicillin-Resistant Staphylococcus aureus (MRSA). Asian J. Pharm. Sci. 2022, 17, 102–119. [Google Scholar] [CrossRef]
- Uhl, P.; Sauter, M.; Hertlein, T.; Witzigmann, D.; Laffleur, F.; Hofhaus, G.; Fidelj, V.; Tursch, A.; Özbek, S.; Hopke, E.; et al. Overcoming the Mucosal Barrier: Tetraether Lipid-Stabilized Liposomal Nanocarriers Decorated with Cell-Penetrating Peptides Enable Oral Delivery of Vancomycin. Adv. Ther. 2021, 4, 2000247. [Google Scholar] [CrossRef]
- Singla, A.; Simbassa, S.B.; Chirra, B.; Gairola, A.; Southerland, M.R.; Shah, K.N.; Rose, R.E.; Chen, Q.; Basharat, A.; Baeza, J.; et al. Hetero-Multivalent Targeted Liposomal Drug Delivery to Treat Pseudomonas aeruginosa Infections. ACS Appl. Mater. Interfaces 2022, 14, 40724–40737. [Google Scholar] [CrossRef]
- Wang, R.; Song, C.; Gao, A.; Liu, Q.; Guan, W.; Mei, J.; Ma, L.; Cui, D. Antibody-Conjugated Liposomes Loaded with Indocyanine Green for Oral Targeted Photoacoustic Imaging-Guided Sonodynamic Therapy of Helicobacter pylori Infection. Acta Biomater. 2022, 143, 418–427. [Google Scholar] [CrossRef]
- Chen, R.; Ji, Y.; Li, T.; Zhao, B.; Guo, H.; Wang, Z.; Yao, H.; Zhang, Z.; Liu, C.; Du, M. Anti-Porphyromonas gingivalis Nanotherapy for Maintaining Bacterial Homeostasis in Periodontitis. Int. J. Antimicrob. Agents 2023, 61, 106801. [Google Scholar] [CrossRef]
- Chen, X.; Yang, H.; Li, C.; Hu, W.; Cui, H.; Lin, L. Enhancing the Targeting Performance and Prolonging the Antibacterial Effects of Clove Essential Oil Liposomes to Campylobacter Jejuni by Antibody Modification. Food Res. Int. 2023, 167, 112736. [Google Scholar] [CrossRef]
- Krivić, H.; Himbert, S.; Sun, R.; Feigis, M.; Rheinstädter, M.C. Erythro-PmBs: A Selective Polymyxin B Delivery System Using Antibody-Conjugated Hybrid Erythrocyte Liposomes. ACS Infect. Dis. 2022, 8, 2059–2072. [Google Scholar] [CrossRef]
- Moreira, L.; Guimarães, N.M.; Pereira, S.; Santos, R.S.; Loureiro, J.A.; Ferreira, R.M.; Figueiredo, C.; Pereira, M.C.; Azevedo, N.F. Engineered Liposomes to Deliver Nucleic Acid Mimics in Escherichia coli. J. Control. Release 2023, 355, 489–500. [Google Scholar] [CrossRef]
- Shi, Y.; Feng, X.; Lin, L.; Wang, J.; Chi, J.; Wu, B.; Zhou, G.; Yu, F.; Xu, Q.; Liu, D.; et al. Virus-Inspired Surface-Nanoengineered Antimicrobial Liposome: A Potential System to Simultaneously Achieve High Activity and Selectivity. Bioact. Mater. 2021, 6, 3207–3217. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Obuki, M.; Sumi, H.; Klußmann, M.; Morimoto, K.; Nakai, S.; Hashimoto, T.; Fujiwara, D.; Fujii, I.; Yuba, E.; et al. Macropinocytosis-Inducible Extracellular Vesicles Modified with Antimicrobial Protein CAP18-Derived Cell-Penetrating Peptides for Efficient Intracellular Delivery. Mol. Pharm. 2021, 18, 3290–3301. [Google Scholar] [CrossRef] [PubMed]
- Celebi, D.; Celebi, O.; Baser, S.; Taghizadehghalehjoughi, A. Evaluation of Antimicrobial and Antibiofilm Efficacy of Bee Venom and Exosome Against Escherichia coli K99 Strain. Kafkas Üniv. Vet. Fak. Derg. 2023, 29, 239–246. [Google Scholar]
- Cheng, Y.; Schorey, J.S. Exosomes Carrying Mycobacterial Antigens Can Protect Mice against Mycobacterium tuberculosis Infection. Eur. J. Immunol. 2013, 43, 3279–3290. [Google Scholar] [CrossRef] [PubMed]
- Hazrati, A.; Soudi, S.; Hashemi, S.M. Wharton’s Jelly Mesenchymal Stem Cells-Derived Exosomes and Imipenem in Combination Reduce Apoptosis and Inflammatory Responses in E. coli-Infected HepG2 Cells. Iran. J. Allergy Asthma Immunol. 2022, 21, 273. [Google Scholar]
- Yang, X.; Xie, B.; Peng, H.; Shi, G.; Sreenivas, B.; Guo, J.; Wang, C.; He, Y. Eradicating Intracellular MRSA via Targeted Delivery of Lysostaphin and Vancomycin with Mannose-Modified Exosomes. J. Control. Release 2021, 329, 454–467. [Google Scholar] [CrossRef]
- Yang, X.; Shi, G.; Guo, J.; Wang, C.; He, Y. Exosome-Encapsulated Antibiotic against Intracellular Infections of Methicillin-Resistant Staphylococcus aureus. Int. J. Nanomed. 2018, 13, 8095–8104. [Google Scholar] [CrossRef]
- Varela, M.F.; Stephen, J.; Lekshmi, M.; Ojha, M.; Wenzel, N.; Sanford, L.M.; Hernandez, A.J.; Parvathi, A.; Kumar, S.H. Bacterial Resistance to Antimicrobial Agents. Antibiotics 2021, 10, 593. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Manzoor, K.N.; Sultan, A.; Saeed, M.; Rafique, M.; Noushad, S.; Talib, A.; Rentschler, S.; Deigner, H.-P. Pulling the Brakes on Fast and Furious Multiple Drug-Resistant (MDR) Bacteria. Int. J. Mol. Sci. 2021, 22, 859. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Santos, J.V.; da Costa Júnior, S.D.; de Fátima Ramos dos Santos Medeiros, S.M.; Cavalcanti, I.D.L.; de Souza, J.B.; Coriolano, D.L.; da Silva, W.R.C.; Alves, M.H.M.E.; Cavalcanti, I.M.F. Panorama of Bacterial Infections Caused by Epidemic Resistant Strains. Curr. Microbiol. 2022, 79, 175. [Google Scholar] [CrossRef] [PubMed]
- Fodor, A.; Abate, B.A.; Deák, P.; Fodor, L.; Gyenge, E.; Klein, M.G.; Koncz, Z.; Muvevi, J.; Ötvös, L.; Székely, G.; et al. Multidrug Resistance (MDR) and Collateral Sensitivity in Bacteria, with Special Attention to Genetic and Evolutionary Aspects and to the Perspectives of Antimicrobial Peptides—A Review. Pathogens 2020, 9, 522. [Google Scholar] [CrossRef] [PubMed]
- Moo, C.-L.; Yang, S.-K.; Yusoff, K.; Ajat, M.; Thomas, W.; Abushelaibi, A.; Lim, S.-H.-E.; Lai, K.-S. Mechanisms of Antimicrobial Resistance (AMR) and Alternative Approaches to Overcome AMR. Curr. Drug Discov. Technol. 2020, 17, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Health and Economic Impacts of Antimicrobial Resistance in the Western Pacific Region, 2020–2030; World Health Organization (WHO): Geneva, Switzerland, 2023. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).