Competitive Chemical Reaction Kinetic Model of Nucleosome Assembly Using the Histone Variant H2A.Z and H2A In Vitro
Abstract
:1. Introduction
2. Results
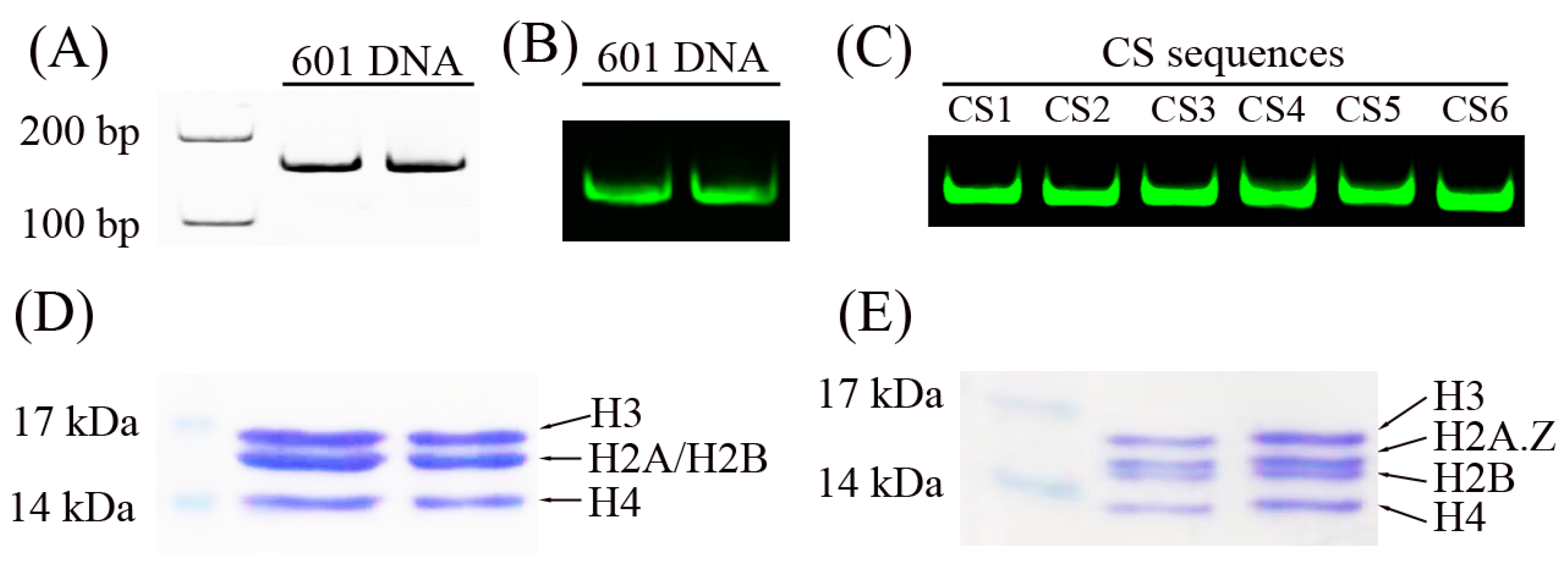
2.1. Preparation of DNA Sequences and Histone Octamers
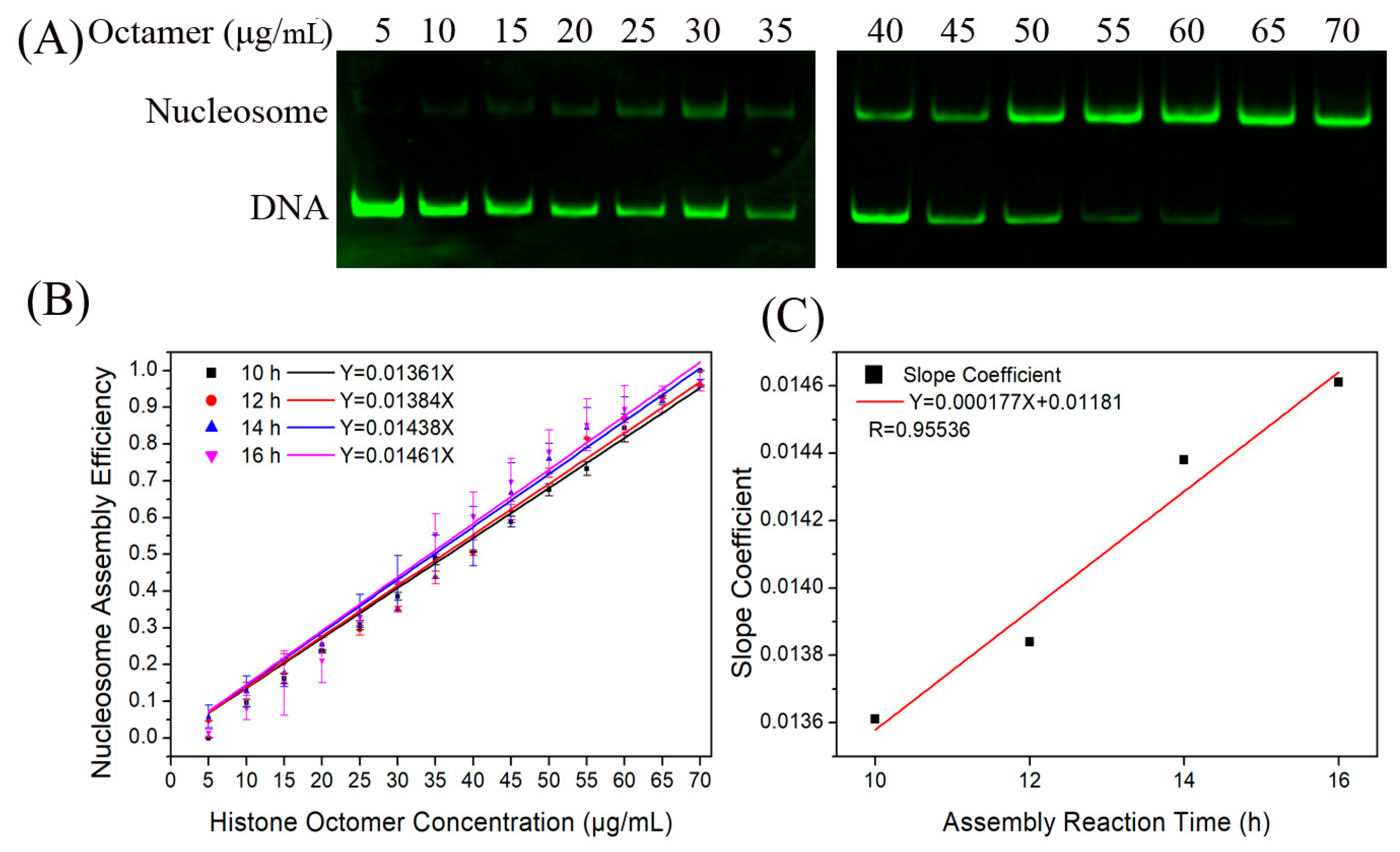
2.2. The H2A.Z-Containing Nucleosome Assembly Can Be Described by a Chemical Kinetic Model
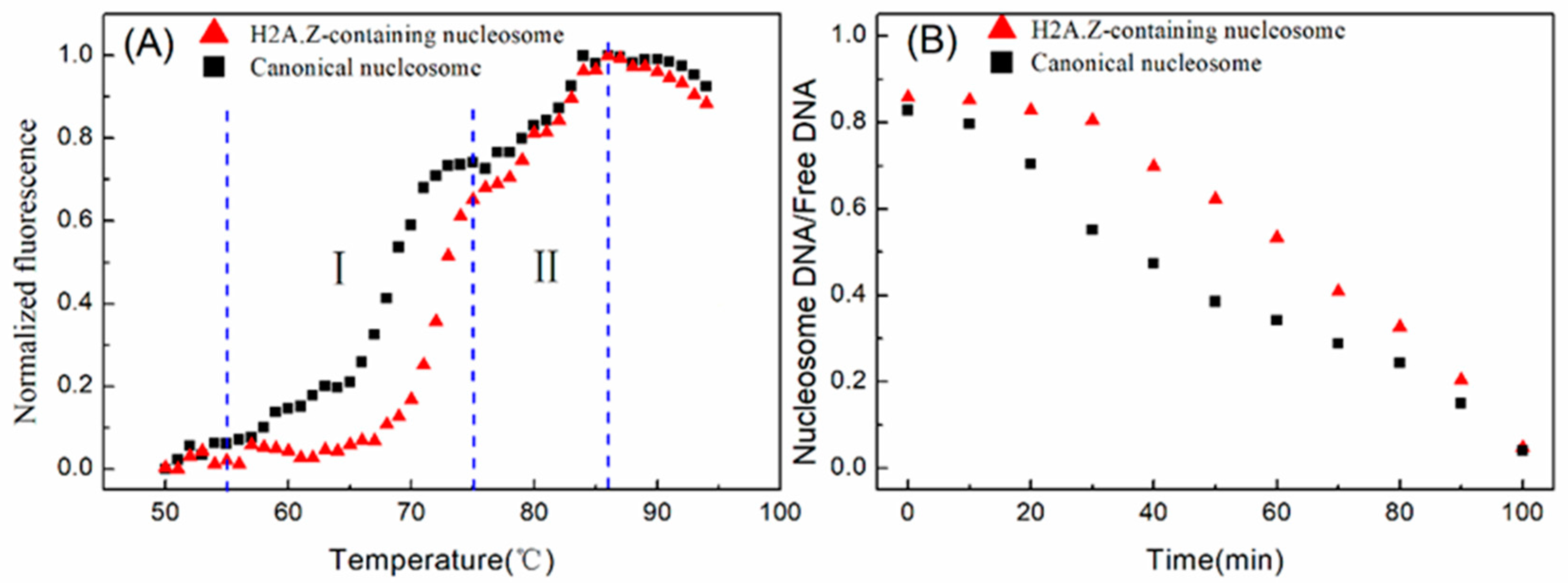
2.3. The Characteristics of the DNA Sequence Can Regulate the Assembly Efficiency of H2A.Z-Containing Nucleosomes In Vitro
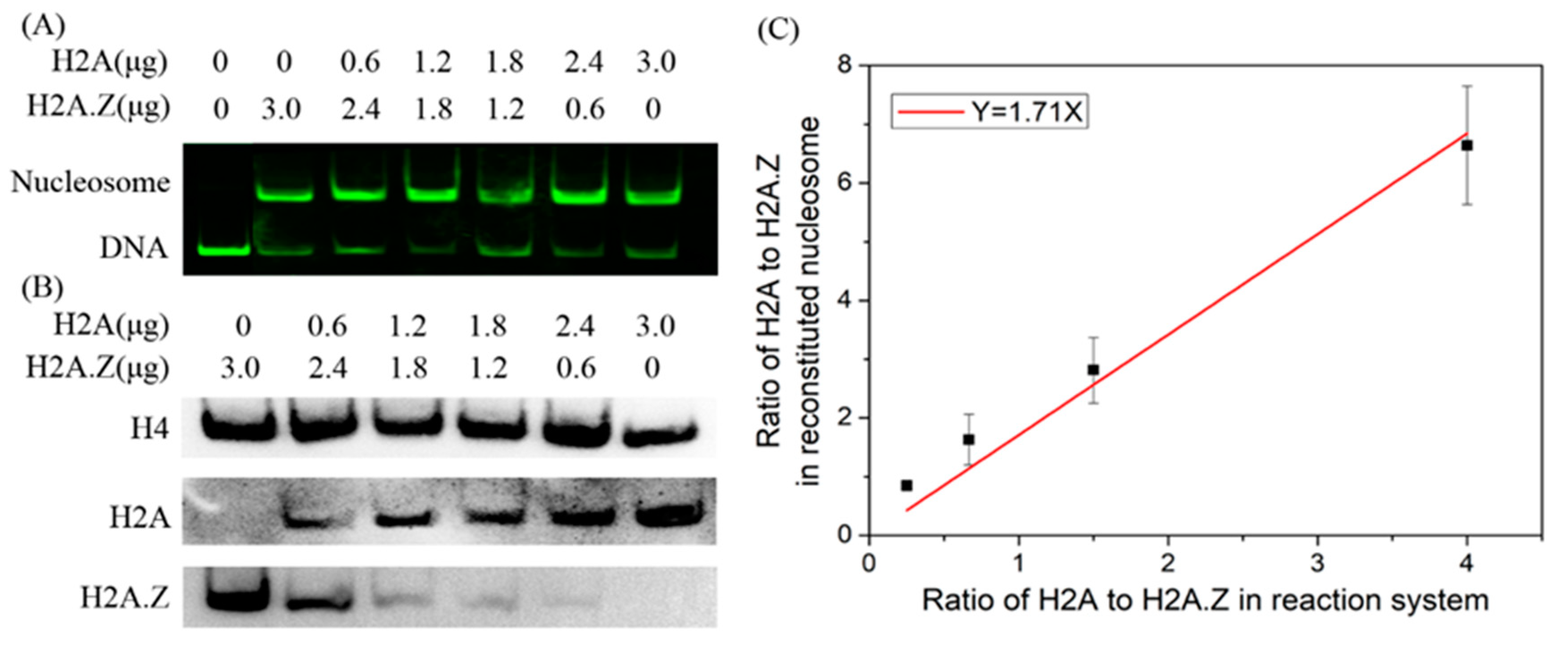
2.4. The Present Model Can Be Used to Describe the Competitive Dynamics of H2A and H2A.Z in Nucleosome Assembly
3. Discussion
4. Materials and Methods
4.1. Preparation of DNA Sequences and Recombinant Histone Octamers
4.2. Nucleosome Assembly Reaction In Vitro
4.3. Gel Analysis of Nucleosome Assembly Efficiency
4.4. Native PAGE/Western Blotting Analysis of the Competitive Ability of the H2A and H2A.Z Histones in Reconstituted Nucleosomes
4.5. Thermal Stability Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Guo, M.; Zhang, F.; Shao, X.; Liu, G.; Xing, Y.; Zhao, X.; Luo, L.; Cai, L. Nucleosome Assembly and Disassembly in vitro Are Governed by Chemical Kinetic Principles. Front. Cell Dev. Biol. 2021, 9, 762571. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Liu, G.J.; Tan, J.X.; Lin, H. DNA physical properties outperform sequence compositional information in classifying nucleosome-enriched and -depleted regions. Genomics 2019, 111, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Jarillo, J.A.; Piñeiro, M. H2A.Z mediates different aspects of chromatin function and modulates flowering responses in Arabidopsis. Plant J. 2015, 83, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Brewis, H.T.; Wang, A.Y.; Gaub, A.; Lau, J.J.; Stirling, P.C.; Kobor, M.S. What makes a histone variant a variant: Changing H2A to become H2A.Z. PLoS Genet. 2021, 17, e1009950. [Google Scholar] [CrossRef]
- Chen, P.; Zhao, J.; Wang, Y.; Wang, M.; Long, H.; Liang, D.; Huang, L.; Wen, Z.; Li, W.; Li, X.; et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes. Dev. 2013, 27, 2109–2124. [Google Scholar] [CrossRef]
- Belotti, E.; Lacoste, N.; Simonet, T.; Papin, C.; Padmanabhan, K.; Scionti, I.; Gangloff, Y.G.; Ramos, L.; Dalkara, D.; Hamiche, A.; et al. H2A.Z is dispensable for both basal and activated transcription in post-mitotic mouse muscles. Nucleic Acids Res. 2020, 48, 4601–4613. [Google Scholar] [CrossRef]
- Giaimo, B.D.; Ferrante, F.; Herchenröther, A.; Hake, S.B.; Borggrefe, T. The histone variant H2A.Z in gene regulation. Epigenetics Chromatin 2019, 12, 37. [Google Scholar] [CrossRef]
- Long, H.; Zhang, L.; Lv, M.; Wen, Z.; Zhang, W.; Chen, X.; Zhang, P.; Li, T.; Chang, L.; Jin, C.; et al. H2A.Z facilitates licensing and activation of early replication origins. Nature 2020, 577, 576–581. [Google Scholar] [CrossRef]
- Sales-Gil, R.; Kommer, D.C.; de Castro, I.J.; Amin, H.A.; Vinciotti, V.; Sisu, C.; Vagnarelli, P. Non-redundant functions of H2A.Z.1 and H2A.Z.2 in chromosome segregation and cell cycle progression. EMBO Rep. 2021, 22, e52061. [Google Scholar] [CrossRef]
- Neves, L.T.; Douglass, S.; Spreafico, R.; Venkataramanan, S.; Kress, T.L.; Johnson, T.L. The histone variant H2A.Z promotes efficient cotranscriptional splicing in S. cerevisiae. Genes Dev. 2017, 31, 702–717. [Google Scholar] [CrossRef]
- Turinetto, V.; Giachino, C. Histone variants as emerging regulators of embryonic stem cell identity. Epigenetics 2015, 10, 563–573. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef]
- Nekrasov, M.; Soboleva, T.A.; Jack, C.; Tremethick, D.J. Histone variant selectivity at the transcription start site: H2A.Z or H2A.Lap1. Nucleus 2013, 4, 431–438. [Google Scholar] [CrossRef]
- Hu, G.; Cui, K.; Northrup, D.; Liu, C.; Wang, C.; Tang, Q.; Ge, K.; Levens, D.; Crane-Robinson, C.; Zhao, K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2013, 12, 180–192. [Google Scholar] [CrossRef]
- Subramanian, V.; Fields, P.A.; Boyer, L.A. H2A.Z: A molecular rheostat for transcriptional control. F1000Prime Rep. 2015, 7, 1. [Google Scholar] [CrossRef]
- Subramanian, V.; Mazumder, A.; Surface, L.E.; Butty, V.L.; Fields, P.A.; Alwan, A.; Torrey, L.; Thai, K.K.; Levine, S.S.; Bathe, M.; et al. H2A.Z acidic patch couples chromatin dynamics to regulation of gene expression programs during ESC differentiation. PLoS Genet. 2013, 9, e1003725. [Google Scholar] [CrossRef]
- Weber, C.M.; Henikoff, J.G.; Henikoff, S. H2A.Z nucleosomes enriched over active genes are homotypic. Nat. Struct. Mol. Biol. 2010, 17, 1500–1507. [Google Scholar] [CrossRef]
- Ranjan, A.; Mizuguchi, G.; FitzGerald, P.C.; Wei, D.; Wang, F.; Huang, Y.; Luk, E.; Woodcock, C.L.; Wu, C. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell 2013, 154, 1232–1245. [Google Scholar] [CrossRef]
- Böhm, V.; Hieb, A.R.; Andrews, A.J.; Gansen, A.; Rocker, A.; Tóth, K.; Luger, K.; Langowski, J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011, 39, 3093–3102. [Google Scholar] [CrossRef]
- Gansen, A.; Valeri, A.; Hauger, F.; Felekyan, S.; Kalinin, S.; Tóth, K.; Langowski, J.; Seidel, C.A. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc. Natl. Acad. Sci. USA 2009, 106, 15308–15313. [Google Scholar] [CrossRef]
- Lowary, P.T.; Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998, 276, 19–42. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, F.; Guo, M.; Xing, Y.; Liu, G.; Zhao, X.; Cai, L. The affinity of DNA sequences containing R5Y5 motif and TA repeats with 10.5-bp periodicity to histone octamer in vitro. J. Biomol. Struct. Dyn. 2019, 37, 1935–1943. [Google Scholar] [CrossRef]
- Arimura, Y.; Ikura, M.; Fujita, R.; Noda, M.; Kobayashi, W.; Horikoshi, N.; Sun, J.; Shi, L.; Kusakabe, M.; Harata, M.; et al. Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome. Nucleic Acids Res. 2018, 46, 10007–10018. [Google Scholar] [CrossRef]
- Gévry, N.; Chan, H.M.; Laflamme, L.; Livingston, D.M.; Gaudreau, L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007, 21, 1869–1881. [Google Scholar] [CrossRef]
- Guillemette, B.; Gaudreau, L. Reuniting the contrasting functions of H2A.Z. Biochem. Cell Biol. 2006, 84, 528–535. [Google Scholar] [CrossRef]
- Dion, M.F.; Kaplan, T.; Kim, M.; Buratowski, S.; Friedman, N.; Rando, O.J. Dynamics of replication-independent histone turnover in budding yeast. Science 2007, 315, 1405–1408. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Halley, J.E.; Rine, J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006, 20, 700–710. [Google Scholar] [CrossRef]
- Rangasamy, D.; Berven, L.; Ridgway, P.; Tremethick, D.J. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 2003, 22, 1599–1607. [Google Scholar] [CrossRef]
- Rangasamy, D.; Greaves, I.; Tremethick, D.J. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 2004, 11, 650–655. [Google Scholar] [CrossRef]
- Colino-Sanguino, Y.; Clark, S.J.; Valdes-Mora, F. The H2A.Z-nucleosome code in mammals: Emerging functions. Trends Genet. 2022, 38, 516. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Hake, S.B. The nucleosome: A little variation goes a long way. Biochem. Cell Biol. 2006, 84, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Zang, C.; Wei, G.; Cui, K.; Peng, W.; Zhao, K.; Felsenfeld, G. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat. Genet. 2009, 41, 941–945. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, M.; Amrichova, J.; Parker, B.J.; Soboleva, T.A.; Jack, C.; Williams, R.; Huttley, G.A.; Tremethick, D.J. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat. Struct. Mol. Biol. 2012, 19, 1076–1083. [Google Scholar] [CrossRef]
- Cole, L.; Kurscheid, S.; Nekrasov, M.; Domaschenz, R.; Vera, D.L.; Dennis, J.H.; Tremethick, D.J. Multiple roles of H2A.Z in regulating promoter chromatin architecture in human cells. Nat. Commun. 2021, 12, 2524. [Google Scholar] [CrossRef]
- Buschbeck, M.; Hake, S.B. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 2017, 18, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Zink, L.M.; Hake, S.B. Histone variants: Nuclear function and disease. Curr. Opin. Genet. Dev. 2016, 37, 82–89. [Google Scholar] [CrossRef]
- Yang, H.D.; Kim, P.J.; Eun, J.W.; Shen, Q.; Kim, H.S.; Shin, W.C.; Ahn, Y.M.; Park, W.S.; Lee, J.Y.; Nam, S.W. Oncogenic potential of histone-variant H2A.Z.1 and its regulatory role in cell cycle and epithelial-mesenchymal transition in liver cancer. Oncotarget 2016, 7, 11412–11423. [Google Scholar] [CrossRef]
- Vardabasso, C.; Gaspar-Maia, A.; Hasson, D.; Pünzeler, S.; Valle-Garcia, D.; Straub, T.; Keilhauer, E.C.; Strub, T.; Dong, J.; Panda, T.; et al. Histone Variant H2A.Z.2 Mediates Proliferation and Drug Sensitivity of Malignant Melanoma. Mol. Cell 2015, 59, 75–88. [Google Scholar] [CrossRef]
- Berta, D.G.; Kuisma, H.; Välimäki, N.; Räisänen, M.; Jäntti, M.; Pasanen, A.; Karhu, A.; Kaukomaa, J.; Taira, A.; Cajuso, T.; et al. Deficient H2A.Z deposition is associated with genesis of uterine leiomyoma. Nature 2021, 596, 398–403. [Google Scholar] [CrossRef]
- Wang, Q.; Qi, Y.; Xiong, F.; Wang, D.; Wang, B.; Chen, Y. The H2A.Z-KDM1A complex promotes tumorigenesis by localizing in the nucleus to promote SFRP1 promoter methylation in cholangiocarcinoma cells. BMC Cancer 2022, 22, 1166. [Google Scholar] [CrossRef] [PubMed]
- Ghiraldini, F.G.; Filipescu, D.; Bernstein, E. Solid tumours hijack the histone variant network. Nat. Rev. Cancer 2021, 21, 257–275. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, R.S.; Long, H.K.; Swigut, T.; Wysocka, J. Single Amino Acid Change Un-derlies Distinct Roles of H2A.Z Subtypes in Human Syndrome. Cell 2019, 178, 1421–1436.e24. [Google Scholar] [CrossRef] [PubMed]
- Chevillard-Briet, M.; Quaranta, M.; Grézy, A.; Mattera, L.; Courilleau, C.; Philippe, M.; Mercier, P.; Corpet, D.; Lough, J.; Ueda, T.; et al. Interplay between chromatin-modifying enzymes controls colon cancer progression through Wnt signaling. Hum. Mol. Genet. 2014, 23, 2120–2131. [Google Scholar] [CrossRef]
- Dai, L.; Xiao, X.; Pan, L.; Shi, L.; Xu, N.; Zhang, Z.; Feng, X.; Ma, L.; Dou, S.; Wang, P.; et al. Recognition of the inherently unstable H2A nucleosome by Swc2 is a major determinant for unidirectional H2A.Z exchange. Cell Rep. 2021, 35, 109183. [Google Scholar] [CrossRef]
- Mizuguchi, G.; Shen, X.; Landry, J.; Wu, W.H.; Sen, S.; Wu, C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 2004, 303, 343–348. [Google Scholar] [CrossRef]
- Ruhl, D.D.; Jin, J.; Cai, Y.; Swanson, S.; Florens, L.; Washburn, M.P.; Conaway, R.C.; Conaway, J.W.; Chrivia, J.C. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 2006, 45, 5671–5677. [Google Scholar] [CrossRef]
- Singh, R.K.; Fan, J.; Gioacchini, N.; Watanabe, S.; Bilsel, O.; Peterson, C.L. Transient Kinetic Analysis of SWR1C-Catalyzed H2A.Z Deposition Unravels the Impact of Nucleosome Dynamics and the Asymmetry of Histone Exchange. Cell Rep. 2019, 27, 374–386.e4. [Google Scholar] [CrossRef]
- Zhao, H.; Xing, Y.; Liu, G.; Chen, P.; Zhao, X.; Li, G.; Cai, L. GAA triplet-repeats cause nucleosome depletion in the human genome. Genomics 2015, 106, 88–95. [Google Scholar] [CrossRef]
- Sueoka, T.; Hayashi, G.; Okamoto, A. Regulation of the Stability of the Histone H2A-H2B Dimer by H2A Tyr57 Phosphorylation. Biochemistry 2017, 56, 4767–4772. [Google Scholar] [CrossRef]

| Reaction Gradient | 2.4 μg Total Proteins | 3 μg Total Proteins | 3.6 μg Total Proteins | |||
|---|---|---|---|---|---|---|
| H2A | H2A.Z | H2A | H2A.Z | H2A | H2A.Z | |
| Gradient 1 | 0 | 2.4 | 0 | 3 | 0 | 3.6 |
| Gradient 2 | 0.48 | 1.92 | 0.6 | 2.4 | 0.72 | 2.88 |
| Gradient 3 | 0.96 | 1.44 | 1.2 | 1.8 | 1.44 | 2.16 |
| Gradient 4 | 1.44 | 0.96 | 1.8 | 1.2 | 2.16 | 1.44 |
| Gradient 5 | 1.92 | 0.48 | 2.4 | 0.6 | 2.88 | 0.72 |
| Gradient 6 | 2.4 | 0 | 3 | 0 | 3.6 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Shao, X.; Guo, M.; Xing, Y.; Wang, J.; Luo, L.; Cai, L. Competitive Chemical Reaction Kinetic Model of Nucleosome Assembly Using the Histone Variant H2A.Z and H2A In Vitro. Int. J. Mol. Sci. 2023, 24, 15846. https://doi.org/10.3390/ijms242115846
Zhao H, Shao X, Guo M, Xing Y, Wang J, Luo L, Cai L. Competitive Chemical Reaction Kinetic Model of Nucleosome Assembly Using the Histone Variant H2A.Z and H2A In Vitro. International Journal of Molecular Sciences. 2023; 24(21):15846. https://doi.org/10.3390/ijms242115846
Chicago/Turabian StyleZhao, Hongyu, Xueqin Shao, Mingxin Guo, Yongqiang Xing, Jingyan Wang, Liaofu Luo, and Lu Cai. 2023. "Competitive Chemical Reaction Kinetic Model of Nucleosome Assembly Using the Histone Variant H2A.Z and H2A In Vitro" International Journal of Molecular Sciences 24, no. 21: 15846. https://doi.org/10.3390/ijms242115846
APA StyleZhao, H., Shao, X., Guo, M., Xing, Y., Wang, J., Luo, L., & Cai, L. (2023). Competitive Chemical Reaction Kinetic Model of Nucleosome Assembly Using the Histone Variant H2A.Z and H2A In Vitro. International Journal of Molecular Sciences, 24(21), 15846. https://doi.org/10.3390/ijms242115846





