Extracellular Vesicles as Possible Plasma Markers and Mediators in Patients with Sepsis-Associated Delirium—A Pilot Study
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
2.2. EV Isolation and Characterization
2.3. Proteomics Candidate Analysis
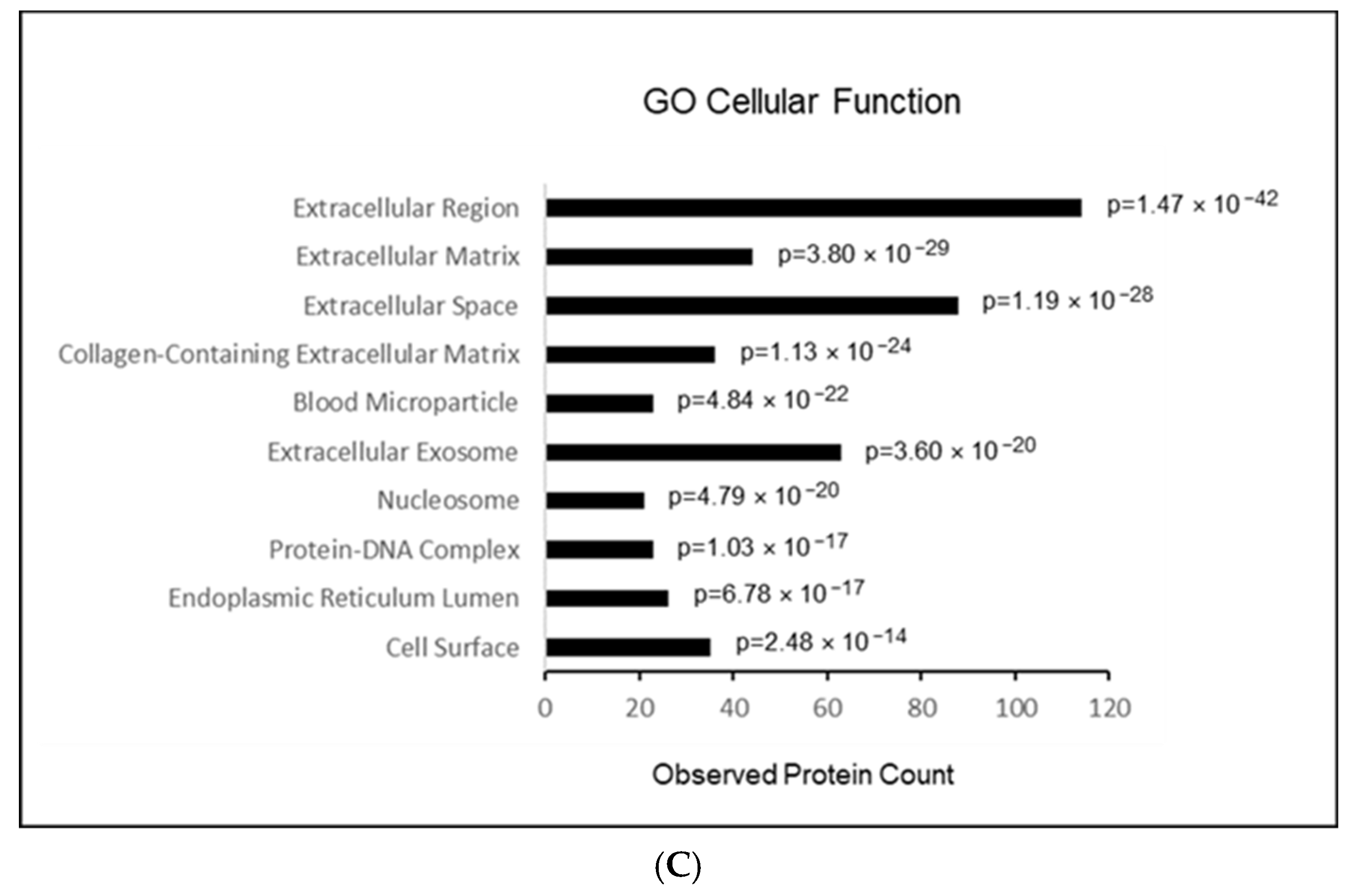
2.4. Results of GO Term Analysis
3. Discussion
3.1. Paraoxonase-1 (PON1)
3.2. Role of Immunological Markers
3.3. Role of Coagulation
3.4. Summary and Relevance
4. Materials and Methods
4.1. Study Design
4.2. Participants and Recruitment Process
4.3. Sample and Data Collection
4.4. Clinical Data Analysis
4.5. Isolation and Characterization of EVs
4.6. Sample Preparation for MS Analysis
4.7. MS Method Orbitrap Exploris 480
4.8. Proteomic Data Analysis
4.9. EV Biomarker Candidate Data Analyses
4.10. GO Term Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uhle, F.; Lichtenstern, C.; Brenner, T.; Weigand, M.A. Pathophysiology of sepsis. Anasthesiol. Intensivmed. Notfallmedizin Schmerzther. 2015, 50, 114–122. [Google Scholar] [CrossRef]
- Fleischmann-Struzek, C.; Schwarzkopf, D.; Reinhart, K. Sepsis incidence in Germany and worldwide. Current knowledge and limitations of research using health claims data. Med. Klin.-Intensivmed. Notfallmedizin 2022, 117, 264–268. [Google Scholar] [CrossRef]
- Gofton, T.E.; Young, G.B. Sepsis-associated encephalopathy. Nat. Rev. Neurol. 2012, 8, 557–566. [Google Scholar] [CrossRef]
- Czempik, P.F.; Pluta, M.P.; Krzych, L.J. Sepsis-associated brain dysfunction: A review of current literature. Int. J. Environ. Res. Public Health 2020, 17, 5852. [Google Scholar] [CrossRef] [PubMed]
- Iwashyna, T.J.; Ely, E.W.; Smith, D.M.; Langa, K.M. Long-term cognitive impairment and functional disability among survivors of severe sepsis. JAMA 2010, 304, 1787–1794. [Google Scholar] [CrossRef]
- Ebersoldt, M.; Sharshar, T.; Annane, D. Sepsis-associated delirium. Intensive Care Med. 2007, 33, 941–950. [Google Scholar] [CrossRef]
- Widmann, C.N.; Heneka, M.T. Long-term cerebral consequences of sepsis. Lancet Neurol. 2014, 13, 630–636. [Google Scholar] [CrossRef]
- Sonneville, R.; de Montmollin, E.; Poujade, J.; Garrouste-Orgeas, M.; Souweine, B.; Darmon, M.; Mariotte, E.; Argaud, L.; Barbier, F.; Goldgran-Toledano, D.; et al. Potentially modifiable factors contributing to sepsis-associated encephalopathy. Intensive Care Med. 2017, 43, 1075–1084. [Google Scholar] [CrossRef]
- Calsavara, A.; Nobre, V.; Barichello, T.; Teixeira, A.L. Post-sepsis cognitive impairment and associated risk factors: A systematic review. Aust. Crit. Care 2018, 31, 242–253. [Google Scholar] [CrossRef]
- Wang, H.E.; Kabeto, M.M.; Gray, M.; Wadley, V.G.; Muntner, P.; Judd, S.E.; Safford, M.M.; Kempker, J.; Levine, D.A. Trajectory of cognitive decline after sepsis. Crit. Care Med. 2021, 49, 1083–1094. [Google Scholar] [CrossRef]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular vesicles in cancer: Cell-to-cell mediators of metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef]
- Witwer, K.W.; Théry, C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell Vesicles 2019, 8, 1648167. [Google Scholar] [CrossRef]
- Stahl, P.D.; Raposo, G. Exosomes and extracellular vesicles: The path forward. Essays Biochem. 2018, 62, 119–124. [Google Scholar] [CrossRef]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-mediated metastasis: Communication from a distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Fricke, F.; Gebert, J.; Kopitz, J.; Plaschke, K. Proinflammatory Extracellular Vesicle-Mediated Signaling Contributes to the Induction of Neuroinflammation in Animal Models of Endotoxemia and Peripheral Surgical Stress. Cell. Mol. Neurobiol. 2021, 41, 1325–1336. [Google Scholar] [CrossRef]
- Liu, W.; Bai, X.; Zhang, A.; Huang, J.; Xu, S.; Zhang, J. Role of exosomes in central nervous system diseases. Front. Mol. Neurosci. 2019, 12, 240. [Google Scholar] [CrossRef]
- Rama Rao, K.V.; Kielian, T. Neuron-astrocyte interactions in neurodegenerative diseases: Role of neuroinflammation. Clin. Exp. Neuroimmunol. 2015, 6, 245–263. [Google Scholar] [CrossRef]
- Gupta, A.; Pulliam, L. Exosomes as mediators of neuroinflammation. J. Neuroinflamm. 2014, 11, 68. [Google Scholar] [CrossRef]
- Pascual, M.; Ibanez, F.; Guerri, C. Exosomes as mediators of neuron-glia communication in neuroinflammation. Neural Regen. Res. 2020, 15, 796–801. [Google Scholar] [CrossRef]
- Upadhya, D.; Shetty, A.K. Extracellular vesicles as therapeutics for brain injury and disease. Curr. Pharm. Res. 2019, 25, 3500–3505. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Wang, B.; Kodali, M.C.; Chen, C.; Kim, E.; Patters, B.J.; Lan, L.; Kumar, S.; Wang, X.; Yue, J.; et al. In vivo evidence for the contribution of peripheral circulating inflammatory exosomes to neuroinflammation. J. Neuroinflamm. 2018, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Sfera, A.; Price, A.I.; Gradini, R.; Cummings, M.; Osorio, C. Proteomic and epigenomic markers of sepsis-induced delirium (SID). Front. Mol. Biosci. 2015, 2, 59. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Noah, A.M.; Almghairbi, D.; Evley, R.; Moppett, I.K. Preoperative inflammatory mediators and postoperative delirium: Systematic review and meta-analysis. Br. J. Anaesth. 2021, 127, 424–434. [Google Scholar] [CrossRef]
- Taler-Verčič, A.; Goličnik, M.; Bavec, A. The Structure and Function of Paraoxonase-1 and Its Comparison to Paraoxonase-2 and -3. Molecules 2020, 25, 5980. [Google Scholar] [CrossRef]
- Reichert, C.O.; Levy, D.; Bydlowski, S.P. Paraoxonase Role in Human Neurodegenerative Diseases. Antioxidants 2021, 10, 11. [Google Scholar] [CrossRef]
- Durrington, P.N.; Bashir, B.; Soran, H. Paraoxonase 1 and atherosclerosis. Front. Cardiovasc. Med. 2023, 16, 1065967. [Google Scholar] [CrossRef]
- Khateeb, J.; Gantman, A.; Kreitenberg, A.J.; Aviram, M.; Fuhrman, B. Paraoxonase 1 (PON1) expression in hepatocytes is upregulated by pomegranate polyphenols: A role for PPAR-gamma pathway. Atherosclerosis 2010, 208, 119–125. [Google Scholar] [CrossRef]
- Reed, B.; Villeneuve, S.; Mack, W.; DeCarli, C.; Chui, H.C.; Jagust, W. Associations between serum cholesterol levels and cerebral amyloidosis. JAMA Neurol. 2014, 71, 195–200. [Google Scholar] [CrossRef]
- Zuliani, G.; Cavalieri, M.; Galvani, M.; Volpato, S.; Cherubini, A.; Bandinelli, S.; Corsi, A.M.; Lauretani, F.; Guralnik, J.M.; Fellin, R.; et al. Relationship between low levels of high-density lipoprotein cholesterol and dementia in the elderly. The InChianti study. J. Gerontol. A Biol. Sci. Med. Sci. 2010, 65, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Singh-Manoux, A.; Gimeno, D.; Kivimaki, M.; Brunner, E.; Marmot, M.G. Low HDL cholesterol is a risk factor for deficit and decline in memory in midlife: The Whitehall II study. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Ronsein, G.E.; Vaisar, T. Deepening our understanding of HDL proteome. Expert Rev. Proteom. 2019, 16, 749–760. [Google Scholar] [CrossRef]
- Yassine, H.N.; Jackson, A.M.; Borges, C.R.; Billheimer, D.; Koh, H.; Smith, D.; Reaven, P.; Lau, S.S.; Borchers, C.H. The application of multiple reaction monitoring and multi-analyte profiling to HDL proteins. Lipids Health Dis. 2014, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Poon, D.C.; Ho, Y.S.; Chiu, K.; Wong, H.L.; Chang, R.C. Sickness: From the focus on cytokines, prostaglandins, and complement factors to the perspectives of neurons. Neurosci. Biobehaviv. Rev. 2015, 57, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Bluthe, R.M.; Walter, V.; Parnet, P.; Laye, S.; Lestage, J.; Verrier, D.; Poole, S.; Stenning, B.E.; Kelley, K.W.; Dantzer, R. Lipopolysaccharide induces sickness behaviour in rats by a vagal mediated mechanism. Comptes Rendus Acad. (Sci. La Vie) 1994, 317, 499–503. [Google Scholar]
- Chiu, I.M.; Heesters, B.A.; Ghasemlou, N.; Von Hehn, C.A.; Zhao, F.; Tran, J.; Wainger, B.; Strominger, A.; Muralidharan, S.; Horswill, A.R.; et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature 2013, 501, 52–57. [Google Scholar] [CrossRef]
- Goehler, L.E.; Gaykema, R.P.; Hansen, M.K.; Anderson, K.; Maier, S.F.; Watkins, L.R. Vagal immune-to brain communication: A visceral chemosensory pathway. Auto Neurosci. 2000, 85, 49–59. [Google Scholar] [CrossRef]
- Wuerfel, E.; Infante-Duarte, C.; Glumm, R.; Wuerfel, J.T. Gadofluorine M-enhanced MRI shows involvement of circumventricular organs in neuroinflammation. J. Neuroinflamm. 2010, 7, 70. [Google Scholar] [CrossRef]
- Rochfort, K.D.; Cummins, P.M. The blood-brain barrier endothelium: A target for proinflammatory cytokines. Biochem. Soc. Trans. 2015, 43, 702–706. [Google Scholar] [CrossRef]
- Chen, C.C.; Liu, L.; Ma, F.; Wong, C.W.; Guo, X.E.; Chacko, J.V.; Farhoodi, H.P.; Zhang, S.X.; Zimak, J.; Segaliny, A.; et al. Elucidation of Exosome Migration across the Blood-Brain Barrier Model In Vitro. Cell. Mol. Bioeng. 2016, 9, 509–529. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, J.; Stewart, T.; Banks, W.A.; Zhang, J. The Transport Mechanism of Extracellular Vesicles at the Blood-Brain Barrier. Curr. Pharm. Des. 2017, 23, 6206–6214. [Google Scholar] [CrossRef] [PubMed]
- Cerejeira, J.; Lagarto, L.; Mukaetova-Ladinska, E.B. The immunology of delirium. Neuroimmunomodulation 2014, 21, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Plaschke, K.; Fichtenkamm, P.; Schramm, C.; Hauth, S.; Martin, E.; Verch, M.; Karck, M.; Kopitz, J. Early postoperative delirium after open-heart cardiac surgery is associated with decreased bispectral EEG and increased cortisol and interleukin-6. Intensive Care Med. 2010, 36, 2081–2089. [Google Scholar] [CrossRef]
- Plaschke, K.; Hill, H.; Engelhardt, R.; Thomas, C.; von Haken, R.; Scholz, M.; Kopitz, J.; Bardenheuer, H.J.; Weisbrod, M.; Weigand, M.A. EEG changes and serum anticholinergic activity measured in patients with delirium in the intensive care unit. Anaesthesia 2007, 62, 1217–1223. [Google Scholar] [CrossRef]
- Marsh, S.E.; Abud, E.M.; Lakatos, A.; Karimzadeh, A.; Yeung, S.T.; Davtyan, H.; Fote, G.M.; Lau, L.; Weinger, J.G.; Lane, T.E.; et al. The adaptive immune system restrains Alzheimer’s disease pathogenesis by modulating microglial function. Proc. Natl. Acad. Sci. USA 2016, 113, E1316–E1325. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.N. Intravenous immunoglobulins for Alzheimer’s disease and mild cognitive impairment due to Alzheimer’s disease: A systematic review with meta-analysis. Expert Rev. Neurother. 2019, 19, 475–480. [Google Scholar] [CrossRef]
- Kile, S.; Au, W.; Parise, C.; Rose, K.; Donnel, T.; Hankins, A.; Chan, M.; Ghassemi, A. IVIG treatment of mild cognitive impairment due to Alzheimer’s disease: A randomised double-blinded exploratory study of the effect on brain atrophy, cognition and conversion to dementia. J. Neurol. Neurosurg. Psychiatry 2017, 88, 106–112. [Google Scholar] [CrossRef]
- Sjöberg, A.P.; Trouw, L.A.; McGrath, F.D.G.; Hack, C.E.; Blom, A.M. Regulation of complement activation by C-reactive protein: Targeting of the inhibitory activity of C4b-binding protein. J. Immunol. 2006, 176, 7612–7620. [Google Scholar] [CrossRef]
- Bíro, A.; Rovó, Z.; Papp, D.; Cervenak, L.; Varga, L.; Füst, G.; Thielens, N.M.; Arlaud, G.J.; Prohászka, Z. Studies on the interactions between C-reactive protein and complement proteins. Immunology 2007, 121, 40–50. [Google Scholar]
- Batema, R.M.; Sharpe, M.D.; Jagger, J.E.; Ellis, C.G.; Solé-Violán, J.; López-Rodríguez, M.; Herrera-Ramos, E.; Ruíz-Hernández, J.; Borderías, L.; Horcajada, J.; et al. 36th International Symposium on Intensive Care and Emergency Medicine: Brussels, Belgium, 15–18 March 2016. Crit. Care 2016, 20 (Suppl. S2), 94. [Google Scholar] [CrossRef] [PubMed]
- Hecke, F.; Schmidt, U.; Kola, A.; Bautsch, W.; Klos, A.; Köhl, J. Circulating complement proteins in multiple trauma patients—Correlation with injury severity, development of sepsis, and outcome. Crit. Care Med. 1997, 25, 2015–2024. [Google Scholar] [CrossRef]
- Chen, L.H.; Liu, J.F.; Lu, Y.; He, X.Y.; Zhang, C.; Zhou, H.H. Complement C1q (C1qA, C1qB, and C1qC) May Be a Potential Prognostic Factor and an Index of Tumor Microenvironment Remodeling in Osteosarcoma. Front. Oncol. 2021, 17, 642144. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, M.I.; Chu, S.H.; Hernandez, M.X.; Fang, M.J.; Modarresi, L.; Selvan, P.; MacGregor, G.R.; Tenner, A.J. Cell-specific deletion of C1qa identifies microglia as the dominant source of C1q in mouse brain. J. Neuroinflamm. 2017, 14, 48. [Google Scholar] [CrossRef] [PubMed]
- Reis, E.S.; Mastellos, D.C.; Hajishengallis, G.; Lambris, J.D. New insights into the immune functions of complement. Nat. Rev. Immunol. 2019, 19, 503–516. [Google Scholar] [CrossRef]
- Merlini, M.; Rafalski, V.A.; Rios Coronado, P.E.; Gill, T.M.; Ellisman, M.; Muthukumar, G.; Subramanian, K.S.; Ryu, J.K.; Syme, C.A.; Davalos, D.; et al. Fibrinogen induces microglia-mediated spine elimination and cognitive impairment in an Alzheimer’s disease model. Neuron 2019, 101, 1099–1108. [Google Scholar] [CrossRef]
- Adams, R.A.; Bauer, J.; Flick, M.J.; Sikorski, S.L.; Nuriel, T.; Lassmann, H.; Degen, J.L.; Akassoglou, K. The fibrin-derived γ377−395 peptide inhibits microglia activation and suppresses relapsing paralysis in central nervous system autoimmune disease. J. Exp. Med. 2007, 204, 571–582. [Google Scholar] [CrossRef]
- Wang, P.; Velagapudi, R.; Kong, C.; Rodriguiz, R.M.; Wetsel, W.C.; Yang, T.; Berger, M.; Gelbard, H.A.; Colton, C.A.; Terrando, N. Neurovascular and immune mechanisms that regulate postoperative delirium superimposed on dementia. Alzheimer’s Dement 2020, 16, 734–749. [Google Scholar] [CrossRef]
- Petersen, M.A.; Ryu, J.K.; Akassoglou, K. Fibrinogen in neurological diseases: Mechanisms, imaging and therapeutics. Nat. Rev. Neurosci. 2018, 19, 283–301. [Google Scholar] [CrossRef]
- McNeil, J.B.; Hughes, C.G.; Girard, T.; Ware, L.B.; Ely, E.W.; Chandrasekhar, R.; Han, J.H. Plasma biomarkers of inflammation, coagulation, and brain injury as predictors of delirium duration in older hospitalized patients. PLoS ONE 2019, 14, e0226412. [Google Scholar] [CrossRef]
- Khan, B.A.; Perkins, A.J.; Prasad, N.K.; Shekhar, A.; Campbell, N.L.; Gao, S.; Wang, S.; Khan, S.H.; Marcantonio, E.R.; Twigg, H.L., 3rd; et al. Biomarkers of delirium duration and delirium severity in the ICU. Crit. Care Med. 2020, 48, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Radtke, F.M.; Franck, M.; Oppermann, S.; Lütz, A.; Seeling, M.; Heymann, A.; Kleinwächter, R.; Kork, F.; Skrobik, Y.; Spies, C.D. The Intensive Care Delirium Screening Checklist (ICDSC)—Translation and validation of intensive care delirium checklist in accordance with guidelines. Anasthesiol. Intensivmed. Notfallmedizin Schmerzther. 2009, 44, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Ely, E.W.; Inouye, S.K.; Bernard, G.R.; Francis, J.; May, L.; Truman, B.; Speroff, T.; Gautam, S.; Margolin, R.; Hart, R.P.; et al. Confusion Assessment Method for the Intensive Care Unit (CAM-ICU). Confusion Assessment Method for the Intensive Care Unit. APA PsycTests 2001. APA PsycNet Direct© 2023 American Psychological Association. 750 First Street NE, Washington, DC 20002-4242. [Google Scholar] [CrossRef]
- Plaschke, K.; von Haken, R.; Scholz, M.; Engelhardt, R.; Brobeil, A.; Martin, E.; Weigand, M.A. Comparison of the confusion assessment method for the intensive care unit (CAM-ICU) with the Intensive Care Delirium Screening Checklist (ICDSC) for delirium in critical care patients gives high agreement rate(s). Intensive Care Med. 2008, 34, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef]
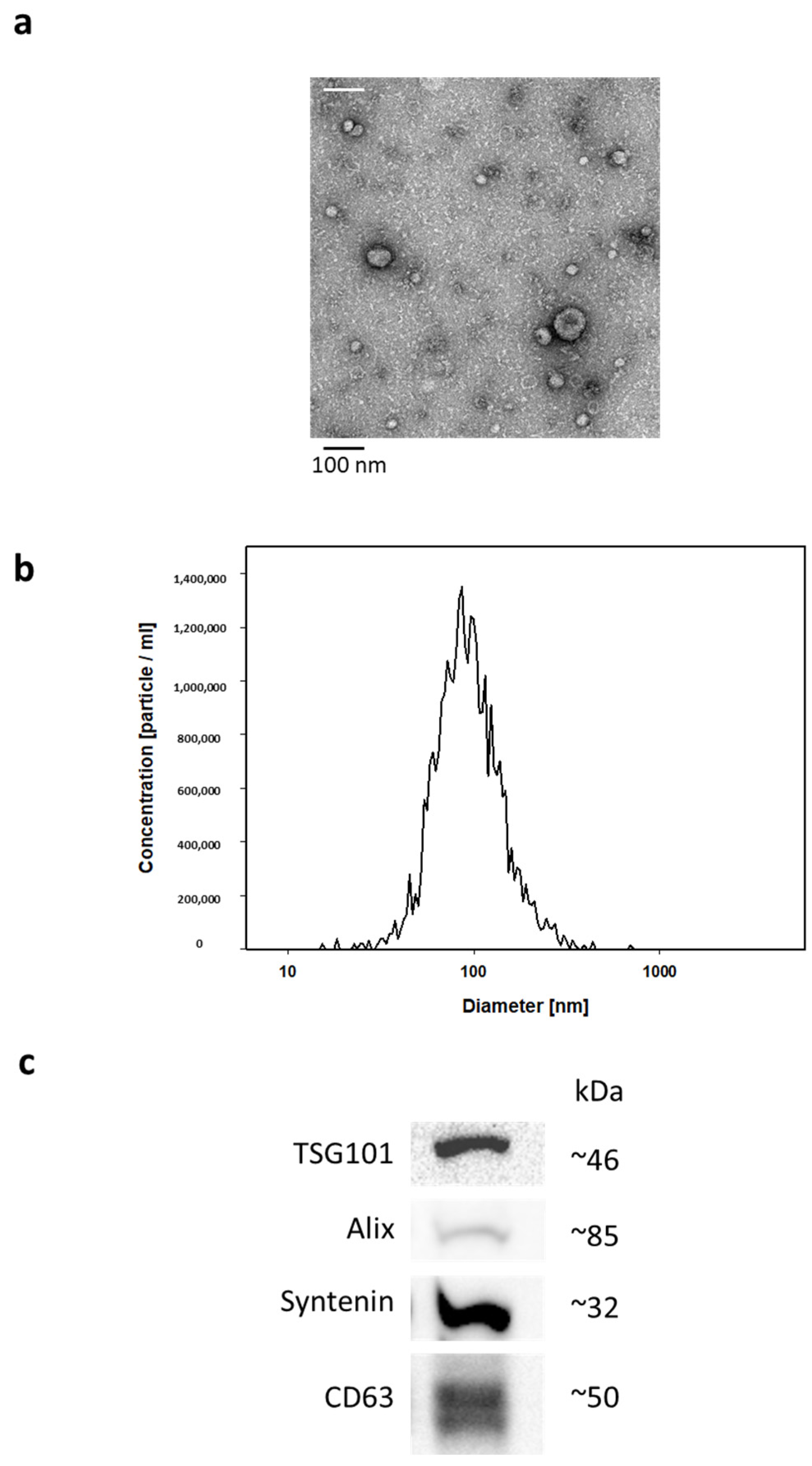


| SAD N = 11 | Non-SAD N = 11 | p-Value | |
|---|---|---|---|
| Age | 75.1 ± 6.9 | 76.3 ± 5.7 | 0.330 |
| Sex (male:female) | 8:3 | 8:3 | 0.987 |
| Length of mechanical ventilation | 24.2 ± 62.4 | 12.2 ± 12.2 | 0.269 |
| Length of ICU stay | 37.3 ± 63.1 | 22.8 ± 16.5 | 0.235 |
| Length of hospital stay | 68.1 ± 72.4 | 36.6 ± 27.7 | 0.096 |
| RASS | 0.0 ± 0.5 | −0.3 ± 1.1 | 0.251 |
| GCS | 14.5 ± 0.7 * | 13.4 ± 1.2 | 0.014 |
| SOFA score | 3.0 ± 2.8 * | 6.6 ± 2.9 | 0.005 |
| 28-day mortality | 1/11 | 2/11 | 0.876 |
| SAD | Non-SAD | p-Values | |
|---|---|---|---|
| pH values | 7.4 ± 0.1 | 7.4 ± 0.1 | 0.235 |
| Blood glucose (mg/dL) | 156.2 ± 68.4 | 129.4 ± 26.5 | 0.119 |
| C-reactive protein (mg/L) | 127.9 ± 80.5 | 148.2 ± 98.4 | 0.304 |
| HCO3− (mmol/L) | 25.6 ± 4.7 | 23.3 ± 2.7 | 0.084 |
| Platelets (/nL) | 244.6 ± 129.8 | 187.2 ± 85.7 | 0.117 |
| Creatinine (mg/dL) | 1.3 ± 1.1 | 1.7 ± 1.0 | 0.176 |
| Bilirubin (mg/dL) | 0.9 ± 0.7 | 1.1 ± 1.3 | 0.325 |
| Lactate (mg/dL) | 11.5 ± 4.6 | 11.5 ± 3.2 | 0.489 |
| Leucocytes (/nL) | 11.5 ± 4.6 | 13.2 ± 7.3 | 0.259 |
| iBAQ-Values | SAD | Non-SAD | Ratio SAD/Non-SAD | p-Values |
|---|---|---|---|---|
| PON1 | 0.017 (0.010) ** | 0.007 (0.005) | 2.5 | 0.005 |
| IgHV3 | 0.074 (0.050) ** | 0.154 (0.094) | 0.5 | 0.011 |
| C1QC | 0.512 (0.092) ** | 0.761 (0.357) | 0.7 | 0.018 |
| THBS1 | 0.0028 (0.004) ** | 0.0005 (0.0008) | 5.5 | 0.026 |
| FGG | 6.422 (3.129) ** | 4.526 (1.924) | 1.4 | 0.050 |
| AZGP1 | 0.006 (0.006) * | 0.016 (0.018) | 0.4 | 0.059 |
| SAA1 | 0.418 (0.483) * | 0.164 (0.184) | 2.6 | 0.059 |
| HbA1/HbA2 | 0.513 (0.644) * | 0.206 (0.143) | 2.5 | 0.067 |
| CRP | 1.088 (0.976) * | 0.589 (0.440) | 1.8 | 0.067 |
| HRNR | 0.003 (0.005) * | 0.0002 (0.0002) | 9.8 | 0.069 |
| IgHV3OR16-12 | 0.018 (0.016) * | 0.031 (0.023) | 0.6 | 0.072 |
| IgLV3-9 | 0.005 (0.004) * | 0.018 (0.016) | 0.4 | 0.074 |
| Ig lambda | 0.213 (0.105) * | 0.268 (0.016) | 0.8 | 0.077 |
| DSG1 | 0.004 (0.005) * | 0.002 (0.002) | 2.1 | 0.084 |
| F13A1 | 0.005 (0.005) * | 0.002 (0.002) | 1.9 | 0.088 |
| IgLC2 | 2.867 (1.235) * | 3.702 (1.575) | 0.8 | 0.090 |
| DSC1 | 0.003 (0.003) * | 0.001 (0.001) | 2.2 | 0.092 |
| LGALS3BP | 0.106 (0.084) * | 0.174 (0.144) | 1.6 | 0.097 |
| IgKV3D-11 | 0.498 (0.395) * | 0.770 (0.547) | 0.6 | 0.098 |
| IgKV2-40 | 0.017 (0.020) * | 0.031 (0.028) | 0.5 | 0.098 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plaschke, K.; Brenner, T.; Fiedler, M.O.; Hölle, T.; von der Forst, M.; Wolf, R.C.; Kopitz, J.; Gebert, J.; Weigand, M.A. Extracellular Vesicles as Possible Plasma Markers and Mediators in Patients with Sepsis-Associated Delirium—A Pilot Study. Int. J. Mol. Sci. 2023, 24, 15781. https://doi.org/10.3390/ijms242115781
Plaschke K, Brenner T, Fiedler MO, Hölle T, von der Forst M, Wolf RC, Kopitz J, Gebert J, Weigand MA. Extracellular Vesicles as Possible Plasma Markers and Mediators in Patients with Sepsis-Associated Delirium—A Pilot Study. International Journal of Molecular Sciences. 2023; 24(21):15781. https://doi.org/10.3390/ijms242115781
Chicago/Turabian StylePlaschke, Konstanze, Thorsten Brenner, Mascha O. Fiedler, Tobias Hölle, Maik von der Forst, Robert Christian Wolf, Jürgen Kopitz, Johannes Gebert, and Markus A. Weigand. 2023. "Extracellular Vesicles as Possible Plasma Markers and Mediators in Patients with Sepsis-Associated Delirium—A Pilot Study" International Journal of Molecular Sciences 24, no. 21: 15781. https://doi.org/10.3390/ijms242115781
APA StylePlaschke, K., Brenner, T., Fiedler, M. O., Hölle, T., von der Forst, M., Wolf, R. C., Kopitz, J., Gebert, J., & Weigand, M. A. (2023). Extracellular Vesicles as Possible Plasma Markers and Mediators in Patients with Sepsis-Associated Delirium—A Pilot Study. International Journal of Molecular Sciences, 24(21), 15781. https://doi.org/10.3390/ijms242115781








