TRA2B Gene Splice Variant Linked to Seizures and Neurodevelopmental Delay: A Second Case Study
Abstract
:1. Introduction
2. Results
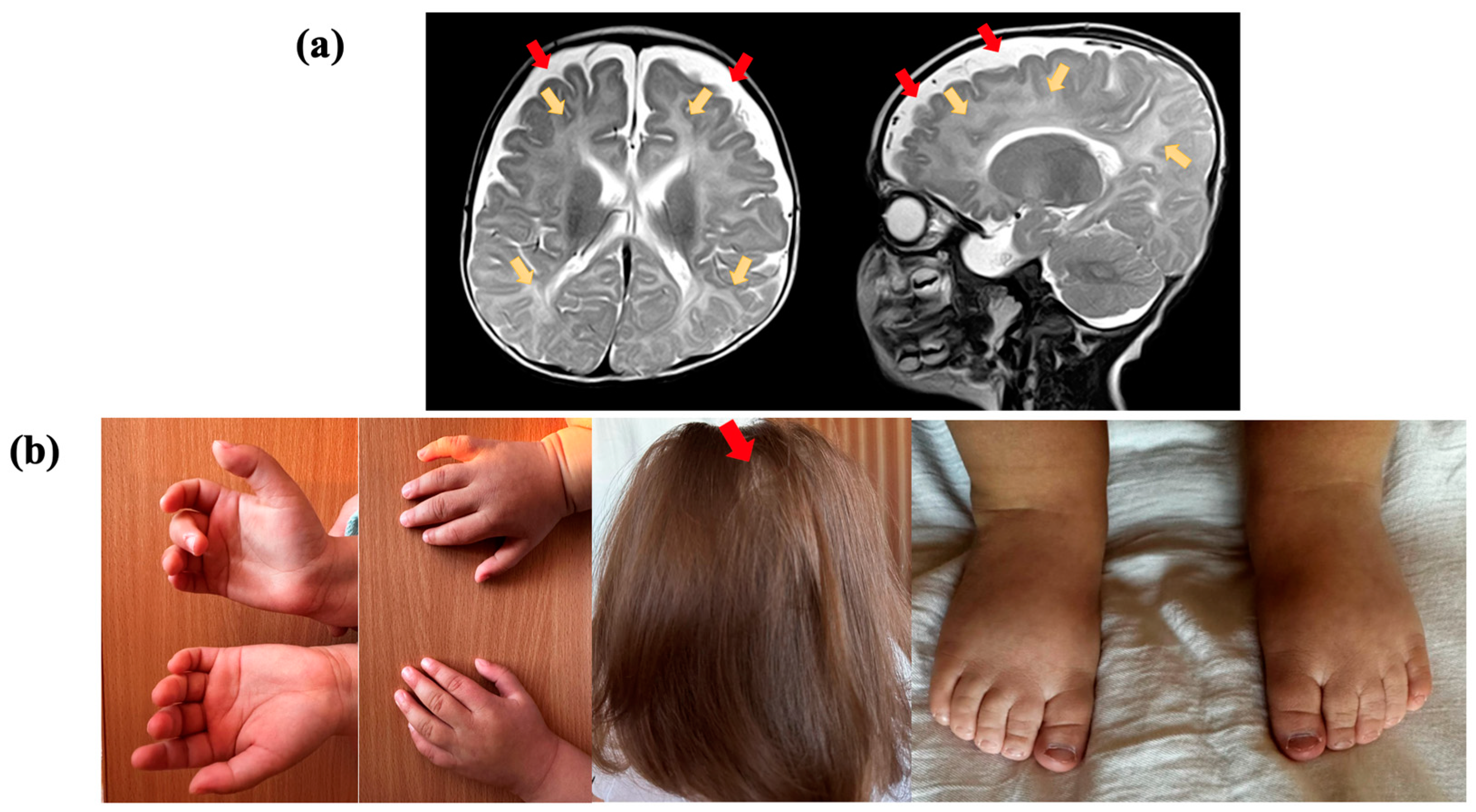
2.1. Clinical Evaluation
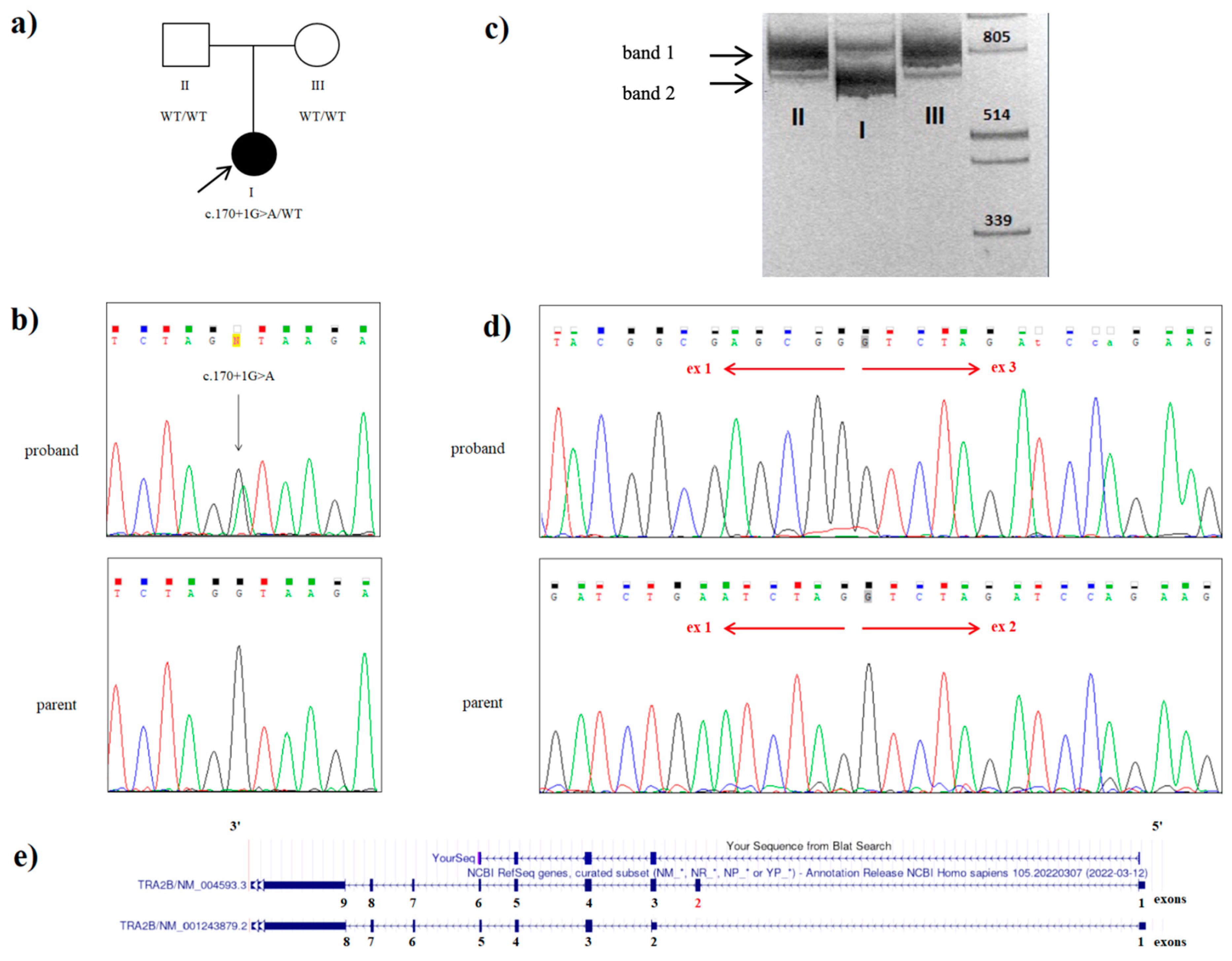
2.2. Whole Genome Sequencing
2.3. mRNA Analysis
3. Discussion
4. Materials and Methods
4.1. Clinical Data
4.2. Genome Sequencing
4.3. RNA Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parenti, I.; Rabaneda, L.G.; Schoen, H.; Novarino, G. Neurodevelopmental Disorders: From Genetics to Functional Pathways. Trends Neurosci. 2020, 43, 608–621. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.K.; Black, D.L.; Zheng, S. The Neurogenetics of Alternative Splicing. Nat. Rev. Neurosci. 2016, 17, 265–281. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.M.; Ennajdaoui, H.; Edmondson, C.; Wirth, B.; Sanford, J.R.; Chen, B. Splicing Factor TRA2B Is Required for Neural Progenitor Survival: TRA2B Is Required for Cortical Development. J. Comp. Neurol. 2014, 522, 372–392. [Google Scholar] [CrossRef] [PubMed]
- Cléry, A.; Jayne, S.; Benderska, N.; Dominguez, C.; Stamm, S.; Allain, F.H.-T. Molecular Basis of Purine-Rich RNA Recognition by the Human SR-like Protein Tra2-Β1. Nat. Struct. Mol. Biol. 2011, 18, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Grellscheid, S.; Dalgliesh, C.; Storbeck, M.; Best, A.; Liu, Y.; Jakubik, M.; Mende, Y.; Ehrmann, I.; Curk, T.; Rossbach, K.; et al. Identification of Evolutionarily Conserved Exons as Regulated Targets for the Splicing Activator Tra2β in Development. PLoS Genet. 2011, 7, e1002390. [Google Scholar] [CrossRef] [PubMed]
- Ramond, F.; Dalgliesh, C.; Grimmel, M.; Wechsberg, O.; Vetro, A.; Guerrini, R.; FitzPatrick, D.; Poole, R.L.; Lebrun, M.; Bayat, A.; et al. Clustered Variants in the 5′ Coding Region of TRA2B Cause a Distinctive Neurodevelopmental Syndrome. Genet. Med. 2023, 25, 100003. [Google Scholar] [CrossRef] [PubMed]
- Nayler, O.; Cap, C.; Stamm, S. Human Transformer-2-Beta Gene (SFRS10): Complete Nucleotide Sequence, Chromosomal Localization, and Generation of a Tissue-Specific Isoform. Genomics 1998, 53, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Pitton Rissardo, J.; Fornari Caprara, A.L.; Casares, M.; Skinner, H.J.; Hamid, U. Antiseizure Medication-Induced Alopecia: A Literature Review. Medicines 2023, 10, 35. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shatokhina, O.; Kovalskaia, V.; Sparber, P.; Sharkova, I.; Mishina, I.; Kuznetsova, V.; Ryzhkova, O. TRA2B Gene Splice Variant Linked to Seizures and Neurodevelopmental Delay: A Second Case Study. Int. J. Mol. Sci. 2023, 24, 15572. https://doi.org/10.3390/ijms242115572
Shatokhina O, Kovalskaia V, Sparber P, Sharkova I, Mishina I, Kuznetsova V, Ryzhkova O. TRA2B Gene Splice Variant Linked to Seizures and Neurodevelopmental Delay: A Second Case Study. International Journal of Molecular Sciences. 2023; 24(21):15572. https://doi.org/10.3390/ijms242115572
Chicago/Turabian StyleShatokhina, Olga, Valeriia Kovalskaia, Peter Sparber, Inna Sharkova, Irina Mishina, Vera Kuznetsova, and Oxana Ryzhkova. 2023. "TRA2B Gene Splice Variant Linked to Seizures and Neurodevelopmental Delay: A Second Case Study" International Journal of Molecular Sciences 24, no. 21: 15572. https://doi.org/10.3390/ijms242115572
APA StyleShatokhina, O., Kovalskaia, V., Sparber, P., Sharkova, I., Mishina, I., Kuznetsova, V., & Ryzhkova, O. (2023). TRA2B Gene Splice Variant Linked to Seizures and Neurodevelopmental Delay: A Second Case Study. International Journal of Molecular Sciences, 24(21), 15572. https://doi.org/10.3390/ijms242115572






