Tryptophan-Starved Human Cells Overexpressing Tryptophanyl-tRNA Synthetase Enhance High-Affinity Tryptophan Uptake via Enzymatic Production of Tryptophanyl-AMP
Abstract
1. Introduction
2. Results
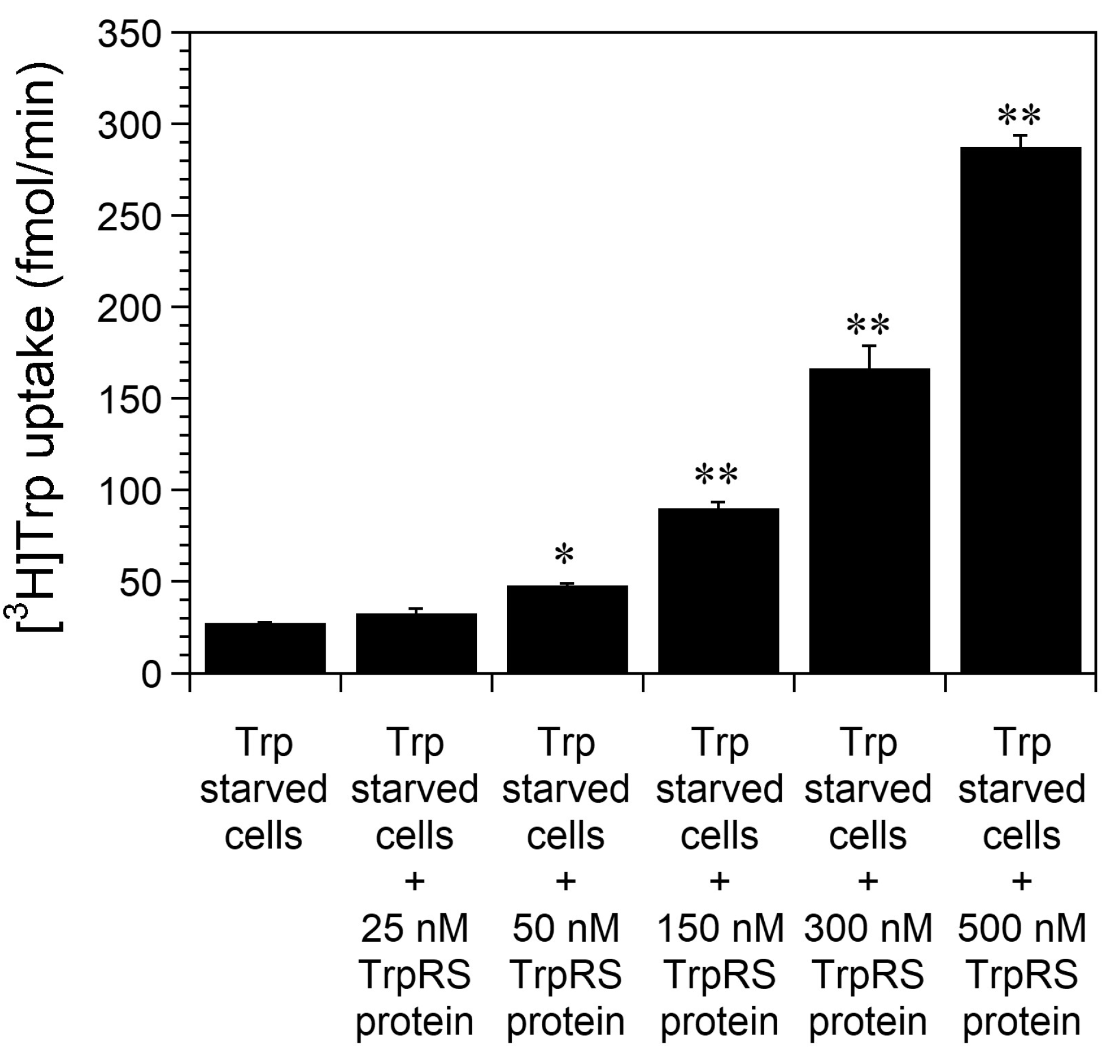
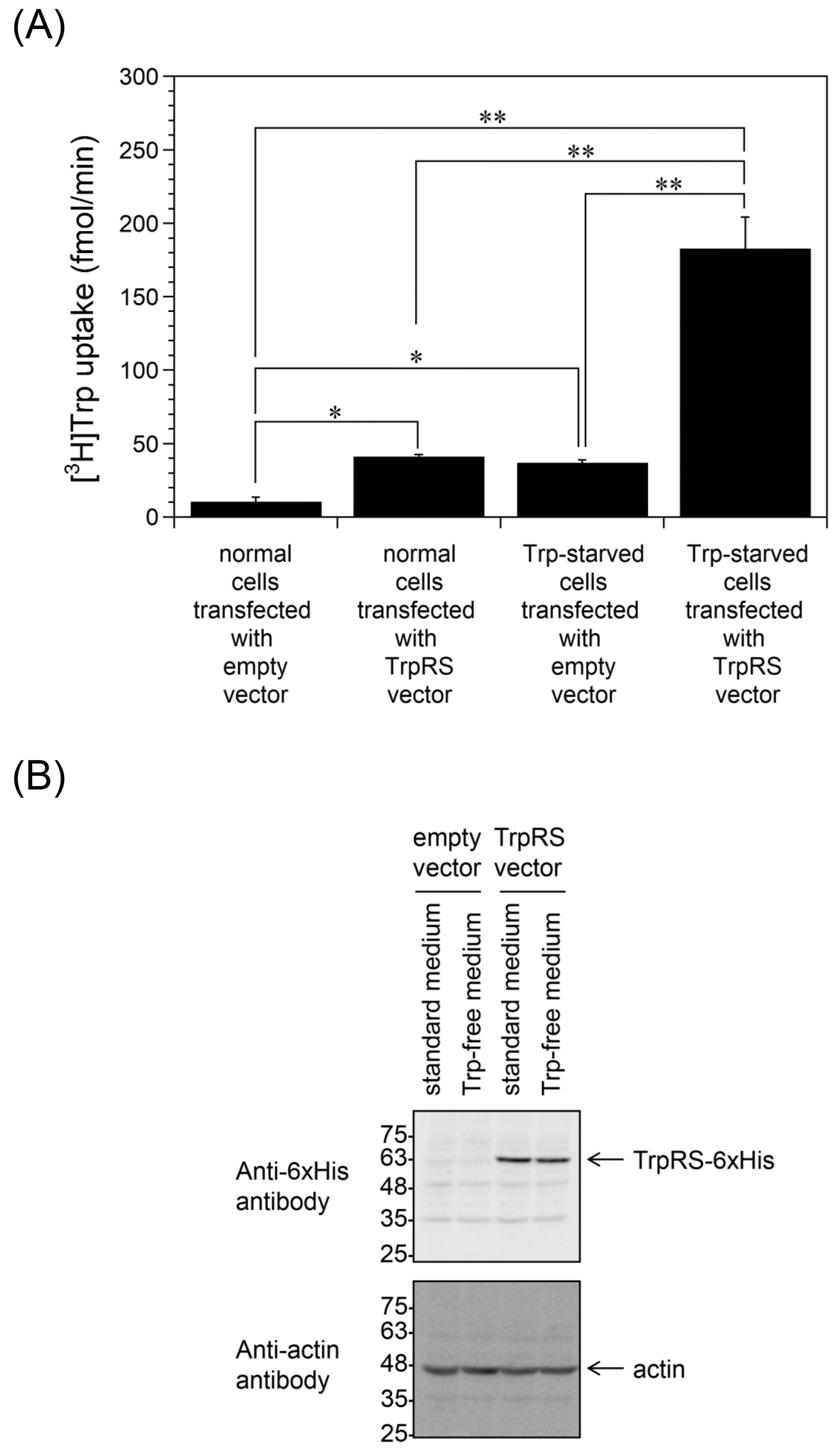
2.1. Trp Depletion Enhances Trp uptake into TrpRS-Overexpressing HeLa Cells
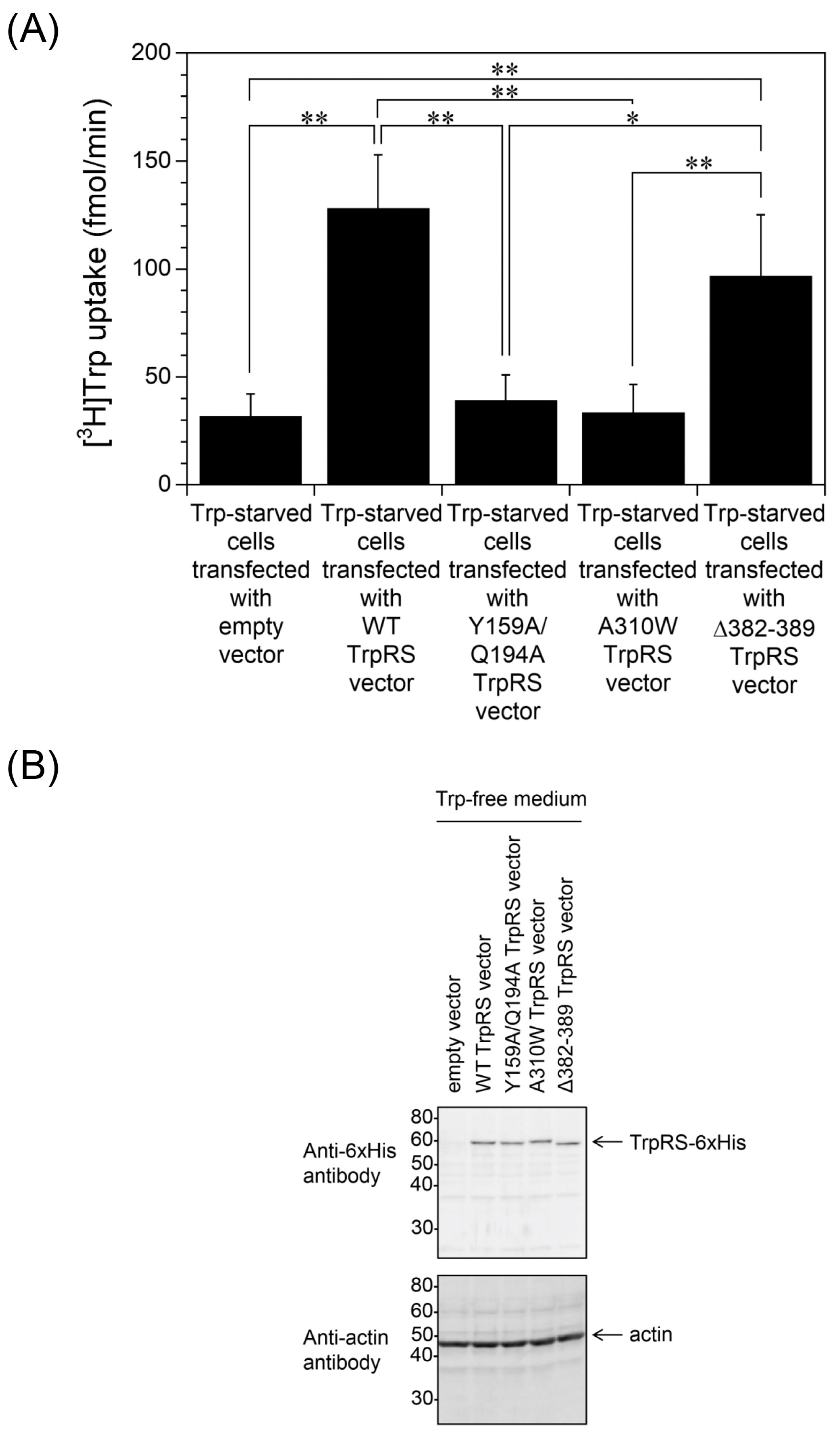
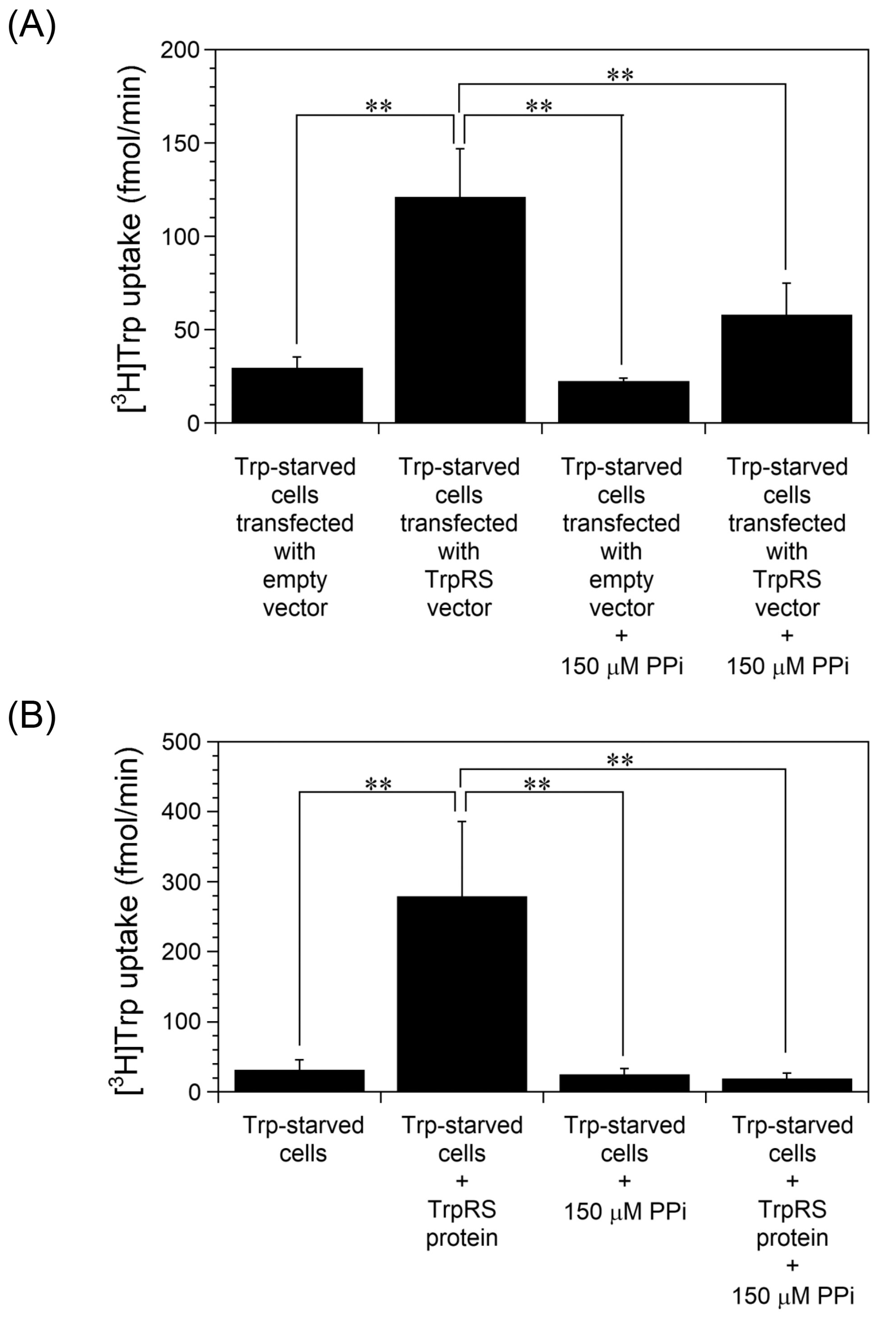
2.2. TrpRS Facilitates High-Affinity Trp Uptake by Production of Tryptophanyl-AMP
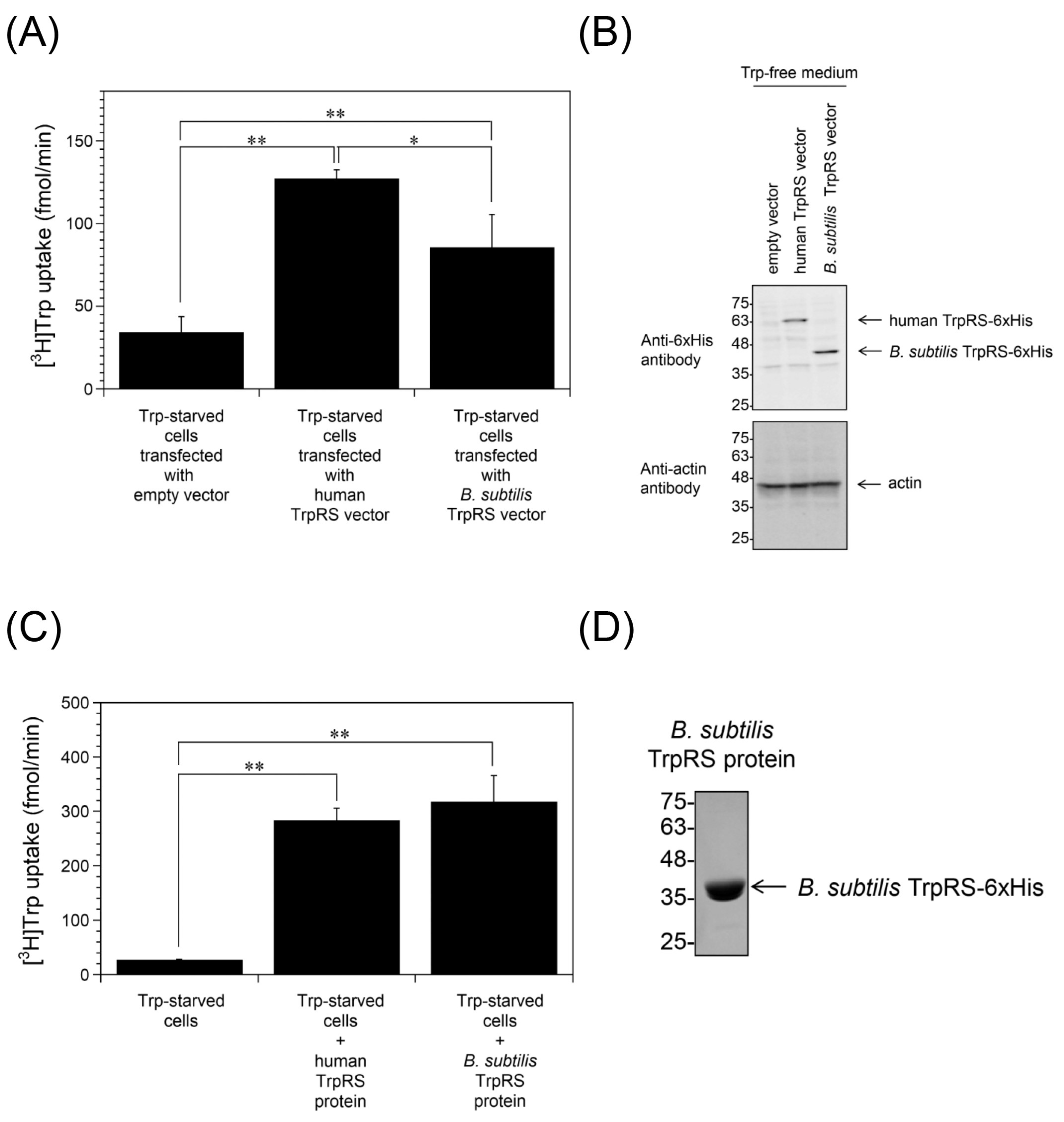
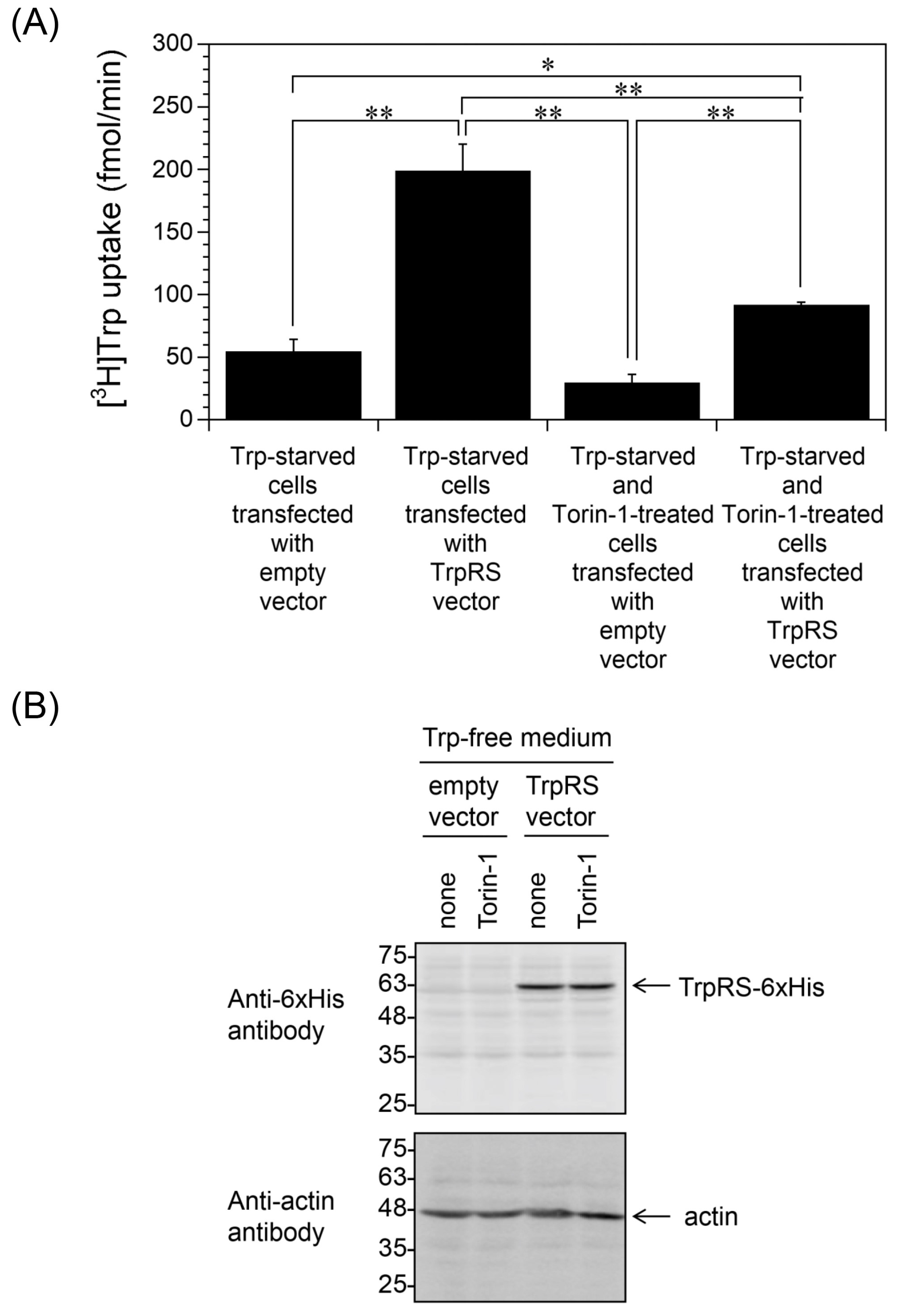
2.3. Inhibition of Trp Uptake into Trp-Depleted TrpRS-Overexpressing HeLa Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. Western Blot Analyses
4.4. Quantification of Trp Uptake
4.5. Plasmids
4.6. Plasmid Transfection Procedure
4.7. Trp Starvation of HeLa Cells
4.8. Preparation of TrpRS Proteins from E. coli
4.9. Statistics
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Munn, D.H.; Zhou, M.; Attwood, J.T.; Bondarev, I.; Conway, S.J.; Marshall, B.; Brown, C.; Mellor, A.L. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science 1998, 281, 1191–1193. [Google Scholar] [CrossRef]
- Mellor, A.L.; Munn, D.H. Tryptophan catabolism and T-cell tolerance: Immunosuppression by starvation? Immunol. Today 1999, 20, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Munn, D.H.; Shafizadeh, E.; Attwood, J.T.; Bondarev, I.; Pashine, A.; Mellor, A.L. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J. Exp. Med. 1999, 189, 1363–1372. [Google Scholar] [CrossRef]
- Hwu, P.; Du, M.X.; Lapointe, R.; Do, M.; Taylor, M.W.; Young, H.A. Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. J. Immunol. 2000, 164, 3596–3599. [Google Scholar] [CrossRef] [PubMed]
- Friberg, M.; Jennings, R.; Alsarraj, M.; Dessureault, S.; Cantor, A.; Extermann, M.; Mellor, A.L.; Munn, D.H.; Antonia, S.J. Indoleamine 2,3-dioxygenase contributes to tumor cell evasion of T cell-mediated rejection. Int. J. Cancer 2002, 101, 151–155. [Google Scholar] [CrossRef]
- Mellor, A.L.; Keskin, D.B.; Johnson, T.; Chandler, P.; Munn, D.H. Cells expressing indoleamine 2,3-dioxygenase inhibit T cell responses. J. Immunol. 2002, 168, 3771–3776. [Google Scholar] [CrossRef]
- Munn, D.H.; Sharma, M.D.; Lee, J.R.; Jhaver, K.G.; Johnson, T.S.; Keskin, D.B.; Marshall, B.; Chandler, P.; Antonia, S.J.; Burgess, R.; et al. Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science 2002, 297, 1867–1870. [Google Scholar] [CrossRef] [PubMed]
- Terness, P.; Bauer, T.M.; Röse, L.; Dufter, C.; Watzlik, A.; Simon, H.; Opelz, G. Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: Mediation of suppression by tryptophan metabolites. J. Exp. Med. 2002, 196, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Uyttenhove, C.; Pilotte, L.; Théate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; Van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef]
- Meisel, R.; Zibert, A.; Laryea, M.; Göbel, U.; Däubener, W.; Dilloo, D. Human bone marrow stromal cells inhibit allogeneic T-cell responses by indoleamine 2,3-dioxygenase-mediated tryptophan degradation. Blood 2004, 103, 4619–4621. [Google Scholar] [CrossRef]
- Mellor, A.L.; Munn, D.H. IDO expression by dendritic cells: Tolerance and tryptophan catabolism. Nat. Rev. Immunol. 2004, 4, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Munn, D.H.; Mellor, A.L. IDO and tolerance to tumors. Trends Mol. Med. 2004, 10, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Pilotte, L.; Larrieu, P.; Stroobant, V.; Colau, D.; Dolusic, E.; Frédérick, R.; De Plaen, E.; Uyttenhove, C.; Wouters, J.; Masereel, B.; et al. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc. Natl. Acad. Sci. USA 2012, 109, 2497–2502. [Google Scholar] [CrossRef] [PubMed]
- van Baren, N.; Van den Eynde, B.J. Tryptophan-degrading enzymes in tumoral immune resistance. Front. Immunol. 2015, 6, 34. [Google Scholar] [CrossRef]
- Munn, D.H.; Mellor, A.L. IDO in the tumor microenvironment: Inflammation, counter-regulation, and tolerance. Trends Immunol. 2016, 37, 193–207. [Google Scholar] [CrossRef]
- Wei, H.; Liu, S.; Lian, R.; Huang, C.; Li, Y.; Chen, L.; Zeng, Y. Abnormal expression of indoleamine 2, 3-dioxygenase in human recurrent miscarriage. Reprod. Sci. 2020, 27, 1656–1664. [Google Scholar] [CrossRef]
- Zhao, Y.; Tao, F.; Jiang, J.; Chen, L.; Du, J.; Cheng, X.; He, Q.; Zhong, S.; Chen, W.; Wu, X.; et al. Tryptophan 2, 3 dioxygenase promotes proliferation, migration and invasion of ovarian cancer cells. Mol. Med. Rep. 2021, 23, 445. [Google Scholar] [CrossRef]
- Bhutia, Y.D.; Babu, E.; Ganapathy, V. Interferon-γ induces a tryptophan-selective amino acid transporter in human colonic epithelial cells and mouse dendritic cells. Biochim. Biophys. Acta 2015, 1848, 453–462. [Google Scholar] [CrossRef]
- Miyanokoshi, M.; Yokosawa, T.; Wakasugi, K. Tryptophanyl-tRNA synthetase mediates high-affinity tryptophan uptake into human cells. J. Biol. Chem. 2018, 293, 8428–8438. [Google Scholar] [CrossRef] [PubMed]
- Seymour, R.L.; Ganapathy, V.; Mellor, A.L.; Munn, D.H. A high-affinity, tryptophan-selective amino acid transport system in human macrophages. J. Leukoc. Biol. 2006, 80, 1320–1327. [Google Scholar] [CrossRef]
- Silk, J.D.; Lakhal, S.; Laynes, R.; Vallius, L.; Karydis, I.; Marcea, C.; Boyd, C.A.; Cerundolo, V. IDO induces expression of a novel tryptophan transporter in mouse and human tumor cells. J. Immunol. 2011, 187, 1617–1625. [Google Scholar] [CrossRef] [PubMed]
- Fleckner, J.; Rasmussen, H.H.; Justesen, J. Human interferon γ potently induces the synthesis of a 55-kDa protein (γ2) highly homologous to rabbit peptide chain release factor and bovine tryptophanyl-tRNA synthetase. Proc. Natl. Acad. Sci. USA 1991, 88, 11520–11524. [Google Scholar] [CrossRef]
- Rubin, B.Y.; Anderson, S.L.; Xing, L.; Powell, R.J.; Tate, W.P. Interferon induces tryptophanyl-tRNA synthetase expression in human fibroblasts. J. Biol. Chem. 1991, 266, 24245–24248. [Google Scholar] [CrossRef]
- Kisselev, L.; Frolova, L.; Haenni, A.L. Interferon inducibility of mammalian tryptophanyl-tRNA synthetase: New perspectives. Trends Biochem. Sci. 1993, 18, 263–267. [Google Scholar] [CrossRef]
- Reano, A.; Richard, M.H.; Denoroy, L.; Viac, J.; Benedetto, J.P.; Schmitt, D. γ interferon potently induces tryptophanyl-tRNA synthetase expression in human keratinocytes. J. Investig. Dermatol. 1993, 100, 775–779. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fleckner, J.; Martensen, P.M.; Tolstrup, A.B.; Kjeldgaard, N.O.; Justesen, J. Differential regulation of the human, interferon inducible tryptophanyl-tRNA synthetase by various cytokines in cell lines. Cytokine 1995, 7, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.C.; Larsen, M.R.; Roepstorff, P.; Justesen, J.; Christiansen, G.; Birkelund, S. Mapping and identification of interferon γ-regulated HeLa cell proteins separated by immobilized pH gradient two-dimensional gel electrophoresis. Electrophoresis 1999, 20, 984–993. [Google Scholar] [CrossRef]
- Schimmel, P.R.; Söll, D. Aminoacyl-tRNA synthetases: General features and recognition of transfer RNAs. Annu. Rev. Biochem. 1979, 48, 601–648. [Google Scholar] [CrossRef] [PubMed]
- Schimmel, P. Aminoacyl-tRNA synthetases: General scheme of structure-functional relationships in the polypeptides and recognition of transfer RNAs. Annu. Rev. Biochem. 1987, 56, 125–158. [Google Scholar] [CrossRef]
- Tolstrup, A.B.; Bejder, A.; Fleckner, J.; Justesen, J. Transcriptional regulation of the interferon-γ-inducible tryptophanyl-tRNA synthetase includes alternative splicing. J. Biol. Chem. 1995, 270, 397–403. [Google Scholar] [CrossRef]
- Turpaev, K.T.; Zakhariev, V.M.; Sokolova, I.V.; Narovlyansky, A.N.; Amchenkova, A.M.; Justesen, J.; Frolova, L.Y. Alternative processing of the tryptophanyl-tRNA synthetase mRNA from interferon treated human cells. Eur. J. Biochem. 1996, 240, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, K.; Slike, B.M.; Hood, J.; Otani, A.; Ewalt, K.L.; Friedlander, M.; Cheresh, D.A.; Schimmel, P. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.H.; Park, S.; Choi, J.J.; Park, B.K.; Rhee, K.H.; Kang, E.; Ahn, S.; Lee, C.H.; Lee, J.S.; Inn, K.S.; et al. Secreted tryptophanyl-tRNA synthetase as a primary defence system against infection. Nat. Microbiol. 2016, 2, 16191. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, K.; Yokosawa, T. Non-canonical functions of human cytoplasmic tyrosyl-, tryptophanyl- and other aminoacyl-tRNA synthetases. Enzymes 2020, 48, 207–242. [Google Scholar]
- Kapoor, M.; Zhou, Q.; Otero, F.; Myers, C.A.; Bates, A.; Belani, R.; Liu, J.; Luo, J.K.; Tzima, E.; Zhang, D.E.; et al. Evidence for annexin II-S100A10 complex and plasmin in mobilization of cytokine activity of human TrpRS. J. Biol. Chem. 2008, 283, 2070–2077. [Google Scholar] [CrossRef]
- Gioelli, N.; Neilson, L.J.; Wei, N.; Villari, G.; Chen, W.; Kuhle, B.; Ehling, M.; Maione, F.; Willox, S.; Brundu, S.; et al. Neuropilin 1 and its inhibitory ligand mini-tryptophanyl-tRNA synthetase inversely regulate VE-cadherin turnover and vascular permeability. Nat. Commun. 2022, 13, 4188. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Choi, Y.H.; Lee, W.-K.; Ji, Y.; Chun, E.; Kim, Y.H.; Lee, J.-E.; Jung, H.S.; Suh, J.H.; Kim, S.; et al. Tryptophan-dependent and -independent secretions of tryptophanyl- tRNA synthetase mediate innate inflammatory responses. Cell Rep. 2023, 42, 111905. [Google Scholar] [CrossRef] [PubMed]
- Yokosawa, T.; Sato, A.; Wakasugi, K. Tryptophan depletion modulates tryptophanyl-tRNA synthetase-mediated high-affinity tryptophan uptake into human cells. Genes 2020, 11, 1423. [Google Scholar] [CrossRef]
- Zhou, Q.; Kapoor, M.; Guo, M.; Belani, R.; Xu, X.; Kiosses, W.B.; Hanan, M.; Park, C.; Armour, E.; Do, M.H.; et al. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat. Struct. Mol. Biol. 2010, 17, 57–61. [Google Scholar] [CrossRef]
- Yang, X.L.; Otero, F.J.; Ewalt, K.L.; Liu, J.; Swairjo, M.A.; Köhrer, C.; RajBhandary, U.L.; Skene, R.J.; McRee, D.E.; Schimmel, P. Two conformations of a crystalline human tRNA synthetase-tRNA complex: Implications for protein synthesis. EMBO J. 2006, 25, 2919–2929. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Chen, X.; Xin, L.; Chen, L.; Jin, Y.; Wang, D. Species-specific differences in the operational RNA code for aminoacylation of tRNATrp. Nucleic Acids Res. 2001, 29, 4125–4133. [Google Scholar] [CrossRef] [PubMed]
- Dignam, J.D.; Deutscher, M.P. Aminoacyl-tRNA synthetase stimulatory factors and inorganic pyrophosphatase. Biochemistry 1979, 18, 3165–3170. [Google Scholar] [CrossRef] [PubMed]
- Bartok, O.; Pataskar, A.; Nagel, R.; Laos, M.; Goldfarb, E.; Hayoun, D.; Levy, R.; Körner, P.R.; Kreuger, I.Z.M.; Champagne, J.; et al. Anti-tumour immunity induces aberrant peptide presentation in melanoma. Nature 2021, 590, 332–337. [Google Scholar] [CrossRef]
- Champagne, J.; Pataskar, A.; Blommaert, N.; Nagel, R.; Wernaart, D.; Ramalho, S.; Kenski, J.; Bleijerveld, O.B.; Zaal, E.A.; Berkers, C.R.; et al. Oncogene-dependent sloppiness in mRNA translation. Mol. Cell 2021, 81, 4709–4721. [Google Scholar] [CrossRef] [PubMed]
- Pataskar, A.; Champagne, J.; Nagel, R.; Kenski, J.; Laos, M.; Michaux, J.; Pak, H.S.; Bleijerveld, O.B.; Mordente, K.; Navarro, J.M.; et al. Tryptophan depletion results in tryptophan-to-phenylalanine substitutants. Nature 2022, 603, 721–727. [Google Scholar] [CrossRef] [PubMed]
- He, X.D.; Gong, W.; Zhang, J.N.; Nie, J.; Yao, C.F.; Guo, F.S.; Lin, Y.; Wu, X.H.; Li, F.; Li, J.; et al. Sensing and transmitting intracellular amino acid signals through reversible lysine aminoacylations. Cell Metab. 2018, 27, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Qin, R.; Zhao, C.; Wang, C.J.; Xu, W.; Zhao, J.Y.; Lin, Y.; Yuan, Y.Y.; Lin, P.C.; Li, Y.; Zhao, S.; et al. Tryptophan potentiates CD8+ T cells against cancer cells by TRIP12 tryptophanylation and surface PD-1 downregulation. J. Immunother. Cancer 2021, 9, e002840. [Google Scholar] [CrossRef]
- Holmgaard, R.B.; Zamarin, D.; Munn, D.H.; Wolchok, J.D.; Allison, J.P. Indoleamine 2,3-dioxygenase is a critical resistance mechanism in antitumor T cell immunotherapy targeting CTLA-4. J. Exp. Med. 2013, 210, 1389–1402. [Google Scholar] [CrossRef] [PubMed]
- Ladomersky, E.; Zhai, L.; Lenzen, A.; Lauing, K.L.; Qian, J.; Scholtens, D.M.; Gritsina, G.; Sun, X.; Liu, Y.; Yu, F.; et al. IDO1 Inhibition Synergizes with Radiation and PD-1 Blockade to Durably Increase Survival Against Advanced Glioblastoma. Clin. Cancer Res. 2018, 24, 2559–2573. [Google Scholar] [CrossRef]
- Wakasugi, K.; Nakano, T.; Morishima, I. Oxidative stress-responsive intracellular regulation specific for the angiostatic form of human tryptophanyl-tRNA synthetase. Biochemistry 2005, 44, 225–232. [Google Scholar] [CrossRef]
- Wakasugi, K. Human tryptophanyl-tRNA synthetase binds with heme to enhance its aminoacylation activity. Biochemistry 2007, 46, 11291–11298. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, K. An exposed cysteine residue of human angiostatic mini tryptophanyl-tRNA synthetase. Biochemistry 2010, 49, 3156–3160. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, K. Species-specific differences in the regulation of the aminoacylation activity of mammalian tryptophanyl-tRNA synthetases. FEBS Lett. 2010, 584, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, T.; Miyanokoshi, M.; Tanaka, T.; Wakasugi, K. Identification of a residue crucial for the angiostatic activity of human mini tryptophanyl-tRNA synthetase by focusing on its molecular evolution. Sci. Rep. 2016, 6, 24750. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yokosawa, T.; Wakasugi, K. Tryptophan-Starved Human Cells Overexpressing Tryptophanyl-tRNA Synthetase Enhance High-Affinity Tryptophan Uptake via Enzymatic Production of Tryptophanyl-AMP. Int. J. Mol. Sci. 2023, 24, 15453. https://doi.org/10.3390/ijms242015453
Yokosawa T, Wakasugi K. Tryptophan-Starved Human Cells Overexpressing Tryptophanyl-tRNA Synthetase Enhance High-Affinity Tryptophan Uptake via Enzymatic Production of Tryptophanyl-AMP. International Journal of Molecular Sciences. 2023; 24(20):15453. https://doi.org/10.3390/ijms242015453
Chicago/Turabian StyleYokosawa, Takumi, and Keisuke Wakasugi. 2023. "Tryptophan-Starved Human Cells Overexpressing Tryptophanyl-tRNA Synthetase Enhance High-Affinity Tryptophan Uptake via Enzymatic Production of Tryptophanyl-AMP" International Journal of Molecular Sciences 24, no. 20: 15453. https://doi.org/10.3390/ijms242015453
APA StyleYokosawa, T., & Wakasugi, K. (2023). Tryptophan-Starved Human Cells Overexpressing Tryptophanyl-tRNA Synthetase Enhance High-Affinity Tryptophan Uptake via Enzymatic Production of Tryptophanyl-AMP. International Journal of Molecular Sciences, 24(20), 15453. https://doi.org/10.3390/ijms242015453





