Circulating microRNA Panels for Detection of Liver Cancers and Liver-Metastasizing Primary Cancers
Abstract
1. Introduction
2. MicroRNA in Cancer
3. Circulating Tumor miRNA
4. Circular miRNA in Primary Liver Cancers
4.1. Hepatocellular Carcinoma
4.1.1. miRNA-Only Panels
4.1.2. miRNA Panels Combined with lncRNA and mRNA
4.1.3. miRNA Panels Combined with AFP
4.2. Cholangiocarcinoma
5. Circular miRNA in Liver-Metastasizing Primary Cancers
5.1. Colorectal Cancer
5.1.1. miRNA-Only Panels
5.1.2. miRNA Panels Combined with lncRNA
5.2. Pancreatic Cancer
5.2.1. miRNA-Only Panels
5.2.2. miRNA Panels Combined with CA19-9
5.2.3. miRNA Panels Combined with Proteins
5.3. Gastric Cancer
5.3.1. miRNA-Only Panels
5.3.2. miRNA Panels Combined with lncRNA
5.4. Lung Cancer
5.4.1. miRNA-Only Panels
5.4.2. miRNA Panels Combined with lncRNA
5.5. Breast Cancer
5.5.1. miRNA-Only Panels
5.5.2. miRNA Panels Combined with lncRNA
6. miRNA Specificity in Cancers
7. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- EASL. Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.; Gogineni, V.; Saeian, K. Epidemiology of primary and secondary liver cancers. Semin. Interv. Radiol. 2006, 23, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Draškovič, T.; Zidar, N.; Hauptman, N. Circulating Tumor DNA Methylation Biomarkers for Characterization and Determination of the Cancer Origin in Malignant Liver Tumors. Cancers 2023, 15, 839. [Google Scholar] [CrossRef] [PubMed]
- Tsilimigras, D.I.; Brodt, P.; Clavien, P.-A.; Muschel, R.J.; D’Angelica, M.I.; Endo, I.; Parks, R.W.; Doyle, M.; de Santibañes, E.; Pawlik, T.M. Liver metastases. Nat. Rev. Dis. Primers 2021, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Centeno, B.A. Pathology of liver metastases. Cancer Control 2006, 13, 13–26. [Google Scholar] [CrossRef]
- de Ridder, J.; de Wilt, J.H.W.; Simmer, F.; Overbeek, L.; Lemmens, V.; Nagtegaal, I. Incidence and origin of histologically confirmed liver metastases: An explorative case-study of 23,154 patients. Oncotarget 2016, 7, 55368–55376. [Google Scholar] [CrossRef]
- van de Wouw, A.J.; Janssen-Heijnen, M.L.; Coebergh, J.W.; Hillen, H.F. Epidemiology of unknown primary tumours; incidence and population-based survival of 1285 patients in Southeast Netherlands, 1984–1992. Eur. J. Cancer 2002, 38, 409–413. [Google Scholar] [CrossRef]
- Ayoub, J.P.; Hess, K.R.; Abbruzzese, M.C.; Lenzi, R.; Raber, M.N.; Abbruzzese, J.L. Unknown primary tumors metastatic to liver. J. Clin. Oncol. 1998, 16, 2105–2112. [Google Scholar] [CrossRef]
- Pavlidis, N.; Pentheroudakis, G. Cancer of unknown primary site. Lancet 2012, 379, 1428–1435. [Google Scholar] [CrossRef]
- Varadhachary, G.R.; Talantov, D.; Raber, M.N.; Meng, C.; Hess, K.R.; Jatkoe, T.; Lenzi, R.; Spigel, D.R.; Wang, Y.; Greco, F.A.; et al. Molecular profiling of carcinoma of unknown primary and correlation with clinical evaluation. J. Clin. Oncol. 2008, 26, 4442–4448. [Google Scholar] [CrossRef]
- Pillai, R.S. MicroRNA function: Multiple mechanisms for a tiny RNA? RNA 2005, 11, 1753–1761. [Google Scholar] [CrossRef] [PubMed]
- Pino, M.S.; Chung, D.C. The chromosomal instability pathway in colon cancer. Gastroenterology 2010, 138, 2059–2072. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Kaikkonen, M.U.; Lam, M.T.; Glass, C.K. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 2011, 90, 430–440. [Google Scholar] [CrossRef]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Zen, K.; Zhang, C.-Y. Circulating MicroRNAs: A novel class of biomarkers to diagnose and monitor human cancers. Med. Res. Rev. 2012, 32, 326–348. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yu, L.; Gao, X.; Hu, J.; Wang, J.; Dai, Z.; Wang, J.F.; Zhang, Z.; Lu, S.; Huang, X.; et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J. Clin. Oncol. 2011, 29, 4781–4788. [Google Scholar] [CrossRef]
- Tan, Y.; Ge, G.; Pan, T.; Wen, D.; Chen, L.; Yu, X.; Zhou, X.; Gan, J. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS ONE 2014, 9, e107986. [Google Scholar] [CrossRef]
- Elemeery, M.N.; Badr, A.N.; Mohamed, M.A.; Ghareeb, D.A. Validation of a serum microRNA panel as biomarkers for early diagnosis of hepatocellular carcinoma post-hepatitis C infection in Egyptian patients. World J. Gastroenterol. 2017, 23, 3864–3875. [Google Scholar] [CrossRef]
- Zhu, H.T.; Liu, R.B.; Liang, Y.Y.; Hasan, A.M.E.; Wang, H.Y.; Shao, Q.; Zhang, Z.C.; Wang, J.; He, C.Y.; Wang, F.; et al. Serum microRNA profiles as diagnostic biomarkers for HBV-positive hepatocellular carcinoma. Liver Int. 2017, 37, 888–896. [Google Scholar] [CrossRef]
- An, Y.; Gao, S.; Zhao, W.C.; Qiu, B.A.; Xia, N.X.; Zhang, P.J.; Fan, Z.P. Novel serum microRNAs panel on the diagnostic and prognostic implications of hepatocellular carcinoma. World J. Gastroenterol. 2018, 24, 2596–2604. [Google Scholar] [CrossRef]
- Cho, H.J.; Baek, G.O.; Seo, C.W.; Ahn, H.R.; Sung, S.; Son, J.A.; Kim, S.S.; Cho, S.W.; Jang, J.W.; Nam, S.W.; et al. Exosomal microRNA-4661-5p-based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 2020, 9, 5459–5472. [Google Scholar] [CrossRef]
- Ali, H.E.A.; Abdel Hameed, R.; Effat, H.; Ahmed, E.K.; Atef, A.A.; Sharawi, S.K.; Ali, M.; Abd Elmageed, Z.Y.; Abdel Wahab, A.H. Circulating microRNAs panel as a diagnostic tool for discrimination of HCV-associated hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2017, 41, e51–e62. [Google Scholar] [CrossRef]
- Jiang, L.; Cheng, Q.; Zhang, B.H.; Zhang, M.Z. Circulating microRNAs as biomarkers in hepatocellular carcinoma screening: A validation set from China. Medicine 2015, 94, e603. [Google Scholar] [CrossRef]
- El-Tawdi, A.H.; Matboli, M.; Shehata, H.H.; Tash, F.; El-Khazragy, N.; Azazy Ael, S.; Abdel-Rahman, O. Evaluation of Circulatory RNA-Based Biomarker Panel in Hepatocellular Carcinoma. Mol. Diagn. Ther. 2016, 20, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Abd El Gwad, A.; Matboli, M.; El-Tawdi, A.; Habib, E.K.; Shehata, H.; Ibrahim, D.; Tash, F. Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J. Cell. Biochem. 2018, 119, 8600–8610. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.S.; Boshra, M.S.; El Meteini, M.S.; Shafei, A.E.; Matboli, M. lncRNA- RP11-156p1.3, novel diagnostic and therapeutic targeting via CRISPR/Cas9 editing in hepatocellular carcinoma. Genomics 2020, 112, 3306–3314. [Google Scholar] [CrossRef]
- Zekri, A.N.; Youssef, A.S.; El-Desouky, E.D.; Ahmed, O.S.; Lotfy, M.M.; Nassar, A.A.; Bahnassey, A.A. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumour Biol. 2016, 37, 12273–12286. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, T.; Qiu, Y.; Zhang, T.; Guo, P.; Ma, X.; Wei, Q.; Han, L. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine 2017, 96, e5642. [Google Scholar] [CrossRef] [PubMed]
- Zuo, D.; Chen, L.; Liu, X.; Wang, X.; Xi, Q.; Luo, Y.; Zhang, N.; Guo, H. Combination of miR-125b and miR-27a enhances sensitivity and specificity of AFP-based diagnosis of hepatocellular carcinoma. Tumour Biol. 2016, 37, 6539–6549. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wan, R.; Ren, L.; Yang, Y.; Ding, Y.; Wang, W. Circulating MicroRNA Panel as a Diagnostic Marker for Hepatocellular Carcinoma. Turk. J. Gastroenterol. 2022, 33, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Trevisani, F.; D’Intino, P.E.; Morselli-Labate, A.M.; Mazzella, G.; Accogli, E.; Caraceni, P.; Domenicali, M.; De Notariis, S.; Roda, E.; Bernardi, M. Serum alpha-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: Influence of HBsAg and anti-HCV status. J. Hepatol. 2001, 34, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Han, L.L.; Lv, Y.; Guo, H.; Ruan, Z.P.; Nan, K.J. Implications of biomarkers in human hepatocellular carcinoma pathogenesis and therapy. World J. Gastroenterol. 2014, 20, 10249–10261. [Google Scholar] [CrossRef]
- Wada, Y.; Shimada, M.; Morine, Y.; Ikemoto, T.; Saito, Y.; Baba, H.; Mori, M.; Goel, A. A blood-based noninvasive miRNA signature for predicting survival outcomes in patients with intrahepatic cholangiocarcinoma. Br. J. Cancer 2022, 126, 1196–1204. [Google Scholar] [CrossRef]
- Horn, S.R.; Stoltzfus, K.C.; Lehrer, E.J.; Dawson, L.A.; Tchelebi, L.; Gusani, N.J.; Sharma, N.K.; Chen, H.; Trifiletti, D.M.; Zaorsky, N.G. Epidemiology of liver metastases. Cancer Epidemiol. 2020, 67, 101760. [Google Scholar] [CrossRef]
- Clark, A.M.; Ma, B.; Taylor, D.L.; Griffith, L.; Wells, A. Liver metastases: Microenvironments and ex-vivo models. Exp. Biol. Med. 2016, 241, 1639–1652. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Vychytilova-Faltejskova, P.; Radova, L.; Sachlova, M.; Kosarova, Z.; Slaba, K.; Fabian, P.; Grolich, T.; Prochazka, V.; Kala, Z.; Svoboda, M.; et al. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis 2016, 37, 941–950. [Google Scholar] [CrossRef]
- Zhu, M.; Huang, Z.; Zhu, D.; Zhou, X.; Shan, X.; Qi, L.W.; Wu, L.; Cheng, W.; Zhu, J.; Zhang, L.; et al. A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget 2017, 8, 17081–17091. [Google Scholar] [CrossRef]
- Li, J.; Liu, Y.; Wang, C.; Deng, T.; Liang, H.; Wang, Y.; Huang, D.; Fan, Q.; Wang, X.; Ning, T.; et al. Serum miRNA expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinoma. Sci. Rep. 2015, 5, 12921. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Pan, B.; Sun, L.; Chen, X.; Zeng, K.; Hu, X.; Xu, T.; Xu, M.; Wang, S. Circulating Exosomal miR-27a and miR-130a Act as Novel Diagnostic and Prognostic Biomarkers of Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2018, 27, 746–754. [Google Scholar] [CrossRef]
- Nakamura, K.; Hernández, G.; Sharma, G.G.; Wada, Y.; Banwait, J.K.; González, N.; Perea, J.; Balaguer, F.; Takamaru, H.; Saito, Y.; et al. A Liquid Biopsy Signature for the Detection of Patients with Early-Onset Colorectal Cancer. Gastroenterology 2022, 163, 1242–1251.e2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, M.; Shan, X.; Zhou, X.; Wang, T.; Zhang, J.; Tao, J.; Cheng, W.; Chen, G.; Li, J.; et al. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene 2019, 687, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, J.; Zhang, C.; Liu, K.; Zhao, L.; Chen, X.; Huang, G.; Lai, Y. A three-miRNA panel in serum as a noninvasive biomarker for colorectal cancer detection. Int. J. Biol. Markers 2020, 35, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Wei, B.; Chen, Z.; Wang, J.; Zhao, L.; Peng, X.; Liu, K.; Lai, Y.; Ni, L. Identification of a four-microRNA panel in serum as promising biomarker for colorectal carcinoma detection. Biomark. Med. 2020, 14, 749–760. [Google Scholar] [CrossRef]
- Luo, X.; Stock, C.; Burwinkel, B.; Brenner, H. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS ONE 2013, 8, e62880. [Google Scholar] [CrossRef]
- Wang, S.; Xiang, J.; Li, Z.; Lu, S.; Hu, J.; Gao, X.; Yu, L.; Wang, L.; Wang, J.; Wu, Y.; et al. A plasma microRNA panel for early detection of colorectal cancer. Int. J. Cancer 2015, 136, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Lin, J.J.; Yang, X.; Gou, D.M.; Fu, L.; Li, F.R.; Yu, X.F. A panel of three plasma microRNAs for colorectal cancer diagnosis. Cancer Epidemiol. 2019, 60, 67–76. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, Z.; Ni, S.; Xiao, X.; Xu, Q.; Wang, L.; Huang, D.; Tan, C.; Sheng, W.; Du, X. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS ONE 2012, 7, e44398. [Google Scholar] [CrossRef]
- Li, S.Q.; Xie, L.Y.; Cai, Z.M.; Wei, H.T.; Xie, M.Z.; Hu, B.L.; Ning, S.F. Systematic analyzing a five- miRNA panel and its diagnostic value of plasma expression in colorectal cancer. Mol. Biol. Rep. 2023, 50, 7253–7261. [Google Scholar] [CrossRef]
- Li, J.; Feng, Y.; Heng, D.; Chen, R.; Wang, Y.; Xu, Z.; Zhang, D.; Zhang, C.; Zhang, Y.; Ji, D.; et al. Circulating non-coding RNA cluster predicted the tumorigenesis and development of colorectal carcinoma. Aging 2020, 12, 23047–23066. [Google Scholar] [CrossRef]
- Matboli, M.; Shafei, A.E.; Ali, M.A.; El-Din Ahmed, T.S.; Naser, M.; Abdel-Rahman, T.; Anber, N.; Ali, M. Role of extracellular LncRNA-SNHG14/miRNA-3940-5p/NAP12 mRNA in colorectal cancer. Arch. Physiol. Biochem. 2021, 127, 479–485. [Google Scholar] [CrossRef]
- Samir, N.; Matboli, M.; El-Tayeb, H.; El-Tawdi, A.; Hassan, M.K.; Waly, A.; El-Akkad, H.A.E.; Ramadan, M.G.; Al-Belkini, T.N.; El-Khamisy, S.; et al. Competing endogenous RNA network crosstalk reveals novel molecular markers in colorectal cancer. J. Cell. Biochem. 2018, 119, 6869–6881. [Google Scholar] [CrossRef]
- Schultz, N.A.; Dehlendorff, C.; Jensen, B.V.; Bjerregaard, J.K.; Nielsen, K.R.; Bojesen, S.E.; Calatayud, D.; Nielsen, S.E.; Yilmaz, M.; Holländer, N.H.; et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA 2014, 311, 392–404. [Google Scholar] [CrossRef]
- Zhou, X.; Lu, Z.; Wang, T.; Huang, Z.; Zhu, W.; Miao, Y. Plasma miRNAs in diagnosis and prognosis of pancreatic cancer: A miRNA expression analysis. Gene 2018, 673, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Zhu, Z.; Roy, S.; Jun, E.; Han, H.; Munoz, R.M.; Nishiwada, S.; Sharma, G.; Cridebring, D.; Zenhausern, F.; et al. An Exosome-based Transcriptomic Signature for Noninvasive, Early Detection of Patients with Pancreatic Ductal Adenocarcinoma: A Multicenter Cohort Study. Gastroenterology 2022, 163, 1252–1266.e2. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Wei, J.; Huang, Z.; Zhou, X.; Lu, Z.; Zhu, W.; Miao, Y. Identification of a six-miRNA panel in serum benefiting pancreatic cancer diagnosis. Cancer Med. 2019, 8, 2810–2822. [Google Scholar] [CrossRef]
- Franklin, O.; Jonsson, P.; Billing, O.; Lundberg, E.; Öhlund, D.; Nyström, H.; Lundin, C.; Antti, H.; Sund, M. Plasma Micro-RNA Alterations Appear Late in Pancreatic Cancer. Ann. Surg. 2018, 267, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Seyed Salehi, A.; Parsa-Nikoo, N.; Roshan-Farzad, F.; Shams, R.; Fathi, M.; Asaszadeh Aghdaei, H.; Behmanesh, A. MicroRNA-125a-3p, -4530, and -92a as a Potential Circulating MicroRNA Panel for Noninvasive Pancreatic Cancer Diagnosis. Dis. Markers 2022, 2022, 8040419. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.S.; Calatayud, D.; Albieri, V.; Schultz, N.A.; Dehlendorff, C.; Werner, J.; Jensen, B.V.; Pfeiffer, P.; Bojesen, S.E.; Giese, N.; et al. The potential diagnostic value of serum microRNA signature in patients with pancreatic cancer. Int. J. Cancer 2016, 139, 2312–2324. [Google Scholar] [CrossRef]
- Liu, J.; Gao, J.; Du, Y.; Li, Z.; Ren, Y.; Gu, J.; Wang, X.; Gong, Y.; Wang, W.; Kong, X. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int. J. Cancer 2012, 131, 683–691. [Google Scholar] [CrossRef]
- Dittmar, R.L.; Liu, S.; Tai, M.C.; Rajapakshe, K.; Huang, Y.; Longton, G.; DeCapite, C.; Hurd, M.W.; Paris, P.L.; Kirkwood, K.S.; et al. Plasma miRNA Biomarkers in Limited Volume Samples for Detection of Early-stage Pancreatic Cancer. Cancer Prev. Res. (Phila.) 2021, 14, 729–740. [Google Scholar] [CrossRef]
- Yang, G.; Qiu, J.; Xu, J.; Xiong, G.; Zhao, F.; Cao, Z.; Chen, G.; Liu, Y.; Tao, J.; Zheng, L.; et al. Using a microRNA panel of circulating exosomes for diagnosis of pancreatic cancer: Multicentre case-control study. Br. J. Surg. 2023, 110, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, B.; Yue, S.; Galli, U.; Rana, S.; Gross, W.; Müller, M.; Giese, N.A.; Kalthoff, H.; Becker, T.; Büchler, M.W.; et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int. J. Cancer 2015, 136, 2616–2627. [Google Scholar] [CrossRef]
- Tsen, A.; Barbara, M.; Rosenkranz, L. Dilemma of elevated CA 19-9 in biliary pathology. Pancreatology 2018, 18, 862–867. [Google Scholar] [CrossRef] [PubMed]
- So, J.B.Y.; Kapoor, R.; Zhu, F.; Koh, C.; Zhou, L.; Zou, R.; Tang, Y.C.; Goo, P.C.K.; Rha, S.Y.; Chung, H.C.; et al. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high-risk population. Gut 2021, 70, 829–837. [Google Scholar] [CrossRef]
- Huang, Z.; Zhu, D.; Wu, L.; He, M.; Zhou, X.; Zhang, L.; Zhang, H.; Wang, W.; Zhu, J.; Cheng, W.; et al. Six Serum-Based miRNAs as Potential Diagnostic Biomarkers for Gastric Cancer. Cancer Epidemiol. Biomark. Prev. 2017, 26, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wang, L.; Yang, Y.; Gong, L.; Xiao, B.; Liu, X. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem. Biophys. Res. Commun. 2017, 493, 1322–1328. [Google Scholar] [CrossRef]
- Zhu, C.; Ren, C.; Han, J.; Ding, Y.; Du, J.; Dai, N.; Dai, J.; Ma, H.; Hu, Z.; Shen, H.; et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br. J. Cancer 2014, 110, 2291–2299. [Google Scholar] [CrossRef]
- Zhao, G.; Jiang, T.; Liu, Y.; Huai, G.; Lan, C.; Li, G.; Jia, G.; Wang, K.; Yang, M. Droplet digital PCR-based circulating microRNA detection serve as a promising diagnostic method for gastric cancer. BMC Cancer 2018, 18, 676. [Google Scholar] [CrossRef]
- Huang, S.; Wang, J.; Li, J.; Luo, Q.; Zhao, M.; Zheng, L.; Dong, X.; Chen, C.; Che, Y.; Liu, P.; et al. Serum microRNA expression profile as a diagnostic panel for gastric cancer. Jpn. J. Clin. Oncol. 2016, 46, 811–818. [Google Scholar] [CrossRef]
- Song, M.Y.; Pan, K.F.; Su, H.J.; Zhang, L.; Ma, J.L.; Li, J.Y.; Yuasa, Y.; Kang, D.; Kim, Y.S.; You, W.C. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS ONE 2012, 7, e33608. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.L.; Ren, L.F.; Wang, H.P.; Bai, Z.T.; Zhang, L.; Meng, W.B.; Zhu, K.X.; Ding, F.H.; Miao, L.; Yan, J.; et al. Plasma microRNAs as potential new biomarkers for early detection of early gastric cancer. World J. Gastroenterol. 2019, 25, 1580–1591. [Google Scholar] [CrossRef]
- Ghaedi, H.; Mozaffari, M.A.N.; Salehi, Z.; Ghasemi, H.; Zadian, S.S.; Alipoor, S.; Hadianpour, S.; Alipoor, B. Co-expression profiling of plasma miRNAs and long noncoding RNAs in gastric cancer patients. Gene 2019, 687, 135–142. [Google Scholar] [CrossRef]
- GASTROClear Blood Test for Early Detection. Available online: https://gastroclear.mirxes.com (accessed on 10 October 2023).
- Ying, L.; Du, L.; Zou, R.; Shi, L.; Zhang, N.; Jin, J.; Xu, C.; Zhang, F.; Zhu, C.; Wu, J.; et al. Development of a serum miRNA panel for detection of early stage non-small cell lung cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 25036–25042. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Kong, H.; Hou, Y.; Ge, D.; Huang, W.; Ou, J.; Yang, D.; Zhang, L.; Wu, G.; Song, Y.; et al. Two plasma microRNA panels for diagnosis and subtype discrimination of lung cancer. Lung Cancer 2018, 123, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Qu, S.; Hu, R.; Gao, W.; Jin, S.; Liu, M.; Zhao, Q. A panel of miRNAs derived from plasma extracellular vesicles as novel diagnostic biomarkers of lung adenocarcinoma. FEBS Open Bio 2019, 9, 2149–2158. [Google Scholar] [CrossRef]
- Nadal, E.; Truini, A.; Nakata, A.; Lin, J.; Reddy, R.M.; Chang, A.C.; Ramnath, N.; Gotoh, N.; Beer, D.G.; Chen, G. A Novel Serum 4-microRNA Signature for Lung Cancer Detection. Sci. Rep. 2015, 5, 12464. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, X.; Li, J.; Wang, J.; Binang, H.; Shi, S.; Duan, W.; Zhao, Y.; Zhang, Y. Serum exosomal miR-1269a serves as a diagnostic marker and plays an oncogenic role in non-small cell lung cancer. Thorac. Cancer 2020, 11, 3436–3447. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Q.; Zhang, M.; Su, W.; Wang, Z.; Li, Y.; Zhang, J.; Beer, D.G.; Yang, S.; Chen, G. Serum microRNA Signature Is Capable of Early Diagnosis for Non-Small Cell Lung Cancer. Int. J. Biol. Sci. 2019, 15, 1712–1722. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Jing, H.; Zhang, J. MicroRNA-340 and MicroRNA-450b-5p: Plasma Biomarkers for Detection of Non-Small-Cell Lung Cancer. J. Environ. Public. Health 2022, 2022, 8024700. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yang, D.; Zhang, H.; Wei, X.; Ma, T.; Cheng, Z.; Hong, Q.; Hu, J.; Zhuo, H.; Song, Y.; et al. Early Detection of Lung Cancer in Serum by a Panel of MicroRNA Biomarkers. Clin. Lung Cancer 2015, 16, 313–319.e1. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, C.; Peng, X.; Liu, K.; Zhao, L.; Chen, X.; Yu, H.; Lai, Y. A combination of four serum miRNAs for screening of lung adenocarcinoma. Hum. Cell 2020, 33, 830–838. [Google Scholar] [CrossRef]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef]
- Geng, X.; Tsou, J.H.; Stass, S.A.; Jiang, F. Utilizing MiSeq Sequencing to Detect Circulating microRNAs in Plasma for Improved Lung Cancer Diagnosis. Int. J. Mol. Sci. 2023, 24, 10277. [Google Scholar] [CrossRef]
- Zhong, Y.; Ding, X.; Bian, Y.; Wang, J.; Zhou, W.; Wang, X.; Li, P.; Shen, Y.; Wang, J.J.; Li, J.; et al. Discovery and validation of extracellular vesicle-associated miRNAs as noninvasive detection biomarkers for early-stage non-small-cell lung cancer. Mol. Oncol. 2021, 15, 2439–2452. [Google Scholar] [CrossRef]
- El-Toukhy, S.E.; El-Daly, S.M.; Kamel, M.M.; Nabih, H.K. The diagnostic significance of circulating miRNAs and metabolite profiling in early prediction of breast cancer in Egyptian women. J. Cancer Res. Clin. Oncol. 2023, 149, 5437–5451. [Google Scholar] [CrossRef]
- Bedard, E.L.R.; Abraham, A.G.; Joy, A.A.; Ghosh, S.; Wang, X.; Lim, A.; Shao, D.; Loebenberg, R.; Roa, W.H. A Novel Composite Biomarker Panel For Detection Of Early Stage Non-small Cell Lung Cancer. Clin. Investig. Med. 2021, 44, E15–E24. [Google Scholar] [CrossRef]
- Peng, H.; Wang, J.; Li, J.; Zhao, M.; Huang, S.K.; Gu, Y.Y.; Li, Y.; Sun, X.J.; Yang, L.; Luo, Q.; et al. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016, 151, 235–242. [Google Scholar] [CrossRef]
- Baran, K.; Kordiak, J.; Jabłoński, S.; Brzeziańska-Lasota, E. Panel of miR-150 and linc00673, regulators of CCR6/CCL20 may serve as non-invasive diagnostic marker of non-small cell lung cancer. Sci. Rep. 2023, 13, 9642. [Google Scholar] [CrossRef]
- Zou, R.; Loke, S.Y.; Tang, Y.C.; Too, H.P.; Zhou, L.; Lee, A.S.G.; Hartman, M. Development and validation of a circulating microRNA panel for the early detection of breast cancer. Br. J. Cancer 2022, 126, 472–481. [Google Scholar] [CrossRef]
- Li, M.; Zou, X.; Xia, T.; Wang, T.; Liu, P.; Zhou, X.; Wang, S.; Zhu, W. A five-miRNA panel in plasma was identified for breast cancer diagnosis. Cancer Med. 2019, 8, 7006–7017. [Google Scholar] [CrossRef]
- Cuk, K.; Zucknick, M.; Madhavan, D.; Schott, S.; Golatta, M.; Heil, J.; Marmé, F.; Turchinovich, A.; Sinn, P.; Sohn, C.; et al. Plasma microRNA panel for minimally invasive detection of breast cancer. PLoS ONE 2013, 8, e76729. [Google Scholar] [CrossRef]
- Li, M.; Zhou, Y.; Xia, T.; Zhou, X.; Huang, Z.; Zhang, H.; Zhu, W.; Ding, Q.; Wang, S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat. 2018, 170, 257–270. [Google Scholar] [CrossRef]
- Chan, M.; Liaw, C.S.; Ji, S.M.; Tan, H.H.; Wong, C.Y.; Thike, A.A.; Tan, P.H.; Ho, G.H.; Lee, A.S. Identification of circulating microRNA signatures for breast cancer detection. Clin. Cancer Res. 2013, 19, 4477–4487. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, X.; Wang, J.; Zhao, L.; Peng, X.; Zhang, C.; Liu, K.; Huang, G.; Lai, Y. Breast invasive ductal carcinoma diagnosis with a three-miRNA panel in serum. Biomark. Med. 2021, 15, 951–963. [Google Scholar] [CrossRef]
- Kim, M.W.; Park, S.; Lee, H.; Gwak, H.; Hyun, K.A.; Kim, J.Y.; Jung, H.I.; Il Kim, S. Multi-miRNA panel of tumor-derived extracellular vesicles as promising diagnostic biomarkers of early-stage breast cancer. Cancer Sci. 2021, 112, 5078–5087. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhao, Q.; Zhang, W.; Zhang, Z.; Gao, J.; Zhang, C.; Li, Y.; Tian, Y. A novel panel of microRNAs provides a sensitive and specific tool for the diagnosis of breast cancer. Mol. Med. Rep. 2014, 10, 785–791. [Google Scholar] [CrossRef]
- Itani, M.M.; Nassar, F.J.; Tfayli, A.H.; Talhouk, R.S.; Chamandi, G.K.; Itani, A.R.S.; Makoukji, J.; Boustany, R.N.; Hou, L.; Zgheib, N.K.; et al. A Signature of Four Circulating microRNAs as Potential Biomarkers for Diagnosing Early-Stage Breast Cancer. Int. J. Mol. Sci. 2021, 22, 6121. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, P.M.M.; Vieira, E.; Lemos, D.S.; Souza, I.L.M.; Zanata, S.M.; Pankievicz, V.C.; Tuleski, T.R.; Souza, E.M.; Wowk, P.F.; Urban, C.A.; et al. Identification of miRNAs Enriched in Extracellular Vesicles Derived from Serum Samples of Breast Cancer Patients. Biomolecules 2020, 10, 150. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.K.; Luo, Q.; Peng, H.; Li, J.; Zhao, M.; Wang, J.; Gu, Y.Y.; Li, Y.; Yuan, P.; Zhao, G.H.; et al. A Panel of Serum Noncoding RNAs for the Diagnosis and Monitoring of Response to Therapy in Patients with Breast Cancer. Med. Sci. Monit. 2018, 24, 2476–2488. [Google Scholar] [CrossRef] [PubMed]
- Hulsen, T. DeepVenn—a web application for the creation of area-proportional Venn diagrams using the deep learning framework Tensorflow.js. arXiv 2022. [Google Scholar]
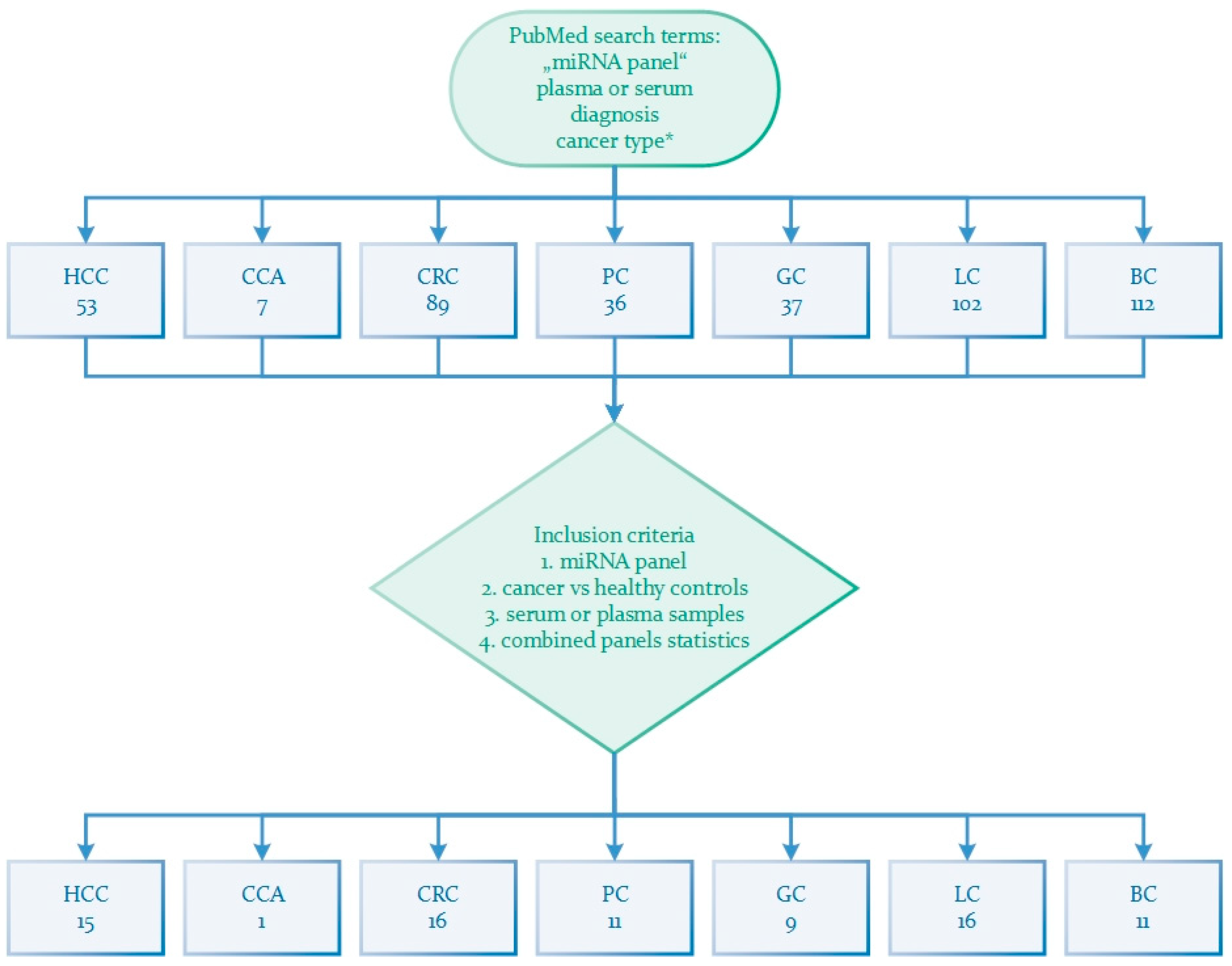
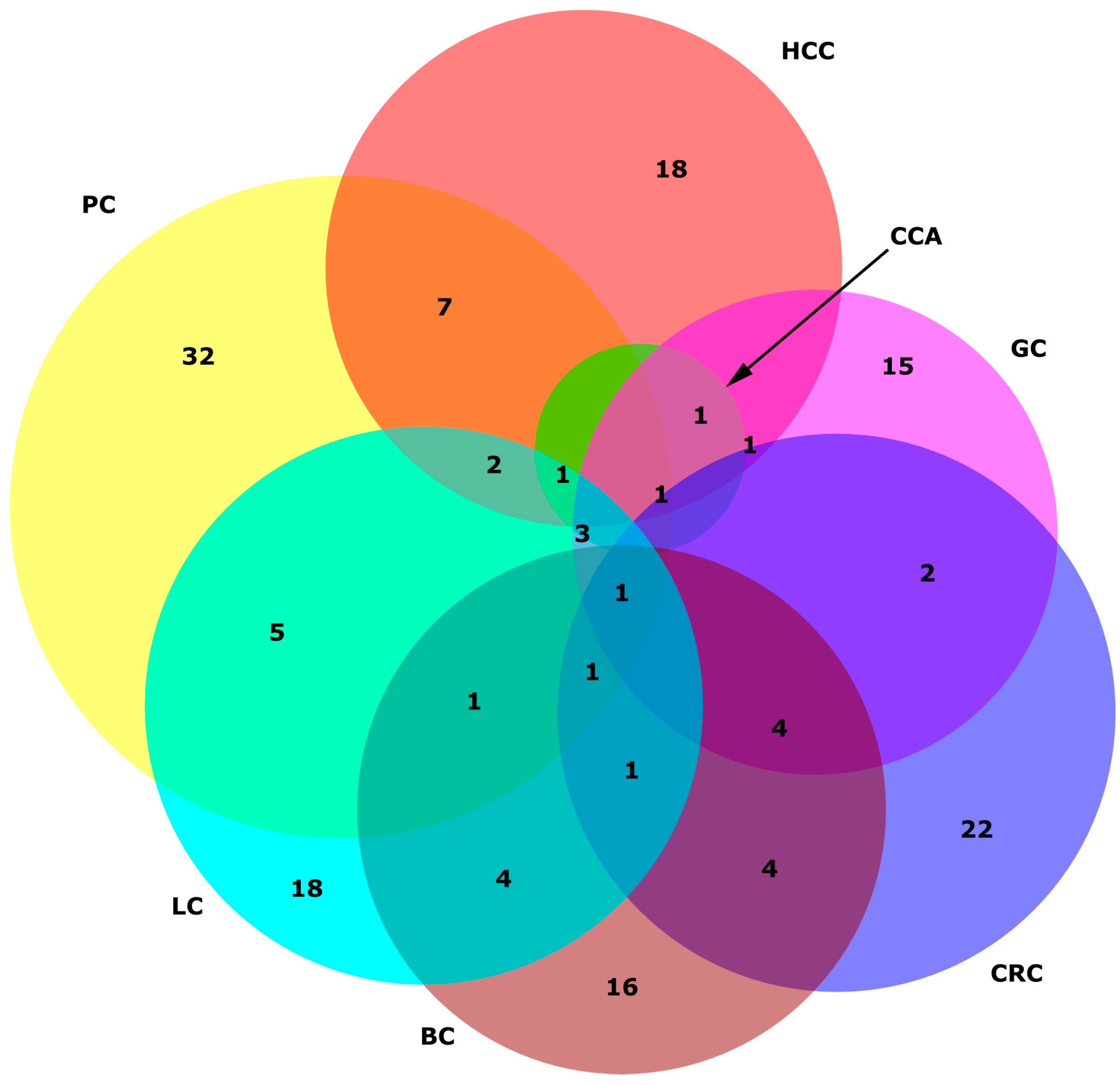
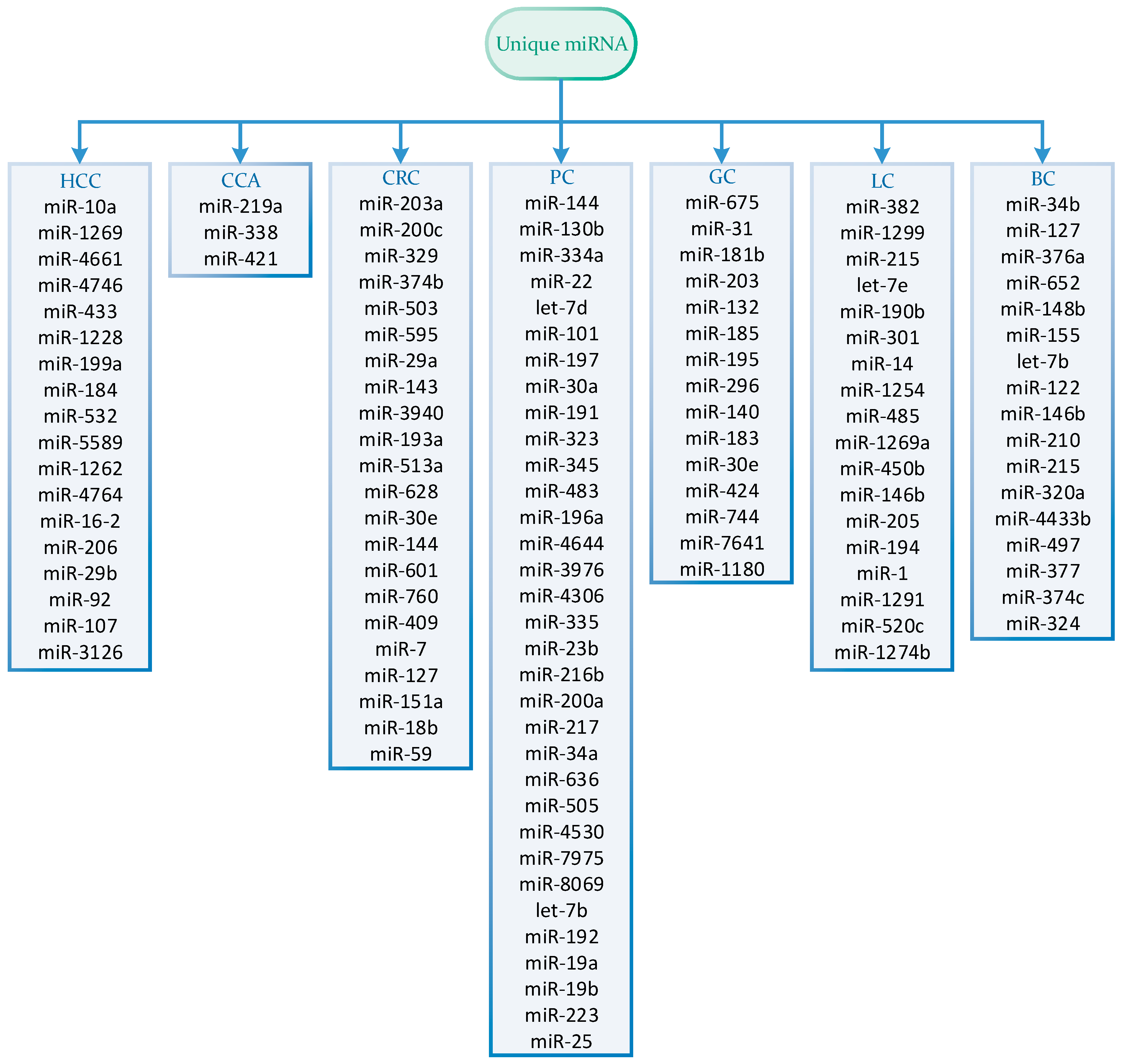
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA Panels | |||||
| miR-122 | Plasma | 457 HCC, 167 HC | ↑ | AUC = 0.941 | [18] |
| miR-192 | ↑ | ||||
| miR-21 | ↑ | ||||
| miR-223 | ↓ | ||||
| miR-26a | ↓ | ||||
| miR-27a | ↓ | ||||
| miR-801 | ↓ | ||||
| miR-206 | Serum | 261 HCC, 173 HC | ↑ | AUC = 0.887 (95% CI = 0.850–0.918) Sensitivity = 85.55% Specificity = 73.3% | [19] |
| miR-141-3p | ↑ | ||||
| miR-433-3p | ↑ | ||||
| miR-1228-5p | ↑ | ||||
| miR-199a-5p | ↓ | ||||
| miR-122-5p | ↓ | ||||
| miR-192-5p | ↓ | ||||
| miR-26a-5p | ↓ | ||||
| miR-214-5p | Serum | 224 HCC, 84 HC | ↓ | AUC = 0.95 with 95% CI Sensitivity = 83.2% Specificity = 96.9% Accuracy = 86.8% | [20] |
| miR-125b | ↓ | ||||
| miR-1269 | ↑ | ||||
| miR-375 | ↓ | ||||
| miR-27b-3p | Serum | 212 HCC, 110 HC | ↑ | AUC = 0.823 (p < 0.0001) | [21] |
| miR-192-5p | ↑ | ||||
| miR-375 | Serum | 149 HCC, 149 HC | ↑ | AUC = 0.995 (95% CI: 0.985–1) | [22] |
| miR-10a | ↑ | ||||
| miR-122 | ↑ | ||||
| miR-423 | ↑ | ||||
| miR-4661-5p | Serum exosomes | 84 HCC, 26 HC | ↑ | AUC = 0.942 (95% CI = 0.895–0.972) Sensitivity = 84.5% Specificity = 89.3% PPV = 88.8% NPV = 85.2% | [23] |
| miR-4746-5p | ↑ | ||||
| miR-126 | Serum | 34 HCC, 25 HC | ↑ | AUC = 1.00 SE = 0 p-value < 0.001 | [24] |
| miR-21 | ↑ | ||||
| miR-30c | ↑ | ||||
| miR-193b | ↑ | ||||
| miR-122 | ↑ | ||||
| miR-222 | ↑ | ||||
| miR-125b | ↑ | ||||
| miR-10b | Serum | 27 HCC, 50 HC | ↑ | AUC = 0.94 (95% CI: 0.89–0.99) | [25] |
| miR-181a | ↓ | ||||
| miR-106b | ↑ | ||||
| miRNA + lncRNA + mRNA panels | |||||
| miR-16-2 | Serum | 78 HCC, 42 HC | ↑ | Sensitivity = 79.5% Specificity = 100% | [26] |
| miR-21-5p | ↑ | ||||
| lncRNA-CTBP | ↑ | ||||
| mRNA LAMP2 | ↑ | ||||
| miR-1262 | Serum exosomes | 60 HCC, 18 HC | ↓ | Sensitivity = 100% Specificity = 76.7% PPV = 81.1% NPV = 100% Accuracy = 88.3% | [27] |
| lncRNA-RP11-513I15.6 | ↓ | ||||
| mRNA RAB11A | ↓ | ||||
| miR-4764-5p | Serum | 49 HCC, 36 HC | ↓ | Sensitivity = 100% Specificity = 76.7% PPV = 81.1% NPV = 100% Accuracy = 88.3% | [28] |
| lncRNA-RP11-156p1.3 | ↓ | ||||
| mRNA RFTN1 | ↓ | ||||
| miRNA + AFP panels | |||||
| miR-122 | Serum | 192 HCC, 95 HC | ↑ | AUC = 1 | [29] |
| miR-885-5p | ↑ | ||||
| miR-29b | ↓ | ||||
| AFP | |||||
| miR-92-3p | Serum | 115 HCC, 40 HC | ↑ | AUC = 0.988 | [30] |
| miR-107 | ↑ | ||||
| miR-3126-5p | ↓ | ||||
| AFP | |||||
| miR-125b | Serum | 90 HCC, 30 HC | ↓ | AUC = 0.936 (CI = 0.878ߝ0.995) Sensitivity = 0.907 Specificity = 0.933 | [31] |
| miR-223 | ↓ | ||||
| miR-27a | ↓ | ||||
| miR-26a | ↓ | ||||
| AFP | |||||
| miR-206 | Plasma | 38 HCC, 20 HC | ↑ | AUC = 0.989 (CI = 0.919-1.000) | [32] |
| miR-126 | ↓ | ||||
| AFP | |||||
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miR-10b-3p | plasma | 48 CCA, 20 HC | ↑ | AUC = 0.781 (95% CI: 0.585–0.914) Sensitivity = 83.3% Specificity = 75.0% PPV = 71.4% NPV = 85.7% | [35] |
| miR-26b-3p | ↑ | ||||
| miR-27a-3p | ↑ | ||||
| miR-106b-3p | ↑ | ||||
| miR-219a-3p | ↑ | ||||
| miR-338-5p | ↑ | ||||
| miR-421 | ↑ |
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA panels | |||||
| miR-23a-3p | Serum | 427 CRC, 276 HC | ↑ | AUC = 0.877 Sensitivity = 81.4% Specificity = 81% PPV = 80.6% NPV = 81.8% Accuracy = 81.2% | [39] |
| miR-27a-3p | ↑ | ||||
| miR-142-5p | ↑ | ||||
| miR-376c-3p | ↑ | ||||
| miR-19a-3p | Serum | 196 CRC, 138 HC | ↑ | AUC = 0.87 | [40] |
| miR-21-5p | ↑ | ||||
| miR-425-5p | ↑ | ||||
| miR-145 | Serum | 175 CRC, 130 HC | ↓ | AUC = 0.886 (95% CI = 0.850–0.921) | [41] |
| miR-106a | ↑ | ||||
| miR-17-3p | ↑ | ||||
| miR-27a | Exosomes | 170 CRC, 130 HC | ↑ | AUC = 0.801 | [42] |
| miR-130a | ↑ | ||||
| miR-193a-5p | Plasma | 149 CRC, 110 HC | ↑ | AUC = 0.88 (95% CI = 0.82–0.93) | [43] |
| miR-210 | ↑ | ||||
| miR-513a-5p | ↑ | ||||
| miR-628-3p | ↑ | ||||
| miR-103a-3p | Plasma | 139 CRC, 132 HC | ↑ | AUC = 0.895 | [44] |
| miR-127-3p | ↑ | ||||
| miR-151a-5p | ↑ | ||||
| miR-17-5p | ↑ | ||||
| miR-181a-5p | ↑ | ||||
| miR-18b-5p | ↑ | ||||
| miR-30e-3p | Serum | 137 CRC, 145 HC | ↑ | AUC = 0.883 Sensitivity = 0.800 Specificity = 0.787 | [45] |
| miR-146a-5p | ↑ | ||||
| miR-148a-3p | ↓ | ||||
| miR-203a-3p | Serum | 135 CRC, 135 HC | ↑ | AUC = 0.893 Sensitivity = 81.25% Specificity = 73.33% | [46] |
| miR-145-5p | ↓ | ||||
| miR-375-3p | ↓ | ||||
| miR-200c-3p | ↓ | ||||
| miR-18a | Plasma | 130 CRC, 244 HC | ↑ | AUC = 0.745 (95% CI = 0.708–0.846) | [47] |
| miR-20a | ↑ | ||||
| miR-21 | ↑ | ||||
| miR-29a | ↑ | ||||
| miR-92a | ↑ | ||||
| miR-106b | ↑ | ||||
| miR-133a | ↑ | ||||
| miR-143 | ↑ | ||||
| miR-145 | ↑ | ||||
| miR-409-3p | Plasma | 124 CRC, 117 HC | ↑ | AUC = 0.897 | [48] |
| miR-7 | ↓ | ||||
| miR-93 | ↓ | ||||
| miR-144-3p | Plasma | 101 CRC, 134 HC | ↓ | Sensitivity = 93.8% Specificity = 91.3% | [49] |
| miR-425-5p | ↓ | ||||
| miR-1260b | ↓ | ||||
| miR-601 | Plasma | 90 CRC, 58 HC | ↓ | AUC = 0.792 Sensitivity = 83.3% Specificity = 69.1% | [50] |
| miR-760 | ↓ | ||||
| miR-126 | Plasma | 50 CRC, 150 HC | ↓ | AUC = 0.906 | [51] |
| miR-139 | ↓ | ||||
| miR-143 | ↓ | ||||
| miR-595 | ↑ | ||||
| miRNA + lncRNA + mRNA panels | |||||
| miR-20b-5p | Plasma | 597 CRC, 585 HC | ↑ | AUC = 0.954 (95% CI = 0.913–0.994) | [52] |
| miR-329-3p | ↑ | ||||
| miR-374b-5p | ↑ | ||||
| miR-503-5p | ↑ | ||||
| lncRNA-XLOC_001120 | ↑ | ||||
| lncRNA-ENSG00000243766.2 | ↑ | ||||
| miR-3940-5p | Plasma | 70 CRC * | ↓ | Sensitivity = 100% Specificity = 88.6% PPV = 100% NPV = 85% Accuracy = 93.07% | [53] |
| lncRNA-SNHG14 | ↑ | ||||
| mRNA-NAP1L2 | ↑ | ||||
| miR-59 | Serum | 70 CRC, 20 HC | ↓ | Sensitivity = 100% Specificity = 61.7% PPV = 75.3% NPV = 100% Accuracy = 83.1% | [54] |
| lncRNA-RP11-909B2.1 | ↑ | ||||
| mRNA L3MBTL1 | ↑ | ||||
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA panels | |||||
| miR-122 | Plasma | 409 PC, 312 HC | ↑ | AUC = 0.93 (95% CI = 0.90–0.96) | [55] |
| miR-34a | ↑ | ||||
| miR-145 | ↑ | ||||
| miR-636 | ↑ | ||||
| miR-223 | ↑ | ||||
| miR-26b | ↑ | ||||
| miR-885-5p | ↑ | ||||
| miR-150 | ↑ | ||||
| miR-126 | ↑ | ||||
| miR-505 | ↑ | ||||
| miR-122-5p | Plasma | 216 PC, 220 HC | ↑ | AUC = 0.937 | [56] |
| miR-125b-5p | ↑ | ||||
| miR-192-5p | ↑ | ||||
| miR-193b-3p | ↑ | ||||
| miR-221-3p | ↑ | ||||
| miR-27b-3p | ↑ | ||||
| miR-30c-5p | Plasma | 168 PC, 124 HC | ↑ | AUC = 0.93 | [57] |
| miR-340-5p | ↑ | ||||
| miR-335-5p | ↑ | ||||
| miR-23b-3p | ↑ | ||||
| miR-142-3p | ↑ | ||||
| miR-145-5p | ↑ | ||||
| miR-200b-3p | ↑ | ||||
| miR-429 | ↑ | ||||
| miR-1260b | ↑ | ||||
| miR-145-3p | ↑ | ||||
| miR-216b-5p | ↑ | ||||
| miR-200a-3p | ↑ | ||||
| miR-217-5p | ↑ | ||||
| let-7b-5p | Plasma | 129 PC, 107 HC | ↑ | AUC = 0.910 | [58] |
| miR-192-5p | ↑ | ||||
| miR-19a-3p | ↑ | ||||
| miR-19b-3p | ↑ | ||||
| miR-223-3p | ↑ | ||||
| miR-25-3p | ↑ | ||||
| miR-574-3p | Plasma | 90 PC, 154 HC | ↑ | AUC = 0.96 (95% CI = 0.92–1.00) | [59] |
| miR-885-5p | ↑ | ||||
| miR-144-3p | ↓ | ||||
| miR-130b-3p | ↑ | ||||
| miR-334a-5p | ↑ | ||||
| miR-24-3p | ↑ | ||||
| miR-106b-5p | ↓ | ||||
| miR-22-5p | ↑ | ||||
| miR-451a | ↓ | ||||
| let-7d-3p | ↑ | ||||
| miR-101-3p | ↓ | ||||
| miR-26a-5p | ↓ | ||||
| miR-197-3p | ↑ | ||||
| miR-423-3p | ↑ | ||||
| miR-122-5p | ↑ | ||||
| miR-125a-3p | Plasma | 77 PC, 65 HC | ↑ | AUC = 0.862 | [60] |
| miR-4530 | ↑ | ||||
| miR-92a-2-5p | ↑ | ||||
| miRNA + CA19-9 panels | |||||
| miR-16 | Serum | 471 PC, 248 HC | * | AUC = 0.94 (95% CI = 0.90–0.97) Sensitivity = 85 %Specificity = 98% Accuracy = 89% | [61] |
| miR-18a | |||||
| miR-20a | |||||
| miR-24 | |||||
| miR-25 | |||||
| miR-27a | |||||
| miR-29c | |||||
| miR-30a-5p | |||||
| miR-191 | |||||
| miR-323-3p | |||||
| miR-345 | |||||
| miR-483-5p | |||||
| CA19-9 | |||||
| miR-16 | Plasma | 138 PC, 68 HC | ↑ | AUC = 0.979 (95% CI = 0.962–0.996) | [62] |
| miR-196a | ↑ | ||||
| CA19-9 | ↑ | ||||
| miR-34a-5p | Plasma | 136 PC, 73 HC | ↑ | AUC = 0.94 (95% CI = 0.89–0.98) | [63] |
| miR-130a-3p | ↑ | ||||
| miR-222-3p | ↑ | ||||
| CA19-9 | ↑ | ||||
| miR-130a-3p | Plasma | 68 PC, 61 HC | ↑ | AUC = 0.986 (95% CI = 0.972–1.000) | [64] |
| miR-21-5p | ↑ | ||||
| miR-223-3p | ↑ | ||||
| miR-7975 | ↑ | ||||
| miR-8069 | ↑ | ||||
| CA19-9 | ↑ | ||||
| miRNA + protein panels | |||||
| miR-1246 | Serum | 131 PC, 20 HC | ↑ | Sensitivity = 100% (95% CI = 95%–100%) Specificity = 80% (95% CI: 67%–90%) | [65] |
| miR-4644 | ↑ | ||||
| miR-3976 | ↑ | ||||
| miR-4306 | ↑ | ||||
| CD44v6 | ↑ | ||||
| Tspan8 | ↑ | ||||
| MET | ↑ | ||||
| CD104 | ↑ | ||||
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA panels | |||||
| miR-140 | Serum | 424 GC, 468 HC | * | AUC = 0.92 (95% CI = 0.88–0.96) Sensitivity = 87.0% (95% CI = 0.794–0.925) Specificity = 68.5% (95% CI = 0.670–0.698) | [67] |
| miR-183 | |||||
| miR-30e | |||||
| miR-103a | |||||
| miR-126 | |||||
| miR-93 | |||||
| miR-142 | |||||
| miR-21 | |||||
| miR-29c | |||||
| miR-424 | |||||
| miR-181a | |||||
| miR-340 | |||||
| miR-10b-5p | Serum/exosomes | 205 GC, 167 HC/30 GC, 28 HC | ↑ | AUC = 0.702 | [68] |
| miR-132-3p | ↑ | ||||
| miR-185-5p | ↑ | ||||
| miR-195-5p | ↑ | ||||
| miR-20a-3p | ↑ | ||||
| miR-296-5p | ↑ | ||||
| miR-19b-3p | Serum exosomes | 130 GC, 130 HC | ↑ | AUC = 0.814 | [69] |
| miR-106a-5p | ↑ | ||||
| miR-16 | Plasma | 124 GC, 160 HC | ↑ | AUC = 0.812 | [70] |
| miR-25 | ↑ | ||||
| miR-92a | ↑ | ||||
| miR-451 | ↑ | ||||
| miR-486-5p | ↑ | ||||
| miR-21 | Plasma | 115 GC, 60 HC | ↑ | AUC = 0.887 (95% CI = 0.83–0.943) Sensitivity = 84.8% Specificity = 79.2% | [71] |
| miR-93 | ↑ | ||||
| miR-106a | ↑ | ||||
| miR-106b | ↑ | ||||
| miR-21 | Serum | 92 GC, 89 HC | ↑ | AUC = 0.919 (95% CI = 0.863-0.975) | [72] |
| miR-31 | ↓ | ||||
| miR-92a | ↓ | ||||
| miR-181b | ↓ | ||||
| miR-203 | ↓ | ||||
| miR-221 | Serum | 82 GC, 82 HC | ↑ | Sensitivity = 82.4% Specificity = 58.8% | [73] |
| miR-744 | ↑ | ||||
| miR-376c | ↑ | ||||
| miR-7641 | Plasma | 62 GC, 90 HC | ↓ | AUC = 0.799 (95% CI = 0.691–0.908) p < 0.001 | [74] |
| miR-425-5p | ↓ | ||||
| miR-1180-3p | ↓ | ||||
| miR-122-5p | ↓ | ||||
| miRNA + lncRNA panels | |||||
| miR-675-5p | Plasma | 62 GC, 40 HC | ↓ | AUC = 0.927 (95% CI = 0.85–0.96) p < 0.0001 Sensitivity = 88.78% Specificity = 85% | [75] |
| H19 | ↑ | ||||
| MEG3 | ↓ | ||||
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA panels | |||||
| let-7a-5p | Serum | 744 NSCLC, 944 HC | ↓ | AUC = 0.973 (95% CI = 0.947–0.987) | [77] |
| miR-375 | ↓ | ||||
| miR-1-3p | ↑ | ||||
| miR-1291 | ↑ | ||||
| miR-214-3p | ↑ | ||||
| miR-17 | Plasma | 676 NSCLC, 456 HC | ↓ | AUC = 0.873 (95% CI = 0.843–0.899) Sensitivity = 81% Specificity = 80% | [78] |
| miR-190b | ↓ | ||||
| miR-19a | ↓ | ||||
| miR-19b | ↓ | ||||
| miR-26b | ↓ | ||||
| miR-375 | ↑ | ||||
| miR-451a | Serum exosomes | 434 LUAD, 149 HC | ↑ | AUC = 0.965 | [79] |
| miR-194-5p | ↑ | ||||
| miR-486-5p | ↑ | ||||
| miR-193b | Serum | 154 NSCLC, 45 HC | ↑ | AUC = 0.993 (95% CI 0.979–1.000) p < 0.001 | [80] |
| miR-301 | ↑ | ||||
| miR-14 | ↑ | ||||
| miR-200b | ↑ | ||||
| miR-9-3p | Serum exosomes | 147 NSCLC, 149 HC | ↑ | AUC = 0.878 | [81] |
| miR-205-5p | ↑ | ||||
| miR-210-5p | ↑ | ||||
| miR-1269a | ↑ | ||||
| miR-146b | Serum | 128 NSCLC, 30 HC | ↑ | AUC = 0.96 Accuracy = 92.005 | [82] |
| miR-205 | ↑ | ||||
| miR-29c | ↑ | ||||
| miR-30b | ↑ | ||||
| miR-340 | Plasma | 120 NSCLC, 120 HC | ↓ | AUC = 0.862 Sensitivity = 78.33% Specificity = 77.5% | [83] |
| miR-450b-5p | ↑ | ||||
| miR-125a-5p | Serum | 118 NSCLC, 135 HC | ↓ | AUC = 0.936 Sensitivity = 87.5% Specificity = 87.5% | [84] |
| miR-25 | ↓ | ||||
| miR-126 | ↓ | ||||
| miR-142-5p | Serum | 112 LUAD, 120 HC | ↑ | AUC = 0.933 (95% CI = 0.884–0.965) Sensitivity = 82.93% Specificity = 96.67% | [85] |
| miR-409-3p | ↓ | ||||
| miR-223-3p | ↑ | ||||
| miR-146a-5p | ↓ | ||||
| let-7b-5p | Plasma | 46 NSCLC, 41 HC | ↓ | AUC = 0.868 Sensitivity = 80% Specificity = 80% | [86] |
| let-7e-5p | ↑ | ||||
| miR-23a-3p | ↓ | ||||
| miR-486-5p | ↓ | ||||
| miR-215-5p | Serum | 39 NSCLC, 32 HC | ↓ | AUC = 0.8013 Sensitivity = 67% Specificity = 68% | [87] |
| miR-1299 | ↓ | ||||
| miR-205-5p | ↑ | ||||
| miR-1246 | ↑ | ||||
| miR-520c-3p | Serum exosomes | 36 NSCLC, 36 HC | ↑ | AUC = 0.857 (95% CI, 0813–0.901) p < 0.0001 | [88] |
| miR-1274b | ↑ | ||||
| miR-145 | Serum | 30 NSCLC, 20 HC | ↑ | AUC = 1 Sensitivity = 100% Specificity 100% | [89] |
| miR-382 | ↑ | ||||
| miR-21 | Serum | 28 NSCLC, 17 HC | ↑ | AUC = 0.91 (95% CI = 0.80–1.0) | [90] |
| miR-223 | ↓ | ||||
| miR-205-5p | Serum | 20 SCLC, 32 HC | ↓ | AUC = 0.948 Sensitivity = 90.00% Specificity = 93.75% | [87] |
| miR-1299 | ↓ | ||||
| miR-215-5p | ↓ | ||||
| miR-141-3p | ↓ | ||||
| miR-200b-5p | ↓ | ||||
| miRNA + lncRNA panels | |||||
| miR-1254 | Serum | 156 NSCLC, 107 HC | ↓ | AUC = 0.844 (95% CI = 0.778–0.91) Sensitivity = 93.3% Specificity = 73.2% | [91] |
| miR-485-5p | ↓ | ||||
| miR-574-5p | ↓ | ||||
| MALAT1 | ↓ | ||||
| miR-150 | Serum | 30 NSCLC, 15 HC | ↓ | AUC = 0.784 Sensitivity = 80% Specificity = 80% | [92] |
| linc00673 | ↑ | ||||
| miRNA Panel | Sample Type | Number of Samples | Expression | Statistics | Reference |
|---|---|---|---|---|---|
| miRNA panels | |||||
| miR-133a-3p | Serum | 540 BC, 502 HC | ↑ | AUC = 0.915 Accuracy = 82.3% Sensitivity = 72.2% Specificity = 91.5% | [93] |
| miR-497-5p | ↑ | ||||
| miR-24-3p | ↑ | ||||
| miR-125b-5p | ↑ | ||||
| miR-377-3p | ↓ | ||||
| miR-374c-5p | ↓ | ||||
| miR-324-5p | ↓ | ||||
| miR-19b-3p | ↓ | ||||
| let-7b-5p | Plasma | 257 BC, 257 HC | ↑ | AUC = 0.978 | [94] |
| miR-122-5p | ↑ | ||||
| miR-146b-5p | ↑ | ||||
| miR-210-3p | ↑ | ||||
| miR-215-5p | ↑ | ||||
| miR-127-3p | Plasma | 247 BC, 140 HC | ↑ | AUC = 0.81 (95% CI = 0.75–0.88) | [95] |
| miR-376a | ↑ | ||||
| miR-652 | ↑ | ||||
| miR-148b | ↑ | ||||
| miR-376c | ↑ | ||||
| miR-409-3p | ↑ | ||||
| miR-801 | ↑ | ||||
| miR-106a-5p | Serum | 204 BC, 202 HC | ↑ | AUC = 0.93 (95% CI = 0.911–0.964) Sensitivity = 87% Specificity = 89% | [96] |
| miR-19b-3p | ↑ | ||||
| miR-20b-5p | ↑ | ||||
| miR-92a-3p | ↑ | ||||
| miR-106a-3p | Plasma | 200 BC, 200 HC | ↑ | AUC = 0.88 (95% CI = 0.855–0.923) Sensitivity = 82% Specificity = 79% | [96] |
| miR-106a-5p | ↑ | ||||
| miR-20b-5p | ↑ | ||||
| miR-92a-2-5p | ↑ | ||||
| miR-92a | Serum | 164 BC, 132 HC | ↑ | AUC = 0.91 | [97] |
| miR-133a | ↑ | ||||
| miR-9-5p | Serum | 135 BC, 125 HC | ↑ | AUC = 0.880 Sensitivity = 86.25% Specificity = 81.25% | [98] |
| miR-34b-3p | ↓ | ||||
| miR-146a-5p | ↓ | ||||
| miR-9 | Plasma | 62 BC, 20 HC | ↑ | AUC = 0.88 (95% CI = 0.78–0.99) Sensitivity = 96.8% Specificity = 80% | [99] |
| miR-16 | ↑ | ||||
| miR-21 | ↑ | ||||
| miR-429 | ↑ | ||||
| miR-451 | Serum | 60 BC, 29 HC | ↓ | AUC = 0.953 Sensitivity = 94.7% Specificity = 82.8% | [100] |
| miR-148a | ↓ | ||||
| miR-27a | ↓ | ||||
| miR-30b | ↓ | ||||
| miR-145 | Plasma | 41 BC, 32 HC | ↑ | AUC = 0.97 (95% CI = 0.929–1.000) Sensitivity 97% Specificity 91% | [101] |
| miR-425-5p | ↑ | ||||
| miR-139-5p | ↑ | ||||
| miR-130a | ↑ | ||||
| miR-142-5p | Serum | 31 BC, 16 HC | ↑ | AUC = 0.8387 Sensitivity = 93.33% Specificity = 68.75% | [102] |
| miR-320a | ↑ | ||||
| miR-4433b-5p | ↑ | ||||
| miRNA + lncRNA panels | |||||
| let-7a | Serum | 158 BC, 107 HC | ↓ | AUC = 0.968 PPV = 0.97 NPV = 0.85 | [103] |
| miR-155 | ↑ | ||||
| miR-574-5p | ↑ | ||||
| MALAT1 | ↑ | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranković, B.; Hauptman, N. Circulating microRNA Panels for Detection of Liver Cancers and Liver-Metastasizing Primary Cancers. Int. J. Mol. Sci. 2023, 24, 15451. https://doi.org/10.3390/ijms242015451
Ranković B, Hauptman N. Circulating microRNA Panels for Detection of Liver Cancers and Liver-Metastasizing Primary Cancers. International Journal of Molecular Sciences. 2023; 24(20):15451. https://doi.org/10.3390/ijms242015451
Chicago/Turabian StyleRanković, Branislava, and Nina Hauptman. 2023. "Circulating microRNA Panels for Detection of Liver Cancers and Liver-Metastasizing Primary Cancers" International Journal of Molecular Sciences 24, no. 20: 15451. https://doi.org/10.3390/ijms242015451
APA StyleRanković, B., & Hauptman, N. (2023). Circulating microRNA Panels for Detection of Liver Cancers and Liver-Metastasizing Primary Cancers. International Journal of Molecular Sciences, 24(20), 15451. https://doi.org/10.3390/ijms242015451






