Peptide-Decorated Degradable Polycarbonate Nanogels for Eliciting Antigen-Specific Immune Responses
Abstract
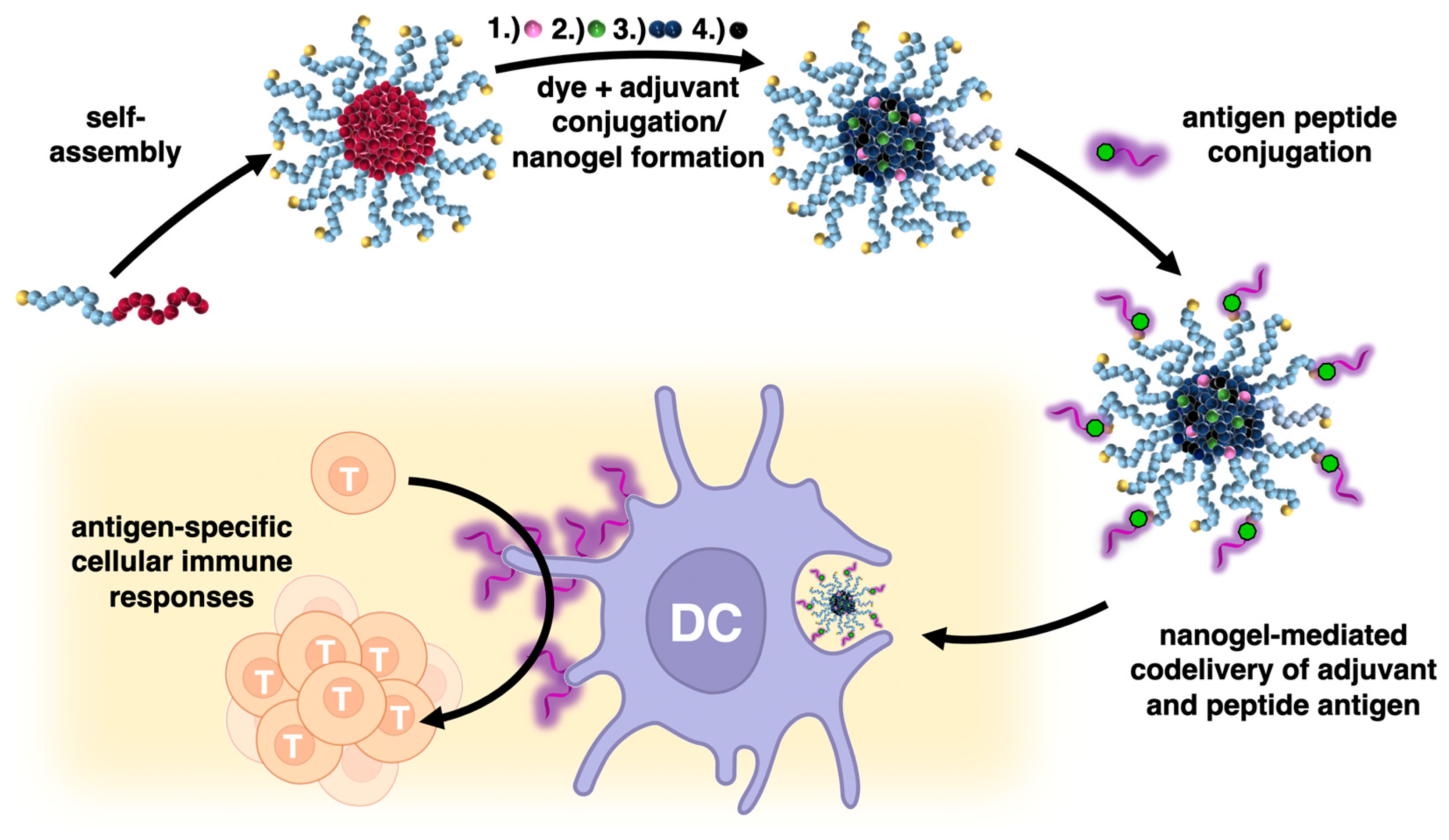
:1. Introduction
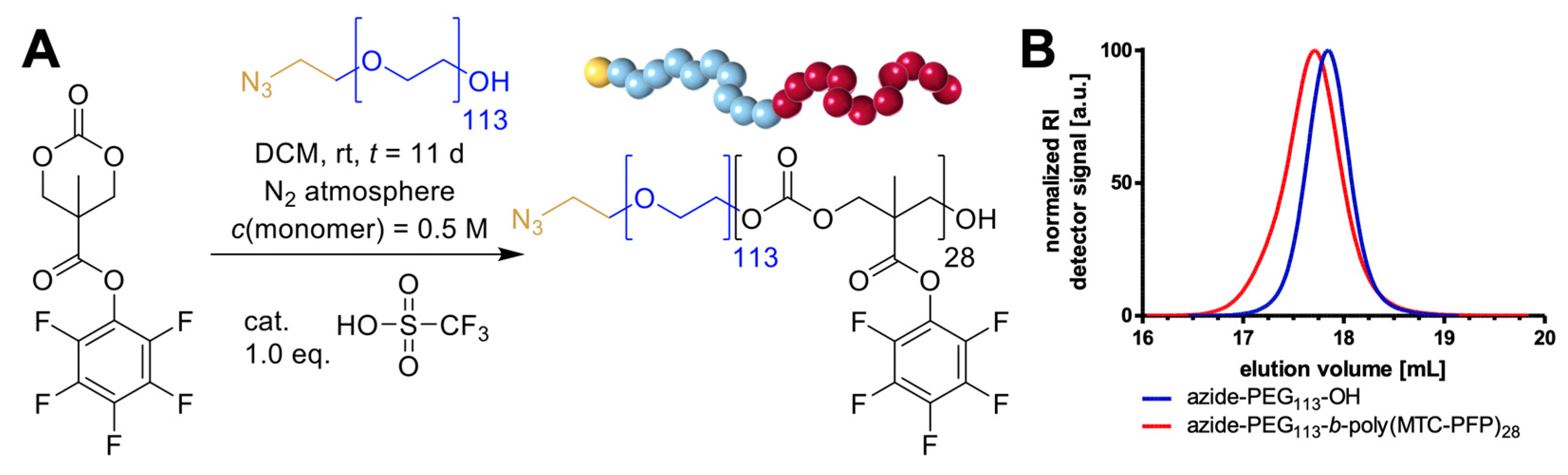
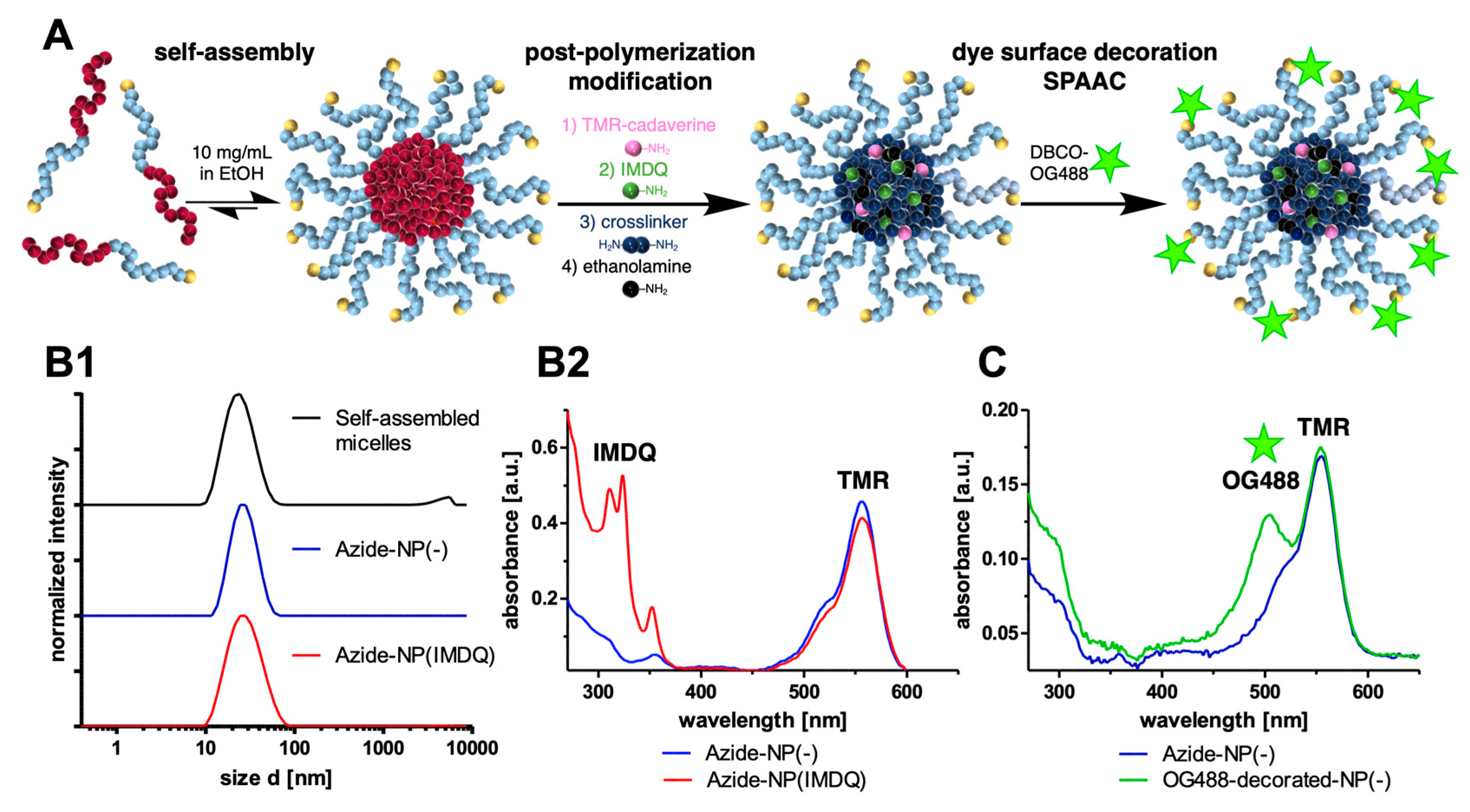
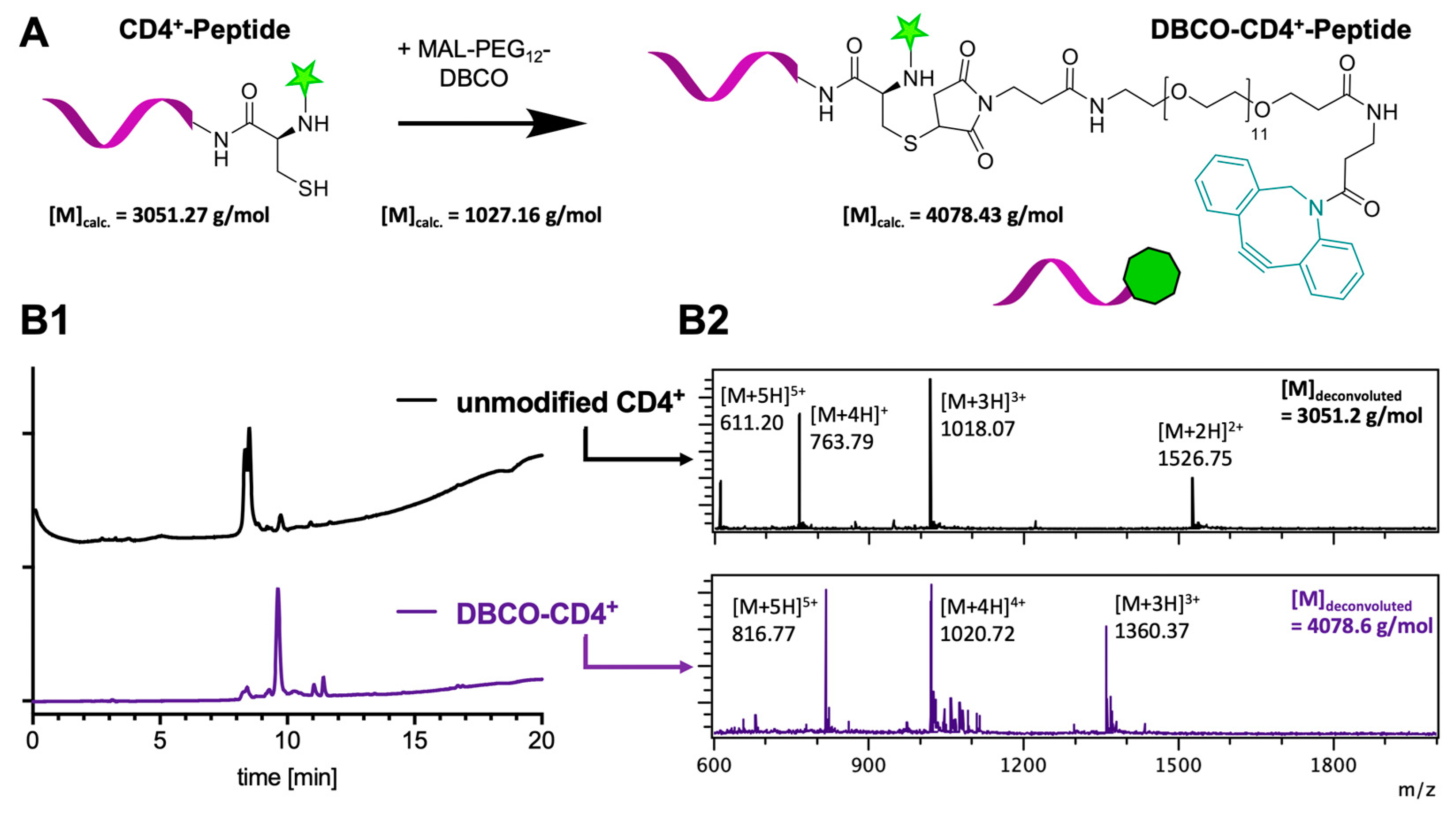
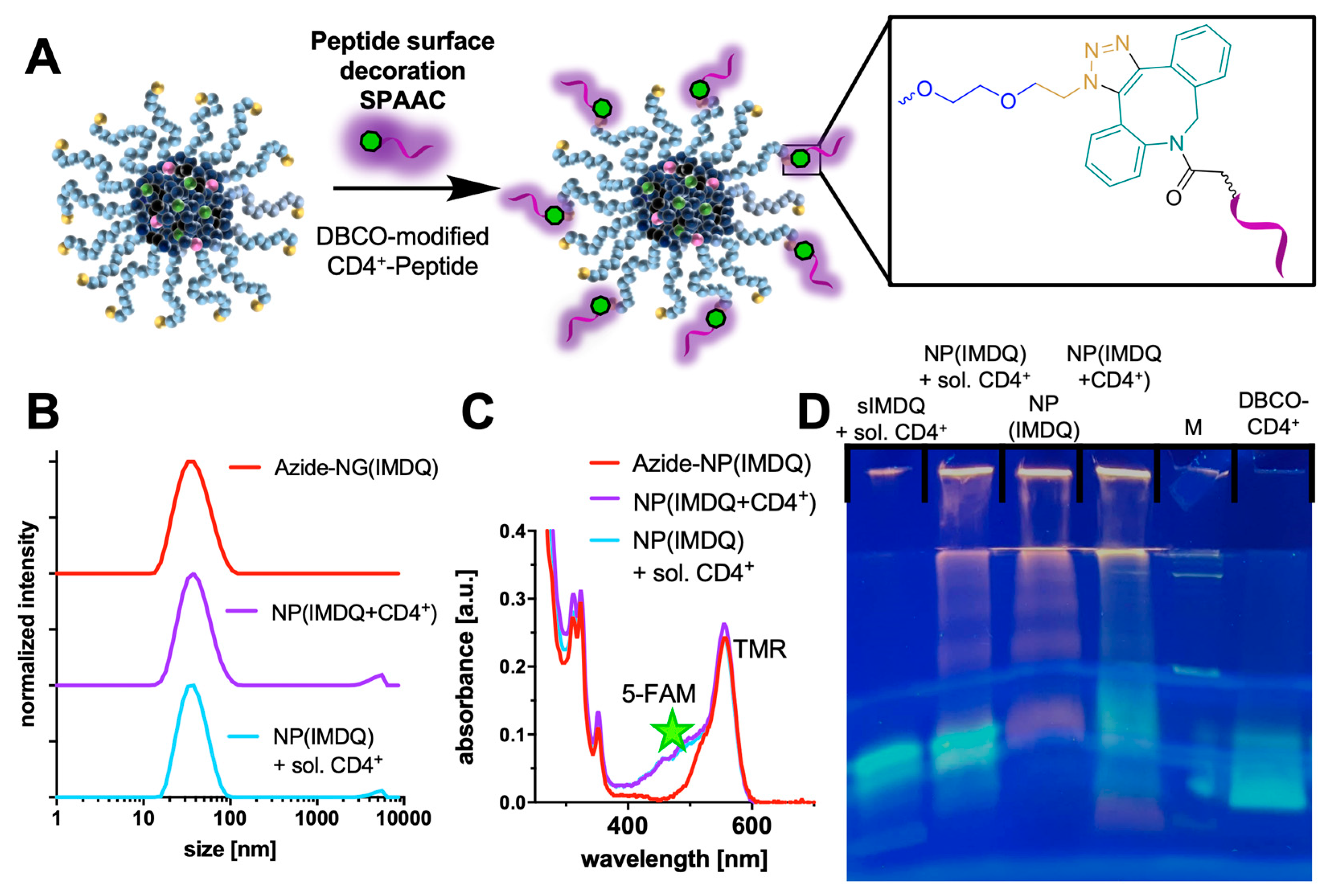
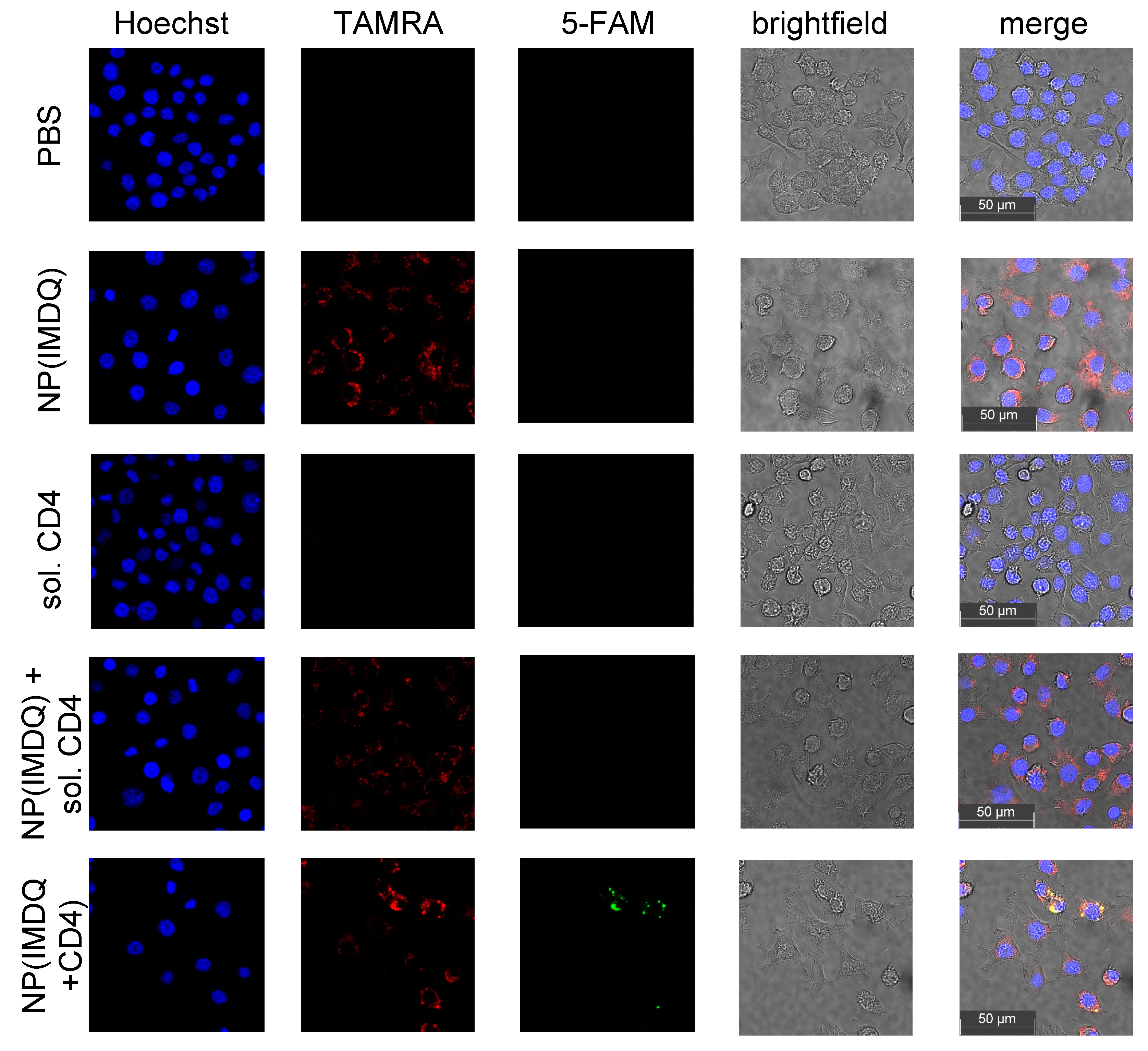
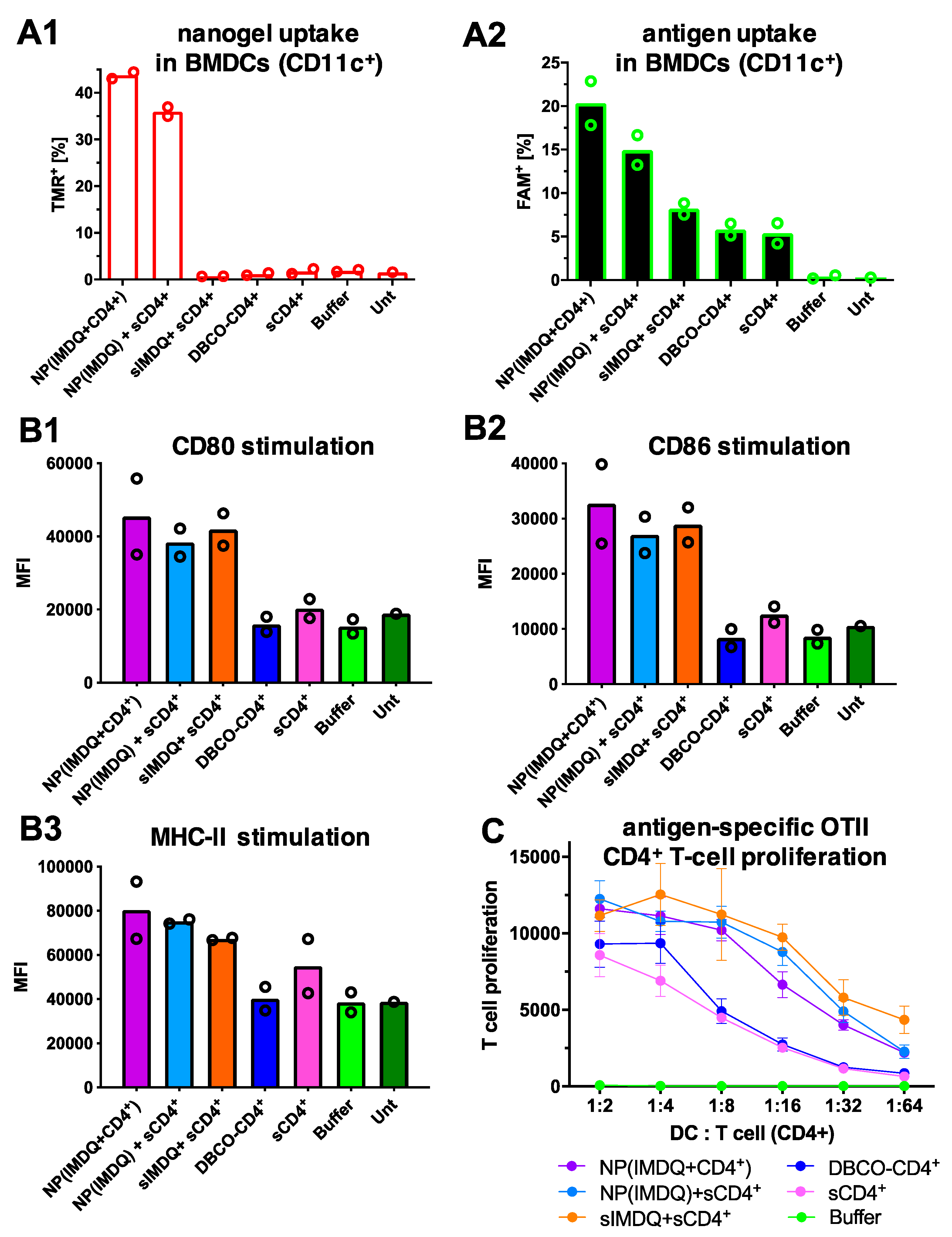
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Instrumentation
3.3. Block Copolymer Synthesis of Azide-PEG113-b-Poly(MTC-PFP)
3.4. Adjuvant-Loaded Azide-Functional Nanogels
3.5. SPAAC of Azide-Functional Nanogels with DBCO-Modified Oregon Green 488
3.6. DBCO-Peptide Modification by Site-Selective Modification of Cysteine
3.7. SPAAC of DBCO-Modified Peptides and Azide-Functional Nanogels
3.8. IMDQ-Loaded Azide-Functionalized Nanogel TLR Activity on RAW-Blue Macrophage Reporter Cell Line
3.9. Influence of the IMDQ-Loaded Azide-Functionalized Nanogels on the Metabolic Activity of the Stimulated Raw Blue Macrophages by MTT Assay
3.10. Confocal Microscopy Studies of Raw Blue Macrophages Stimulated with IMDQ-Loaded and Peptide-Decorated Nanogels
3.11. Bone Marrow-Derived Dendritic Cells (BMDCs) Culture
3.12. Flow Cytometry
3.13. T-Cell Proliferation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Delany, I.; Rappuoli, R.; De Gregorio, E. Vaccines for the 21st century. EMBO Mol. Med. 2014, 6, 708–720. [Google Scholar] [CrossRef]
- Iwasaki, A.; Omer, S.B. Why and how vaccines work. Cell 2020, 183, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Heitmann, J.S.; Bilich, T.; Tandler, C.; Nelde, A.; Maringer, Y.; Marconato, M.; Reusch, J.; Jäger, S.; Denk, M.; Richter, M.; et al. A COVID-19 peptide vaccine for the induction of SARS-CoV-2 T cell immunity. Nature 2022, 601, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Moreira, E.D.; Zerbini, C.; et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.D.; Shukla, S.; Chung, Y.H.; Beiss, V.; Chan, S.K.; Ortega-Rivera, O.A.; Wirth, D.M.; Chen, A.; Sack, M.; Pokorski, J.K.; et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020, 15, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Türeci, Ö. Personalized vaccines for cancer immunotherapy. Science 2018, 359, 1355–1360. [Google Scholar] [CrossRef]
- Malonis, R.J.; Lai, J.R.; Vergnolle, O. Peptide-based vaccines: Current progress and future challenges. Chem. Rev. 2020, 120, 3210–3229. [Google Scholar] [CrossRef]
- Purcell, A.W.; McCluskey, J.; Rossjohn, J. More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 2007, 6, 404–414. [Google Scholar] [CrossRef]
- Matsumoto, M.; Seya, T. TLR3: Interferon induction by double-stranded RNA including poly (I:C). Adv. Drug Deliv. Rev. 2008, 60, 805–812. [Google Scholar] [CrossRef]
- Latz, E.; Schoenemeyer, A.; Visintin, A.; Fitzgerald, K.A.; Monks, B.G.; Knetter, C.F.; Lien, E.; Nilsen, N.J.; Espevik, T.; Golenbock, D.T. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 2004, 5, 190–198. [Google Scholar] [CrossRef]
- Shukla, N.M.; Malladi, S.S.; Mutz, C.A.; Balakrishna, R.; David, S.A. Structure−activity relationships in human Toll-like receptor 7-active imidazoquinoline analogues. J. Med. Chem. 2010, 53, 4450–4456. [Google Scholar] [CrossRef]
- Bhagchandani, S.; Johnson, J.A.; Irvine, D.J. Evolution of Toll-like receptor 7/8 agonist therapeutics and their delivery approaches: From antiviral formulations to vaccine adjuvants. Adv. Drug Deliv. Rev. 2021, 175, 113803. [Google Scholar] [CrossRef]
- Federico, S.; Pozzetti, L.; Papa, A.; Carullo, G.; Gemma, S.; Butini, S.; Campiani, G.; Relitti, N. Modulation of the innate immune response by targeting toll-like receptors: A perspective on their agonists and antagonists. J. Med. Chem. 2020, 63, 13466–13513. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.J.; Burgess-Brown, N.A.; Jiang, S. Beyond just peptide antigens: The complex world of peptide-based cancer vaccines. Front. Immunol. 2021, 12, 696791. [Google Scholar] [CrossRef] [PubMed]
- Palucka, K.; Banchereau, J. Cancer immunotherapy via dendritic cells. Nat. Rev. Cancer 2012, 12, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Merad, M.; Sathe, P.; Helft, J.; Miller, J.; Mortha, A. The dendritic cell lineage: Ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013, 31, 563–604. [Google Scholar] [CrossRef]
- Farhood, B.; Najafi, M.; Mortezaee, K. CD8+ cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell. Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef]
- Desrichard, A.; Snyder, A.; Chan, T.A. Cancer neoantigens and applications for immunotherapy. Clin. Cancer Res. 2016, 22, 807–812. [Google Scholar] [CrossRef]
- Li, W.; Joshi, M.D.; Singhania, S.; Ramsey, K.H.; Murthy, A.K. Peptide vaccine: Progress and challenges. Vaccines 2014, 2, 515–536. [Google Scholar] [CrossRef]
- Khong, H.; Overwijk, W.W. Adjuvants for peptide-based cancer vaccines. J. Immunother. Cancer 2016, 4, 56. [Google Scholar] [CrossRef]
- Lynn, G.M.; Laga, R.; Darrah, P.A.; Ishizuka, A.S.; Balaci, A.J.; Dulcey, A.E.; Pechar, M.; Pola, R.; Gerner, M.Y.; Yamamoto, A.; et al. In vivo characterization of the physicochemical properties of polymer-linked TLR agonists that enhance vaccine immunogenicity. Nat. Biotechnol. 2015, 33, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Hanson, M.C.; Crespo, M.P.; Abraham, W.; Moynihan, K.D.; Szeto, G.L.; Chen, S.H.; Melo, M.B.; Mueller, S.; Irvine, D.J. Nanoparticulate STING agonists are potent lymph node–targeted vaccine adjuvants. J. Clin. Investig. 2015, 125, 2532–2546. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Gao, J.; Shi, Y.; Lammers, T.; Yu, H. Engineering nanoparticles to reprogram the tumor immune microenvironment for improved cancer immunotherapy. Theranostics 2019, 9, 7981–8000. [Google Scholar] [CrossRef]
- Zhu, X.; Nishimura, F.; Sasaki, K.; Fujita, M.; Dusak, J.E.; Eguchi, J.; Fellows-Mayle, W.; Storkus, W.J.; Walker, P.R.; Salazar, A.M.; et al. Toll like receptor-3 ligand poly-ICLC promotes the efficacy of peripheral vaccinations with tumor antigen-derived peptide epitopes in murine CNS tumor models. J. Transl. Med. 2007, 5, 10. [Google Scholar] [CrossRef]
- Hammerich, L.; Marron, T.U.; Upadhyay, R.; Svensson-Arvelund, J.; Dhainaut, M.; Hussein, S.; Zhan, Y.; Ostrowski, D.; Yellin, M.; Marsh, H.; et al. Systemic clinical tumor regressions and potentiation of PD1 blockade with in situ vaccination. Nat. Med. 2019, 25, 814–824. [Google Scholar] [CrossRef]
- Nuhn, L.; De Koker, S.; Van Lint, S.; Zhong, Z.; Catani, J.P.; Combes, F.; Deswarte, K.; Li, Y.; Lambrecht, B.N.; Lienenklaus, S.; et al. Nanoparticle-Conjugate TLR7/8 Agonist Localized Immunotherapy Provokes Safe Antitumoral Responses. Adv. Mater. 2018, 30, 1803397. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, N.; Suh, H.; Irvine, D.J. Nanoparticle anchoring targets immune agonists to tumors enabling anti-cancer immunity without systemic toxicity. Nat. Commun. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Irvine, D.J.; Dane, E.L. Enhancing cancer immunotherapy with nanomedicine. Nat. Rev. Immunol. 2020, 20, 321–344. [Google Scholar] [CrossRef]
- Goldberg, M.S. Improving cancer immunotherapy through nanotechnology. Nat. Rev. Cancer 2019, 19, 587–602. [Google Scholar] [CrossRef]
- O’Neill, C.L.; Shrimali, P.C.; Clapacs, Z.P.; Files, M.A.; Rudra, J.S. Peptide-based supramolecular vaccine systems. Acta Biomater. 2021, 133, 153–167. [Google Scholar] [CrossRef]
- Zacco, E.; Anish, C.; Martin, C.E.; Berlepsch, H.V.; Brandenburg, E.; Seeberger, P.H.; Koksch, B. A self-assembling peptide scaffold for the multivalent presentation of antigens. Biomacromolecules 2015, 16, 2188–2197. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Barz, M.; De Geest, B.G.; Diken, M.; Hennink, W.E.; Kiessling, F.; Lammers, T.; Shi, Y. Nanomedicine and macroscale materials in immuno-oncology. Chem. Soc. Rev. 2019, 48, 351–381. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Maynard, E.; Chiaradia, V.; Arno, M.C.; Dove, A.P. Aliphatic polycarbonates from cyclic carbonate monomers and their application as biomaterials. Chem. Rev. 2021, 121, 10865–10907. [Google Scholar] [CrossRef]
- Brannigan, R.P.; Dove, A.P. Synthesis, properties and biomedical applications of hydrolytically degradable materials based on aliphatic polyesters and polycarbonates. Biomater. Sci. 2017, 5, 9–21. [Google Scholar] [CrossRef]
- Becker, G.; Wurm, F.R. Functional biodegradable polymers via ring-opening polymerization of monomers without protective groups. Chem. Soc. Rev. 2018, 47, 7739–7782. [Google Scholar] [CrossRef]
- Amsden, B. In vivo degradation mechanisms of aliphatic polycarbonates and functionalized aliphatic polycarbonates. Macromol. Biosci. 2021, 21, 2100085. [Google Scholar] [CrossRef]
- Pratt, R.C.; Nederberg, F.; Waymouth, R.M.; Hedrick, J.L. Tagging alcohols with cyclic carbonate: A versatile equivalent of (meth) acrylate for ring-opening polymerization. Chem. Commun. 2008, 2, 114–116. [Google Scholar] [CrossRef]
- Czysch, C.; Ding, T.; Fröder, Y.; Bixenmann, L.; Komforth, P.; Balint, A.; Räder, H.-J.; Naumann, S.; Nuhn, L. Nontoxic N-Heterocyclic Olefin Catalyst Systems for Well-Defined Polymerization of Biocompatible Aliphatic Polycarbonates. ACS Polym. Au 2022, 2, 371–379. [Google Scholar] [CrossRef]
- Sanders, D.P.; Fukushima, K.; Coady, D.J.; Nelson, A.; Fujiwara, M.; Yasumoto, M.; Hedrick, J.L. A simple and efficient synthesis of functionalized cyclic carbonate monomers using a versatile pentafluorophenyl ester intermediate. J. Am. Chem. Soc. 2010, 132, 14724–14726. [Google Scholar] [CrossRef]
- Engler, A.C.; Chan, J.M.W.; Coady, D.J.; O’Brien, J.M.; Sardon, H.; Nelson, A.; Sanders, D.P.; Yang, Y.Y.; Hedrick, J.L. Accessing new materials through polymerization and modification of a polycarbonate with a pendant activated ester. Macromolecules 2013, 46, 1283–1290. [Google Scholar] [CrossRef]
- Engler, A.C.; Ke, X.; Gao, S.; Chan, J.M.W.; Coady, D.J.; Ono, R.J.; Lubbers, R.; Nelson, A.; Yang, Y.Y.; Hedrick, J.L. Hydrophilic polycarbonates: Promising degradable alternatives to poly (ethylene glycol)-based stealth materials. Macromolecules 2015, 48, 1673–1678. [Google Scholar] [CrossRef]
- Chan, J.M.W.; Sardon, H.; Engler, A.C.; García, J.M.; Hedrick, J.L. Tetra-n-butylammonium fluoride as an efficient transesterification catalyst for functionalizing cyclic carbonates and aliphatic polycarbonates. ACS Macro Lett. 2013, 2, 860–864. [Google Scholar] [CrossRef] [PubMed]
- Kockelmann, J.; Stickdorn, J.; Kasmi, S.; De Vrieze, J.; Pieszka, M.; Ng, D.Y.W.; David, S.A.; De Geest, B.G.; Nuhn, L. Control over Imidazoquinoline Immune Stimulation by pH-Degradable Poly(norbornene) Nanogels. Biomacromolecules 2020, 21, 2246–2257. [Google Scholar] [CrossRef] [PubMed]
- Nuhn, L.; Hartmann, S.; Palitzsch, B.; Gerlitzki, B.; Schmitt, E.; Zentel, R.; Kunz, H. Water-soluble polymers coupled with glycopeptide antigens and T-cell epitopes as potential antitumor vaccines. Angew. Chemie Int. Ed. 2013, 52, 10652–10656. [Google Scholar] [CrossRef]
- De Vrieze, J.; Van Herck, S.; Nuhn, L.; De Geest, B.G. Design of pH-Degradable Polymer-Lipid Amphiphiles Using a Ketal-Functionalized RAFT Chain Transfer Agent. Macromol. Rapid Commun. 2020, 41, 2000034. [Google Scholar] [CrossRef] [PubMed]
- Czysch, C.; Medina-Montano, C.; Dal, N.K.; Dinh, T.; Fröder, Y.; Winterwerber, P.; Maxeiner, K.; Räder, H.; Schuppan, D.; Schild, H.; et al. End Group Dye-Labeled Polycarbonate Block Copolymers for Micellar (Immuno-) Drug Delivery. Macromol. Rapid Commun. 2022, 43, 2200095. [Google Scholar] [CrossRef]
- Czysch, C.; Medina-Montano, C.; Zhong, Z.; Fuchs, A.; Stickdorn, J.; Winterwerber, P.; Schmitt, S.; Deswarte, K.; Raabe, M.; Scherger, M.; et al. Transient Lymph Node Immune Activation by Hydrolysable Polycarbonate Nanogels. Adv. Funct. Mater. 2022, 32, 2203490. [Google Scholar] [CrossRef]
- Nuhn, L.; Vanparijs, N.; De Beuckelaer, A.; Lybaert, L.; Verstraete, G.; Deswarte, K.; Lienenklaus, S.; Shukla, N.M.; Salyer, A.C.D.; Lambrecht, B.N.; et al. pH-degradable imidazoquinoline-ligated nanogels for lymph node-focused immune activation. Proc. Natl. Acad. Sci. USA 2016, 113, 8098–8103. [Google Scholar] [CrossRef]
- Stickdorn, J.; Stein, L.; Arnold-Schild, D.; Hahlbrock, J.; Medina-Montano, C.; Bartneck, J.; Ziß, T.; Montermann, E.; Kappel, C.; Hobernik, D.; et al. Systemically administered TLR7/8 agonist and antigen-conjugated nanogels govern immune responses against tumors. ACS Nano 2022, 16, 4426–4443. [Google Scholar] [CrossRef]
- Irvine, D.J.; Swartz, M.A.; Szeto, G.L. Engineering synthetic vaccines using cues from natural immunity. Nat. Mater. 2013, 12, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Hirosue, S.; Kourtis, I.C.; van der Vlies, A.J.; Hubbell, J.A.; Swartz, M.A. Antigen delivery to dendritic cells by poly (propylene sulfide) nanoparticles with disulfide conjugated peptides: Cross-presentation and T cell activation. Vaccine 2010, 28, 7897–7906. [Google Scholar] [CrossRef] [PubMed]
- Lynn, G.M.; Sedlik, C.; Baharom, F.; Zhu, Y.; Ramirez-Valdez, R.A.; Coble, V.L.; Tobin, K.; Nichols, S.R.; Itzkowitz, Y.; Zaidi, N.; et al. Peptide–TLR-7/8a conjugate vaccines chemically programmed for nanoparticle self-assembly enhance CD8 T-cell immunity to tumor antigens. Nat. Biotechnol. 2020, 38, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.S.; Hirosue, S.; Raczy, M.M.; Bonilla-Ramirez, L.; Jeanbart, L.; Wang, R.; Kwissa, M.; Franetich, J.F.; Broggi, M.A.S.; Diaceri, G.; et al. Antigens reversibly conjugated to a polymeric glyco-adjuvant induce protective humoral and cellular immunity. Nat. Mater. 2019, 18, 175–185. [Google Scholar] [CrossRef]
- Zom, G.G.; Khan, S.; Britten, C.M.; Sommandas, V.; Camps, M.G.M.; Loof, N.M.; Budden, C.F.; Meeuwenoord, N.J.; Filippov, D.V.; van der Marel, G.A.; et al. Efficient induction of antitumor immunity by synthetic toll-like receptor ligand–peptide conjugates. Cancer Immunol. Res. 2014, 2, 756–764. [Google Scholar] [CrossRef]
- Meng, J.; Zhang, P.; Chen, Q.; Wang, Z.; Gu, Y.; Ma, J.; Li, W.; Yang, C.; Qiao, Y.; Hou, Y.; et al. Two-Pronged Intracellular Co-Delivery of Antigen and Adjuvant for Synergistic Cancer Immunotherapy. Adv. Mater. 2022, 34, 2202168. [Google Scholar] [CrossRef]
- Silva, J.M.; Zupancic, E.; Vandermeulen, G.; Oliveira, V.G.; Salgado, A.; Videira, M.; Gaspar, M.; Graca, L.; Préat, V.; Florindo, H.F. In vivo delivery of peptides and Toll-like receptor ligands by mannose-functionalized polymeric nanoparticles induces prophylactic and therapeutic anti-tumor immune responses in a melanoma model. J. Control. Release 2015, 198, 91–103. [Google Scholar] [CrossRef]
- Nuhn, L.; Van Hoecke, L.; Deswarte, K.; Schepens, B.; Li, Y.; Lambrecht, B.N.; De Koker, S.; David, S.A.; Saelens, X.; De Geest, B.G. Potent anti-viral vaccine adjuvant based on pH-degradable nanogels with covalently linked small molecule imidazoquinoline TLR7/8 agonist. Biomaterials 2018, 178, 643–651. [Google Scholar] [CrossRef]
- Bolli, E.; Scherger, M.; Arnouk, S.M.; Antunes, A.R.P.; Straßburger, D.; Urschbach, M.; Stickdorn, J.; De Vlaminck, K.; Movahedi, K.; Räder, H.-J.; et al. Targeted repolarization of tumor-associated macrophages via imidazoquinoline-linked nanobodies. Adv. Sci. 2021, 8, 2004574. [Google Scholar] [CrossRef]
- Scherger, M.; Pilger, Y.A.; Stickdorn, J.; Komforth, P.; Schmitt, S.; Koynov, K.; Räder, H.-J.; Nuhn, L. Efficient Self-Immolative RAFT End Group Modification for Macromolecular Immunodrug Delivery. Biomacromolecules 2023, 24, 2380–2391. [Google Scholar] [CrossRef]
- Scherger, M.; Pilger, Y.A.; Stickdorn, J.; Komforth, P.; Schmitt, S.; Arnouk, S.M.; Lebegge, E.; Koynov, K.; Räder, H.-J.; Van Ginderachter, J.A.; et al. Self-Immolative Nanobody-Cysteine Residue Modification for Controlled Immunodrug Delivery. Adv. Ther. 2023, 202300076. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stickdorn, J.; Czysch, C.; Medina-Montano, C.; Stein, L.; Xu, L.; Scherger, M.; Schild, H.; Grabbe, S.; Nuhn, L. Peptide-Decorated Degradable Polycarbonate Nanogels for Eliciting Antigen-Specific Immune Responses. Int. J. Mol. Sci. 2023, 24, 15417. https://doi.org/10.3390/ijms242015417
Stickdorn J, Czysch C, Medina-Montano C, Stein L, Xu L, Scherger M, Schild H, Grabbe S, Nuhn L. Peptide-Decorated Degradable Polycarbonate Nanogels for Eliciting Antigen-Specific Immune Responses. International Journal of Molecular Sciences. 2023; 24(20):15417. https://doi.org/10.3390/ijms242015417
Chicago/Turabian StyleStickdorn, Judith, Christian Czysch, Carolina Medina-Montano, Lara Stein, Lujuan Xu, Maximilian Scherger, Hansjörg Schild, Stephan Grabbe, and Lutz Nuhn. 2023. "Peptide-Decorated Degradable Polycarbonate Nanogels for Eliciting Antigen-Specific Immune Responses" International Journal of Molecular Sciences 24, no. 20: 15417. https://doi.org/10.3390/ijms242015417
APA StyleStickdorn, J., Czysch, C., Medina-Montano, C., Stein, L., Xu, L., Scherger, M., Schild, H., Grabbe, S., & Nuhn, L. (2023). Peptide-Decorated Degradable Polycarbonate Nanogels for Eliciting Antigen-Specific Immune Responses. International Journal of Molecular Sciences, 24(20), 15417. https://doi.org/10.3390/ijms242015417







